Introduction
The cyclin D binding myb-like transcription factor 1
(DMTF1) was first identified as a cyclin D2 binding protein in a
yeast two-hybrid interactive screen study and was thus named cyclin
D-interacting myb-like protein (DMP1) DMTF1 (1). Moreover, the affinity of DMTF1 in
binding consensus DNA sequences CCCG(G/T)ATGT and its activity of
promoting gene expression were also revealed. The ectopic
expression of DMTF1α can cause cell cycle arrest by blocking the S
phase entry. DMTF1α can also bind other D-type cyclins, such as
cyclins D1 and D3. All 3 cyclins can antagonize the function of
DMTF1α in activating gene expression and suppressing cell cycle
progression (2). The investigation
of the molecular mechanisms of DMTF1α-mediated cell cycle arrest
has revealed a canonical DMTF1α recognition site in the ARF
promoter and the transactivation of the ARF gene by DMTF1α
(3). The tumor suppressive role of
DMTF1α was reinforced by the finding that the disruption of DMTF1
can enhance cell immortalization, RAS transformation and
spontaneous tumorigenesis in mice (4). In a previous study, MYC-induced
lymphomas were significantly accelerated, but did not exhibit any
differences between cohorts with either one or both DMTF1 alleles
being deleted, suggesting that DMTF1α is a haplo-insufficient tumor
suppressor (5). The authors have
previously demonstrated that the mammary-specific expression of
DMTF1α in transgenic mice leads to poorly developed mammary glands
and reduced HER2/neu-driven oncogenic transformation (6). The authors have also revealed that
the DMTF1 heterozygous status can significantly accelerate mouse
mammary carcinomas with decreased apoptosis and increased
metastasis at a transgenic background of cyclin D1 or cyclin
D1(T286A) (7). In addition,
microRNA (miRNA/miR)-155 and -675 have been reported to target
DMTF1 mRNA, leading to the enhanced growth of bladder and
colorectal cancer cells, respectively (8,9). All
these studies strongly suggest a tumor suppressive role of DMTF1α
during oncogenic transformation.
Pre-mRNA splicing is an essential step for the
transcript maturation of multi-exon genes. Importantly, it allows
one gene to encode multiple different isoforms that may have
distinct biological functions, which greatly expands the genomic
capacity of eukaryotes (10). The
DMTF1 pre-mRNA consists of 18 exons with its start codon ATG
present in exon 3 and stop codon in exon 18. The alternative
splicing of the DMTF1 pre-mRNA was first reported by Tschan et
al who discovered two alternative acceptor sites (or 3′ splice
sites) in intron 9 that led to the formation of 2 new isoforms,
designated as DMTF1β and DMTF1γ, respectively (11). Thus, DMTF1 with tumor suppressive
activity reported prior to the present study should be named as
DMTF1α. The read-frames of DMTF1β and γ transcripts after the
splicing are coincidently the same, while they encounter a stop
codon 'UAA' inside intron 9. As a result, the β and γ isoforms are
translated into 2 proteins [272 and 285 amino acids (aa),
respectively], much shorter than DMTF1α (760 aa) (10). DMTF1β and γ share the first 273
amino acids with DMTF1α, which embrace the transactivation domain
(TAD) and cyclin D binding site (CBS), but contain just a small
part of the myb-homology region (MHR). Thus, these two short
isoforms lack binding affinity to the consensus DNA element for
DMTF1α. DMTF1β is weakly expressed in a number of cell lines, but
exhibits a high expression in quiescent CD34+ cells and
peripheral blood leukocytes, while DMTF1γ is ubiquitously expressed
at low levels (11). Since the
specific regions for these DMTF1 proteins are very limited, it is
difficult to determine their relative expression levels,
particularly between DMTF1β and γ.
The functions of DMTF1β and γ had remained elusive
for over a decade, since DMTF1 pre-mRNA alternative splicing was
initially discovered in 2003; however, they have begun to be
unraveled in recent years. It has been demonstrated that DMTF1β can
stimulate mammary cell proliferation and promote mammary
oncogenesis using a transgenic mouse model (12). It has also been revealed that
DMTF1β is increasingly expressed in human breast cancer based on
immunohistochemical studies of clinical samples and the analyses of
a breast cancer RNA-seq dataset. In addition, DMTF1β levels are
positively associated with the poor prognosis of breast cancer
patients (12). Consistently,
another group also reported that DMTF1β inhibited the
transactivation of the ARF promoter (13). In addition, increased DMTF1β levels
can desensitize breast cancer cells to cisplatin treatment
(14).
In the present study, the factors that regulate
DMTF1 expression were investigated. The functional interplay of
DMTF1β and γ with DMTF1α was also explored. The data suggest that
both DMTF1β and γ possess oncogenic activity by antagonizing
DMTF1α-mediated ARF transactivation.
Materials and methods
Antibodies, DNA and vectors
The antibodies used herein with their catalog
numbers and vendors include the following: GAPDH (10R-G109A,
Fitzgerald Industries International), Flag (M2; cat. no. F1804,
Sigma-Aldrich; Merck GmbH) and HA (32-6700, Invitrogen; Thermo
Fisher Scientific, Inc.). RAD, a DMTF1 antibody against all 3
isoforms, was generated in our laboratory as previously reported
(15). Oligonucleotides for PCR
and DNA sequencing were synthesized by Genewiz. The pGL4 luciferase
vector used in constructing the ARF promoter reporter was purchased
from Promega Corporation. As collected by the Ensembl Project
Database, the DMTF1 gene can encode 38 splice variants, while only
DMTF1α, β and γ (annotated as ENST00000394703.9, ENST00000579677.5
and ENST00000447863.5, respectively) have been functionally
verified. The DMTF1α and β coding sequences were obtained as
previously described (12). The
DMTF1γ coding sequence was obtained by reverse transcription as
described below using total RNA extracted from MCF-10A cells,
(purchased from ATCC) followed by PCR amplifications using the
primers GTA GGG ATC CAG CAC AGT GGA AGA GGA TTC TG and CCT GGA ATT
CTT ATT CTT CAT TCT TCT TCT TCC C (forward and reverse,
respectively, with BamHI and EcoRI restriction sites
underlined).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA samples were extracted from the cultured
cells using the Tripure™ reagent (Roche Applied Science) according
to the manufacturer's protocol. Reverse transcription was carried
out using the One-Step gDNA Removal and cDNA Synthesis SuperMix
(Transgen Biotech Co., Ltd.). In this process, 1 µg RNA of
each sample was reverse transcribed by a poly(dT) or random primer
at 42°C for 30 min in a total volume of 20 µl. For
quantitative PCR (qPCR), cDNA of each sample was amplified in
triplicate using the LightCycler® 480 SYBR-Green I
Master on LightCycler® 480II Real-Time PCR System (Roche
Diagnostics) with a two-step protocol at 95°C (30 sec) and 60°C (30
sec) for 40 cycles. Expression was quantified according to the
comparative threshold method using the 2−ΔΔCq method
(16). The primer sequences for
qPCR of DMTF1, ARF and GAPDH are listed in Fig. S1.
Cell culture, transfection, lentiviral
production and infection
MCF-10A, HeLa, MDA-MB-231 and MCF-7 cells were
purchased from ATCC and cultured according to the suppliers'
protocols. DMEM was purchased from Corning, Inc. and fetal bovine
serum (FBS) was from ExCell Bio. Generally, transfection was
carried out using the Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the procedure provided by the
manufacturer. The cells were assayed after 48 h of transfection.
Lentiviral production and infection followed a procedure described
by us previously (17). Following
48 h of infection, the cells were selected using 1.0 µl/ml
of puromycin for 48 h prior to use in functional assays.
Flow cytometry
HeLa cells in 6-cm dishes were individually
transfected with 0.5 µg of pSL2-3xFlag-DMTF1α, β and γ
together with 0.6 µg of pSL3-RFP plasmid. The pSL2 vectors,
generated in our laboratory, uses the CMV promoter to drive an
inserted coding sequence and also expresses the ZsGreen located
downstream of an internal ribosomal entry sequence (IRES). A
pSL3-RFP vector that employs the CMV promoter was also generated to
drive the expression of the red fluorescent protein (RFP). After
being transfected for 48 h, the cells were trypsinized, washed
twice with PBS, and then directly analyzed using a flow cytometer
(Accuri C6, BD Biosciences).
Generation of the reporter construct and
reporter assay
The following oligonucleotides were designed to
amplify the -1,000 to -1 region of the human ARF promoter using the
genomic DNA extracted from HeLa cells as a template: CAG
TGG ATC CAG GCG GCG
GGA TCA AGG GGA GTC and CCT GGA ATT CGC GCC CGC CCC CCA CCT
TCA C. The 2 underlined sequences are BamHI and EcoRI
sites, respectively. The PCR fragment was inserted between the
BamHI and EcoRI sites upstream of the Firefly
luciferase (Fluc) coding sequence in the pGL4 luciferase vector.
The ARF promoter region in the reporter construct was sequenced to
confirm its consistency with the data in the NCBI database.
According to the authors' experience, if 2 proteins bind at a 1:1
stoichiometric ratio, they should have at least similar molecular
levels to make it possible for one protein to competitively
interfere with the activities of the other protein (data not
shown). The pcDNA3 vector could express 3 DMTF1 isoforms at
comparable protein levels, but DMTF1α has a molecular weight
2-3-fold higher than that of DMTF1β or γ. Based on this
stoichiometry, the same amount of 3 DMTF1 isoform plasmids should
produce an approximate 1:2:2 molecular ratio of α/β/γ proteins. In
reporter assays, HeLa cells were transfected with 0.3 µg of
pcDNA3-3xFlag-DMTF1α, 0.15 or 0.3 µg of pcDNA3-3xFlag-DMTF1β
or γ, together with 0.2 µg of ARF promoter reporter
construct and 0.1 µg of CMV-SEAP (secreted alkaline
phosphatase, cat# 24595, Addgene Inc.). The activities of Fluc and
SEAP were measured as previously described (18). For each sample, the Fluc reading
was normalized against the corresponding SEAP activity. Each
condition was tested in triplicate and at least repeated 3
times.
Western blot analysis
Whole cell lysates of transfected HeLa cells
prepared in ice-cold lysis buffer (5 mM EDTA, 0.1% NP-40, 150 mM
NaCl, and 50 mM Tris·HCl, pH 7.5) were assayed by Bradford solution
(0.05% Coomassie Brilliant Blue G-250, 23.5% methanol, 50%
phosphoric acid) for protein concentrations. The same amount of
proteins (10-20 µg) of each sample was mixed with an equal
volume of 2X SDS loading buffer, heated at 100°C for 5 min, and
then resolved in a 10% SDS-PAGE gel, followed by a transfer to a
polyvinylidene difluoride (PVDF) membrane at a constant current of
200 mA for 2 h. The membrane was then blocked by 5% skim milk at
room temperature for 1 h, and incubated with a primary antibody
overnight at 4°C. Following extensive washing with PBS, the
membrane was incubated with a secondary antibody [diluted 1:10,000,
HRP-conjugated goat anti-mouse or anti-rabbit IgG (H+L); cat nos.
G-21040 and 31460, respectively, Thermo Fisher Scientific, Inc.] at
room temperature for 1 h. After 3 times of wash by PBS, the
membrane was exposed using an ECL kit (Vazyme Biotech Co., Ltd.)
and the image was captured by ImageQuant LAS500 (GE Health Life
Sciences). The primary antibodies and their corresponding dilutions
were as follows: Flag (1:2,000), HA (1:1,000), RAD (1:300) and
GAPDH (1:2,000, as a protein loading control).
Protein stability assay
The half-lives of the 3 DMTF1 isoforms were
determined following a previously published procedure (17). Briefly, pcDNA3-3xFlag-DMTF1α, β and
γ constructs were individually transfected into HeLa cells. After
24 h, the cells in each dish were trypsinized and replated into 6
dishes (6-cm dishes). Following a further 24 h of growth, one dish
for each DMTF1 isoform was harvested at 'time zero' and
cycloheximide was added to a final concentration of 25 µg/ml
to the remaining dishes. Cells from the dishes were collected at
different time points (1, 2, 4, 8 and 12 h). The collected cells
from each time point were washed and the cell lysates were
normalized followed by western blot analyses using the Flag
antibody. Blotting with the GAPDH antibody was used as the loading
control. The densitometric density of the bands was quantified
using Quantity One software (Bio-Rad Laboratories, Inc.). and the
data were graphed to determine the half-lives of DMTF1 isoform
proteins using linear regression.
Co-immunoprecipitation
A total of 2 µg of pcDNA3-HA- DMTF1α, β and γ
were individually transfected with 2 µg of
pcDNA3-3xFlag-DMTF1α into HeLa cells cultured in 6-cm dishes. After
being transfected for 48 h, the cells were collected and lysed in
lysis buffer containing 5 mM EDTA, 0.1% NP-40, 150 mM NaCl and 50
mM Tris·HCl, pH 7.5. Approximately 300 µg of cell lysates
were incubated with 15 µl of Flag antibody-conjugated
magnetic beads (Cat. no. M8823, Sigma-Aldrich; Merck GmbH) in the
lysis buffer and the tubes were rotated at 4°C overnight. The beads
were then washed 8-10 times using the lysis buffer. During each
step of the washing, each tube was supplemented with 1 ml of the
buffer, inverted 5 times to resuspend the beads, loaded into the
magnetic Particle Concentrator (cat# 123.21D, Invitrogen; Thermo
Fisher Scientific, Inc.) for 30 sec, and aspirated to remove the
buffer. The immuno-precipitated samples were mixed with 20
µl of SDS loading buffer, heated at 100°C for 5 min, and
then examined by western blot analysis.
Immunostaining assay
MCF-7 and MDA-MB-231 cells cultured on sterilized
glass coverslips in 24-well plates overnight were individually
transfected with 500 ng of pcDNA3-3xFlag-DMTF1α, β and γ together
with 200 ng of an EGFP expression vector. After 2 days, cells were
fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton
X-100 and then blocked with 1% bovine serum albumin (catalog no.
001-000-161; Jackson ImmunoResearch Laboratory). The cells were
then incubated with the Flag antibody (1:500, Sigma-Aldrich; Merck
GmbH) at 4°C overnight followed by incubation with a secondary
antibody (1:200, Alexa Fluor® 594 goat anti-mouse IgG;
cat. no. A11032, Invitrogen; Thermo Fisher Scientific, Inc.) for 30
min at room temperature. Subsequently, the coverslips were
individually stained by 200 µl of DAPI (1:20,000; cat. no.
C1006, Beyotime Institute of Biotechnology, Inc.) for 10 min at
room temperature. Images were captured with the DeltaVision Elite
imaging system (GE Healthcare).
Electrophoretic mobility shift assay
(EMSA)
A Cy3-labeled oligonucleotide
(5′-GTCAGGTGACGGATGTAGCTAGG-3′) containing the DMTF1 binding motif
on the human ARF promoter was synthesized, and then annealed with
its complementary oligonucleotide to generate a double-stranded
probe. In a binding reaction, 0.5 pmol of the labeled
double-stranded probe and 1 µg of purified Hisx6-DMTF1α
protein were mixed in a binding buffer (250 mM HEPES, 500 mM KCl,
20 mM MgSO4, 10 mM DTT, pH 8.0) and incubated on ice for
30 min. For competitive binding, unlabeled double-stranded probe
was added to a binding reaction with a mole ratio of 16:1 to the
labeled probe. To evaluate the effects of DMTF1β and γ on the DNA
binding affinity of DMTF1a, increasing amounts of purified
Hisx6-DMTF1β and γ proteins were added to a binding reaction of
DMTF1a with the probe. The samples were analyzed by 8% native
polyacrylamide gel electrophoresis at 100 V for 100 min at 4°C in a
running buffer of 0.5X TBE (with final concentrations of 45 mM for
Tris-Borate and 1 mM for EDTA, pH 8.0). The fluorescent intensity
of the bands was determined using Typhoon FLA7000 (GE
Healthcare).
Analyses of association between DMTF1
transcripts and clinical outcomes
A TCGA dataset of invasive breast carcinoma
containing RNA-seq data (ID: TCGA.BRCA. sampleMap/HiSeqV2) and
matched clinical outcome information (ID:
TCGA.BRCA.sampleMap/BRCA_clinicalMatrix) of breast cancer patients
was downloaded from the UCSC Cancer Browser (https://genome-cancer.ucsc.edu). The information of
DMTF1β/α and DMTF1γ/α ratios was extracted from the TCGA Spliceseq
browser (http://bioinformatics.mdanderson.org/TCGASpliceSeq).
Patients were placed into the 'high' and 'low' groups based on
their DMTF1a, β, γ, DMTF1β/α or DMTF1γ/α ratios higher or lower
than the means, respectively. Kaplan-Meier graphs for the survival
of the breast cancer patients against DMTF1β/α or DMTF1γ/α ratios
in the corresponding groups were then analyzed using MedCalc
software 19.5. The long- and short-term survival outcomes of the
patients were analyzed using the log-rank and Gehan
Breslow-Wilcoxon tests, respectively.
Statistical analysis
Data in reporter assays are presented as the means ±
SD. Comparisons among all groups on a single parameter were
conducted by one-way ANOVA, followed by Tukey's multiple comparison
test. Statistical analyses were performed using SigmaStat 12.5
(Systat Software Inc.). The criterion for statistical significance
was set at P<0.05.
Results
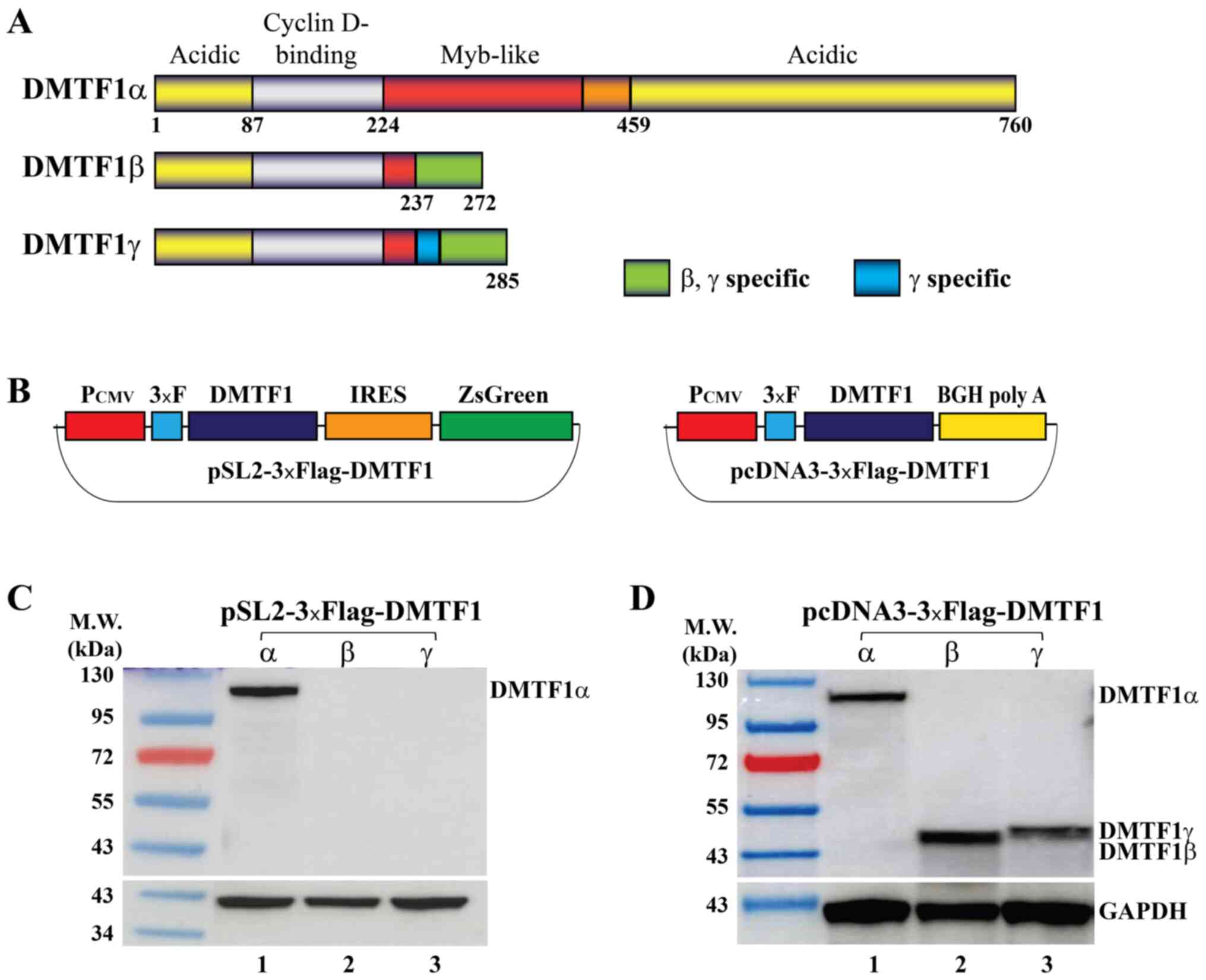
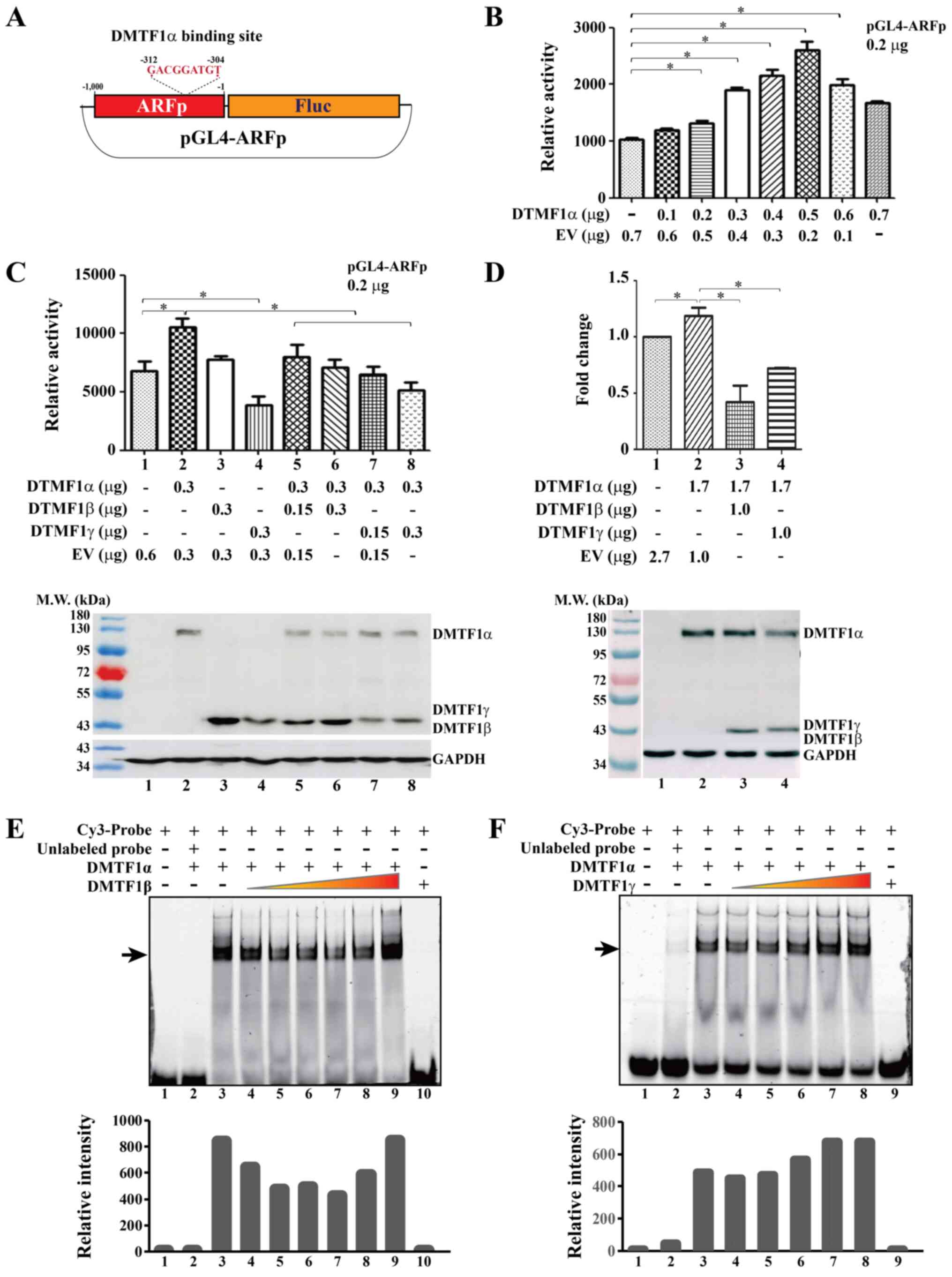
Generation of DMTF1 expression vectors
and determination of their expression
DMTF1α, β and γ transcripts are produced by
alternative splicing of the DMTF1 pre-mRNA and encode 3 proteins
with different lengths (Fig. 1A).
Due to the presence of a stop codon for DMTF1β and γ in intron 9,
these 2 isoform proteins only retain a small portion of the
Myb-like domain and thus have lost the DNA binding affinity that
DMTF1α has.
The endogenous expression of DMTF1β and γ,
particularly that of the latter, is relatively low, while no
antibody is available to discriminate these 2 isoforms. Thus, to
investigate whether any biological process in addition to
alternative splicing could regulate the relative levels of DMTF1
transcripts, the present study designed 2 sets of expression
vectors to express the 3 isoforms. First, the coding sequences of
the 3 DMTF1 isoforms were subcloned into a lentiviral vector
pSL2-3xFlag (Fig. 1B, left panel).
When these plasmids were transfected into HeLa cells,
well-expressed DMTF1α was detected by western blot analysis;
however, no detectable expression of DMTF1β and γ was observed
(Fig. 1C). Second, the 3 DMTF1
coding sequences were inserted into a pcDNA3-3xFlag vector, in
which a BGH poly A site is right downstream of each coding sequence
(Fig. 1B, right panel). These 3
expression vectors, pcDNA3-3xFlag-DMTF1α, β and γ, all steadily
expressed DMTF1 proteins when transfected into HeLa cells (Fig. 1D).
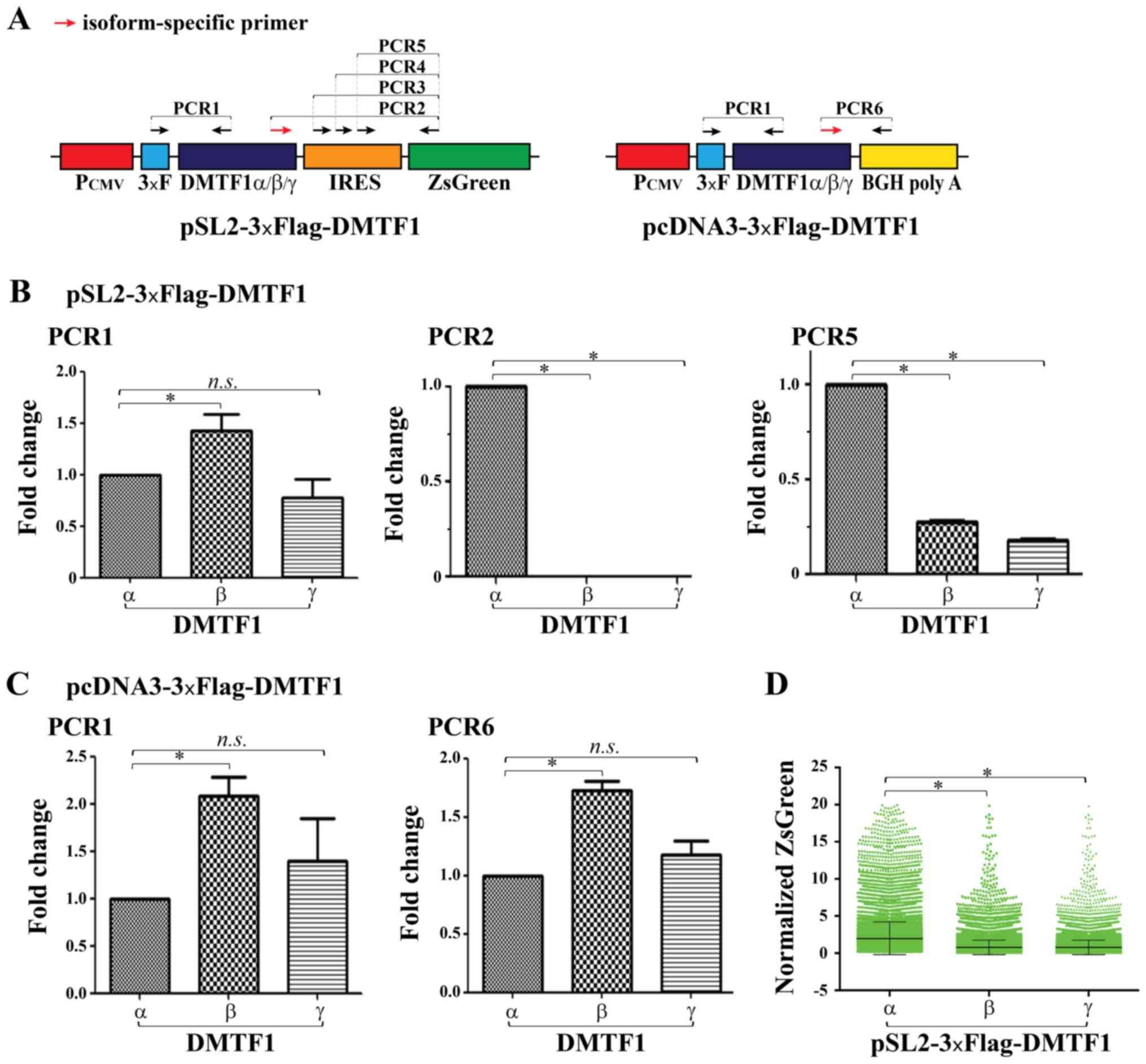
DMTF1β and γ specific regions reduce RNA
stability
The present study investigated the possible reasons
that may contribute to the undetectable DMTF1β and γ expression in
the bicistronic pSL2 lentiviral vector. In pSL2 vectors, the stop
codon of each DMTF1 coding sequence is immediately followed by an
internal ribosome entry site (IRES) and the ZsGreen coding
sequence; however, in pcDNA3 vectors, a BGH poly A sequence is
present right behind each DMTF1 isoform coding sequence (Fig. 1B). Thus, it was hypothesized that a
poly A sequence may be essential to the mRNA stability or integrity
of DMTF1β and γ. To test the cellular levels of 3 DMTF1 isoform
transcripts in a form of '3xFlag-DMTF1-IRES-ZsGreen', random primer
was used as a primer to reverse transcribe RNA extracted from HeLa
cells that was individually transfected with pSL2-3xFlag-DMTF1α, β
and γ plasmids, and several sets of primers for amplification were
designed (Fig. 2A). First,
semi-quantitative PCR was performed using an upstream primer in the
Flag epitope tag and a downstream primer in the region shared by
the 3 DMTF1 isoforms (PCR1 in Fig.
2A) and a band was amplified from each sample (compare RT-PCR
lane 1 of DMTF1α, β and γ in Fig.
S3B; lanes C1 to C5 are 'Plasmid PCR' controls using DMTF1
isoform plasmids as templates). Subsequently, 3 isoform-specific
primers were designed using the sequences at the junction sites of
DMTF1α, β and γ (Fig. S2A and B),
and the specificity to their corresponding isoforms was verified by
RT-qPCR (Fig. S2C). When using
these isoform specific primers together with a downstream primer,
ZG-94L, in ZsGreen, only a specific band from the cDNA containing
DMTF1α could be amplified, but not these with DMTF1β and γ (compare
lane 2 of DMTF1α, β and γ in Fig.
S3B). Moreover, amplifications using 3 different upstream
primers in the IRES together with primer ZG-94L could produce
specific bands for all isoforms; however, the intensity of α was
generally stronger than that of β and γ (compare lanes 3-5 of
DMTF1α, β and γ in Fig. S3B). The
quantification of relative band intensity was shown in Fig. S3C. Consistently, RT-qPCR analyses
revealed comparable transcript quantities among the 3 isoforms when
amplifying the region close to the 5′-ends of DMTF1 coding
sequences (PCR1 in Fig. 2B), but
almost undetectable β and γ transcripts when isoform-specific
primers were used in qPCR (PCR2, Fig.
2B). Furthermore, the amplification of the ZsGreen region
transcribed by pSL2-3xFlag-DMTF1β and γ vectors demonstrated a
markedly reduced signal compared to that of the α vector (PCR5,
Fig. 2B). When using RT-qPCR to
quantify the transcripts of pcDNA3-3xFlag-DMTF1 isoforms, their
relatively comparable levels were detected when amplifying both
common and isoform-specific regions (Fig. 2C). The ZsGreen expressed by
pSL2-3xFlag-DMTF1 vectors could also indirectly reflect mRNA
stability. Thus, ZsGreen protein levels were evaluated in HeLa
cells individually transfected with pSL2-3xFlag-DMTF1α, β and γ
plasmids with a vector expressing RFP as a transfection control.
With co-transfected RFP, the green fluorescent signal could only be
determined in RFP-positive cells by flow cytometry, which would
largely reduce the effects of transfection efficiency difference on
the comparison of the green fluorescent signal among the 3
plasmids. As shown in Figs. 2D and
S4, the ZsGreen signal was
significantly reduced in the pSL2-3xFlag-DMTF1β- and γ-transfected
cells compared to the α-transfected cells. The data presented above
strongly suggested that DMTF1β and γ contain specific regions
vulnerable to break if not immediately followed by a poly A. The
reduced stability or integrity of the transcripts was likely the
cause of undetectable DMTF1β and γ proteins when using pSL2-3xFlag
vectors as an expression vector, and could also contribute to
relatively low levels of downstream ZsGreen, compared to the DMTF1α
vector.
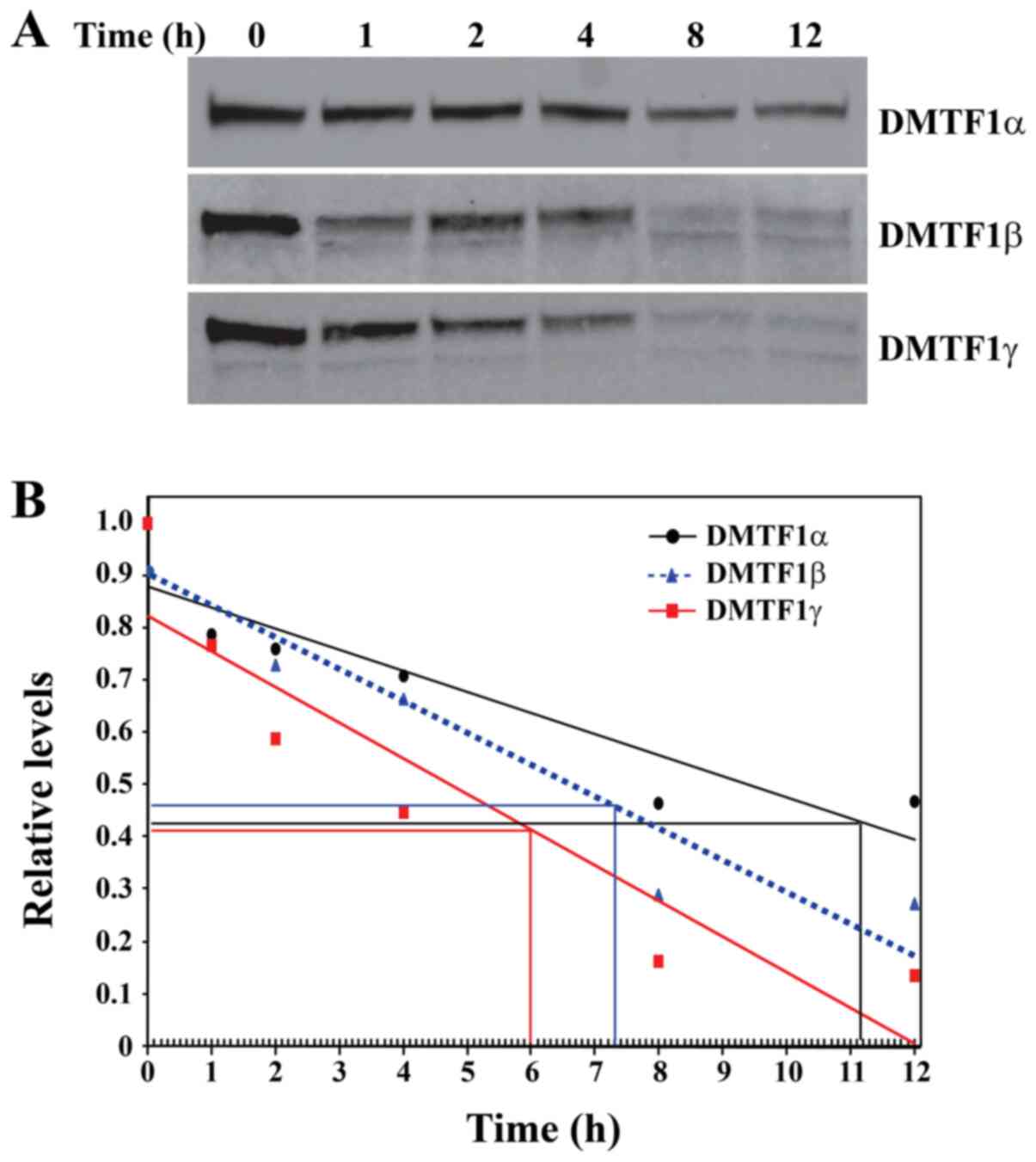
DMTF1β and γ proteins exhibit reduced
stability compared to DMTF1α
The present study also evaluated the relative
stability of the 3 DMTF1 protein isoforms. For this purpose, HeLa
cells were transfected with pcDNA3-3xFlag-DMTF1 plasmids and the
cells were treated with 25 µg/ml cycloheximide followed by
cell harvesting at the time points of 0, 1, 2, 4, 8 and 12 h. After
determining the protein concentrations of the cell lysates, the
same amount of proteins was loaded on SDS-PAGE followed by western
blot analysis. Apparently, DMTF1α exhibited a longer half-life than
DMTF1β and γ based on the gel western blot images (Fig. 3A). The densitometric density of
each band was then quantified using Quantity One software and the
data were graphed as shown in Fig.
3B. The DMTF1β sample collected at 1 h was abandoned due to its
obvious deviation. Based on regression lines of these samples, the
half-lives of DMTF1α, β and γ proteins were approximately
determined as 8.7, 3.5 and 1.9 h, respectively. Therefore, the 2
shorter DMTF1β and γ isoforms clearly exhibited reduced stability
compared to DMTF1α.
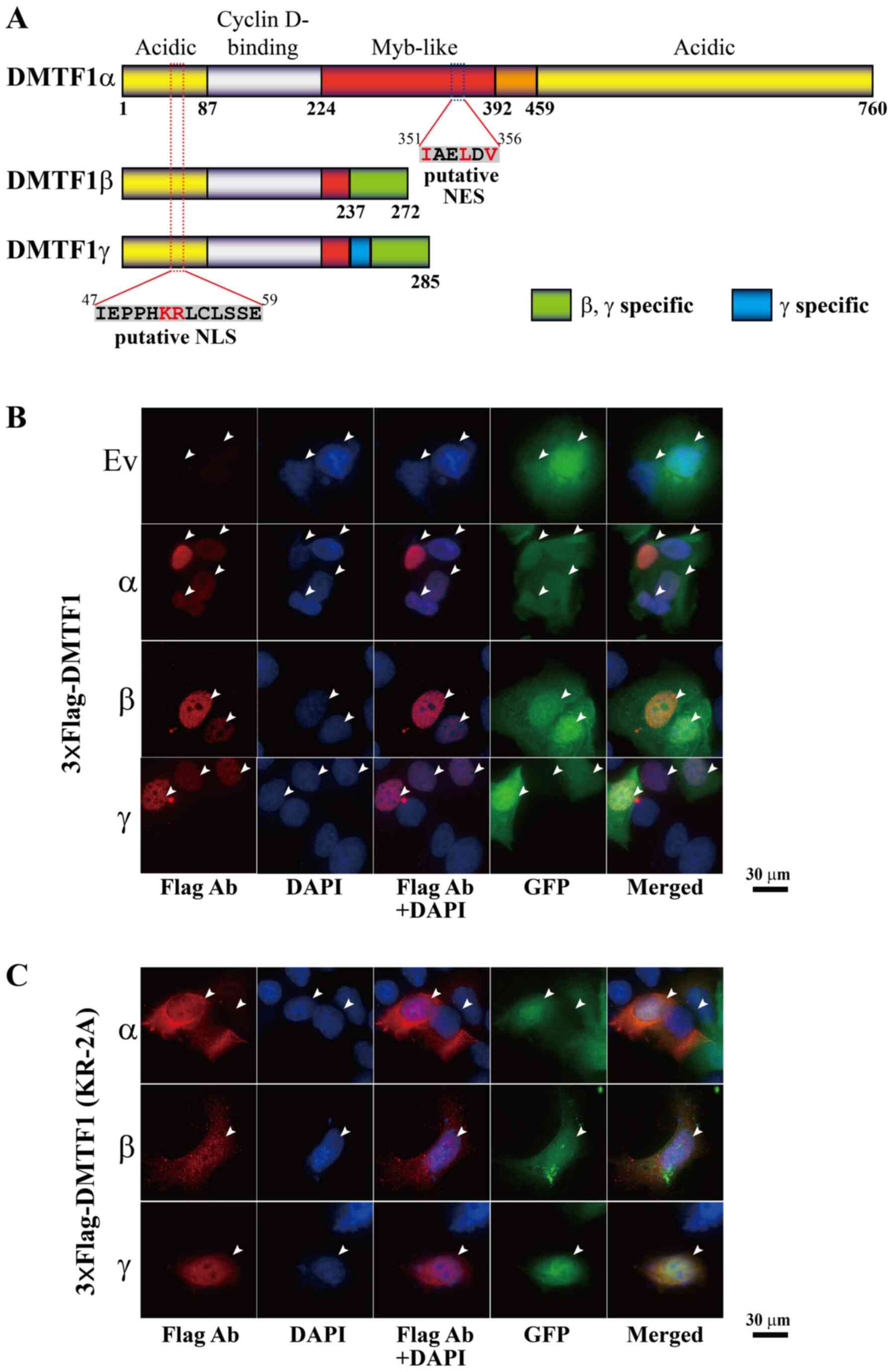
All 3 DMTF1 isoforms are localized in
nucleus
To predict the subcellular localization of the 3
DMTF1 isoforms, their protein sequences were first analyzed using a
previously reported strategy (19)
and a putative nuclear localization signal (NLS) was identified
among the amino acids 47-59 (IEPPHKRLCLSSE), shared by the 3
isoforms (Fig. 4A). Consistent
with this prediction, previous studies have demonstrated the
nuclear localization of DMTF1α (1,2).
Moreover, DMTF1 proteins were also analyzed for their nuclear
export signal (NES) based on a published algorithm (20) and a weak NES sequence (IAELDV, with
crucial residues underlined) was identified among the amino acids
351-356, which is located in the Myb-like domain, only present in
DMTF1α (Fig. 4A). To determine the
localization of DMTF1 proteins, pcDNA3-3xFlag-DMTF1α, β and γ were
individually transfected into MCF-7 and MDA-MB-231 cells, and the
transfected cells were immunostained using a Flag antibody. As
shown in Figs. 4B and S5A, 3xFlag-DMTF1α, β and γ were all
localized in the nuclei of the transfected breast cancer cells. In
these images, the EGFP signal was used to visualize the regions of
both nucleus and cytoplasm. To determine whether the potential NLS
plays a role in nuclear localization of DMTF1 proteins, the K52 and
R53 were replaced by 2 alanines to generate KR-2A mutants of
DMTF1α, β and γ. The immunostaining analyses of MCF-7 and
MDA-MB-231 cells transfected with these 3xFlag-tagged mutants
indicated that the KR-2A mutations led to DMTF1α nuclear exclusion
and DMTF1β, γ distribution in both cytoplasm and nuclei (Figs. 4C and S5B), suggesting that K52 and R53 are
essential residues for the nuclear localization of DMTF1
proteins.
Both DMTF1β and γ interfere with
DMTF1α-mediated trans-activation
The ARF promoter contains a DMTF1α consensus binding
site 'GACGGATGT' in the region of -312 to -304, if the first
nucleotide downstream the transcription start site (TSS) is
designate as '-1' (Fig. 5A)
(3). To determine the effect of
DMTF1 proteins on its target gene ARF, the-1,000 to -1 region of
the ARF promoter (ARFp) was amplified and inserted upstream of the
Fluc coding sequence in the pGL4 vector to generate pGL4-ARFp
reporter construct (Fig. 5A). The
ratio of the transfected pGL4-ARFp plasmid and DMTF1α expression
vector was first optimized. When 0.2 µg of the pGL4-ARFp
reporter was co-transfected with increasing amounts of DMTF1α
vector into HeLa cells, the Fluc activity increased accordingly,
reaching its highest value at 0.5 µg of DMTF1α, and then
decreased (Fig. 5B). CMV-SEAP was
used as a transfection control and Fluc data were normalized
against the SEAP activity. When determining the effects of DMTF1β
and γ on DMTF1α-mediated transcription, 0.3 µg of DMTF1α
vector was used together with 0.15 and 0.3 µg of DMTF1β and
γ expression plasmids to transfect HeLa cells. As shown in the top
panel of Fig. 5C, while DMTF1α
stimulated ARF promoter activity, DMTF1β and γ did not have this
effect (compare columns 3 and 4 to 1 and 2), consistent with their
loss of the DNA binding site due to alternative splicing (10). Actually, DMTF1γ could even decrease
the expression of the ARF promoter reporter. Importantly, it was
observed that both DMTF1β and γ attenuated the DMTF1α-mediated
transactivation (compare columns 5-8 to 2, top panel of Fig. 5C). In the samples for the reporter
assay, transfected DMTF1 isoform plasmids could steadily express
corresponding proteins, as determined by western blot analysis
(bottom panel of Fig. 5C). To
evaluate the effects of DMTF1 proteins on endogenous ARF
expression, DMTF1α was co-transfected with an empty vector, DMTF1β
or DMTF1γ into HeLa cells and endogenous ARF expression was
determined by RT-qPCR. While DMTF1α alone significantly increased
the level of endogenous ARF mRNA, DMTF1β and γ markedly antagonized
this effect (Fig. 5D, top panel).
Western blot analysis confirmed the expression of DMTF1 proteins in
the present study (Fig. 5D, bottom
panel). Thus, the effects of DMTF1 isoform proteins on the
expression of endogenous ARF are consistent with the results of
reporter assays. Since DMTF1β and γ reduced DMTF1α-activated ARF
promoter activity, the present study wished to determine whether
these two short isoforms can alter DMTF1α binding to the ARF
promoter. In EMSA experiments using purified recombinant DMTF1
proteins and a probe based on the DMTF1α-binding element on the ARF
promoter, slowly migrated bands for the complex of DMTF1a and
Cy3-labeled probe were detected on native polyacryl-amide gel
electrophoresis (lanes 3 vs. 1 in Fig.
5E and F), while DMTF1β and γ did not exhibit any binding
affinity to the probe (lanes 10 and 9 in Fig. E and F, respectively). The retarded bands
were diminished when unlabeled probe was excessively added (lanes 2
of Fig. 5E and F). With increasing
amounts of DMTF1β (lanes 4-7 in Fig.
5E), the DMTF1a-probe complex exhibited a reduced intensity.
However, the addition of DMTF1γ (lanes 4-5 in Fig. 5F) exerted a relatively modest
effect. Of note, the further increase in DMTF1β and γ somehow
promoted the DMTF1α-probe complex formation (lanes 7-9 in Fig. 5E and lanes 6-8 in Fig. 5F). Overall, DMTF1α activated ARF
promoter activity, while DMTF1β counteracted this effect, possibly
by interfering with DMTF1α-binding to the promoter. However, the
effects of DMTF1γ on DNA binding affinity of DMTF1α were very
marginal.
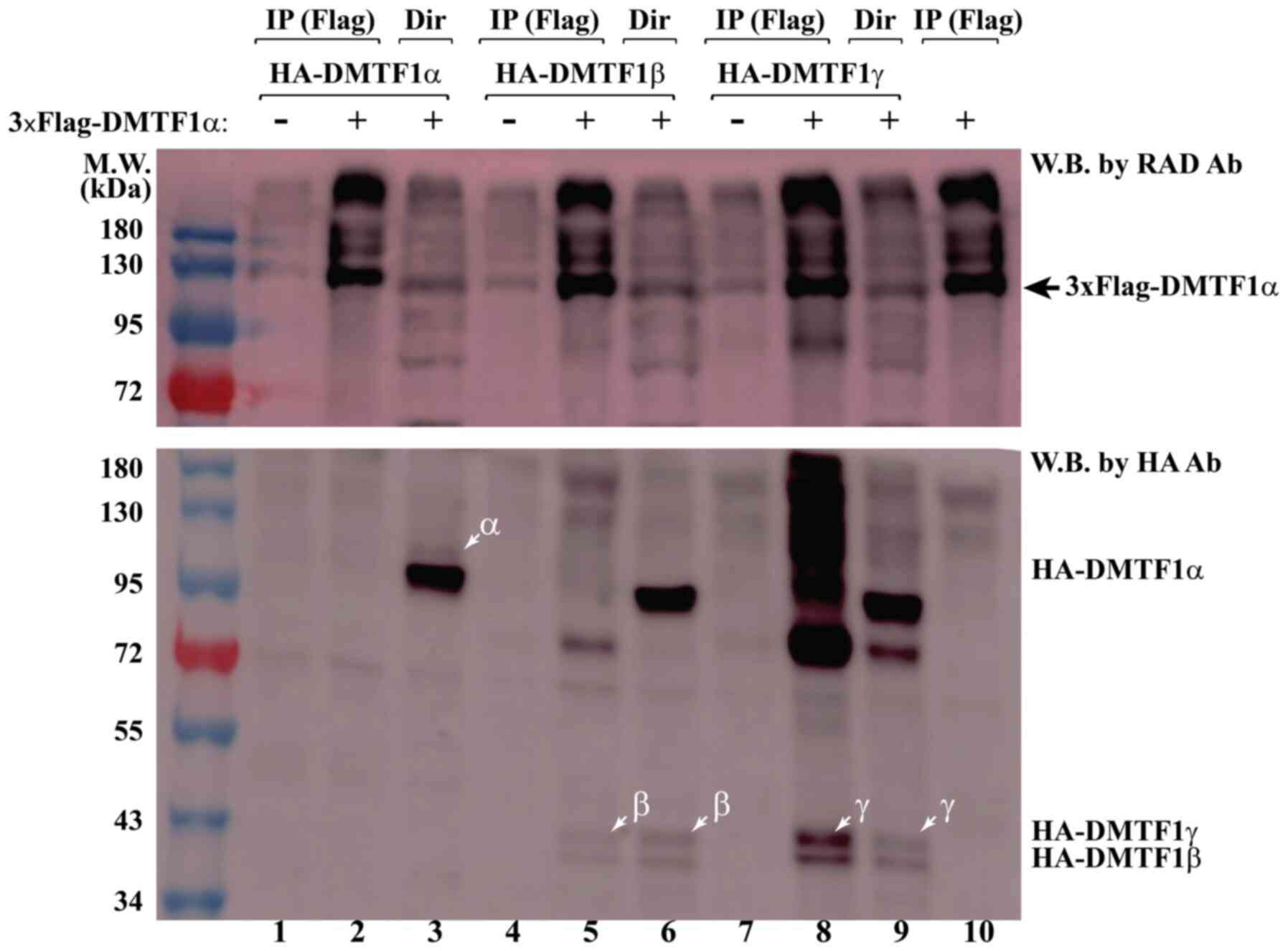
DMTF1α physically associates with both
DMTF1β and γ
To explore the mechanism underlying the antagonistic
effects of DMTF1β and γ on DMTF1α-mediated ARF promoter
transac-tivation, we carried out co-immunoprecipitation experiments
to determine their potential interaction. The 3xFlag-DMTF1α vector
was co-transfected with pcDNA3-HA-DMTF1α, β and γ vectors,
respectively, into HeLa cells, and Flag antibody-conjugated agarose
beads were then used to immu-noprecipitate 3xFlag-DMTF1α (Fig. 6, top labels of lanes 2, 5 and 8).
When blotting the samples by an HA antibody, HA-DMTF1β and γ, but
not α, we could detected, brought down together with 3xFlag-DMTF1α
(compare lanes 5 and 8 with lane 2 in bottom panel of Fig. 6), suggesting that DMTF1β and γ
could physically associate with DMTF1α to interfere with its
mediated transactivation.
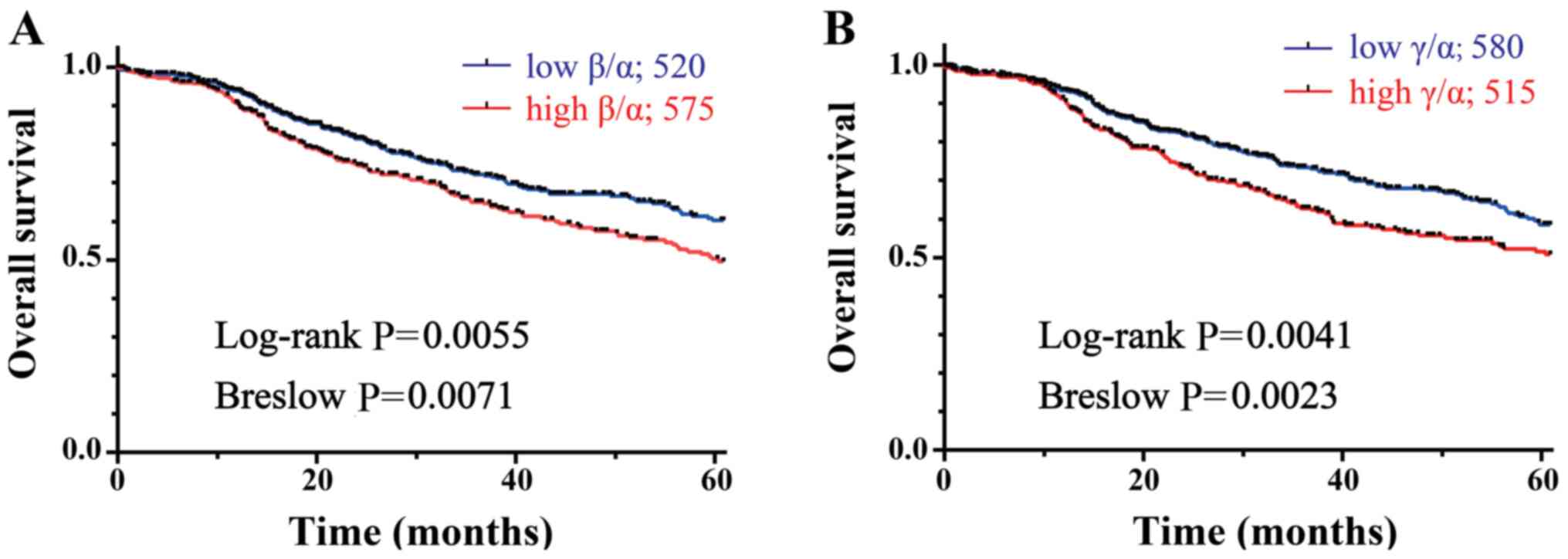
Increased DMTF1β/α and γ/α ratios are
associated with a poor prognoses of breast cancer patients
Previous studies by the authors indicate a tumor
suppressive role of DMTF1α in breast cancer development (6,7);
thus, the activities of DMTF1β and γ in binding and antagonizing
DMTF1α function suggest their oncogenic or proliferative role in
cancer development. It has previously been demonstrated that DMTF1β
can promote mammary oncogenesis in a transgenic mouse model
(12). This led us to determine
whether the ratio of DMTF1β/α or γ/α is associated with the
clinical outcomes of breast cancer patients. Since no antibody was
available to discriminate β and γ isoform proteins, the current
experiments were based on the DMTF1 mRNA levels of breast cancer
patients. In the present study, the Kaplan-Meier survival curve was
employed to analyze the RNA-seq data of 1,095 breast cancer samples
derived from a TCGA SpliceSeq Database (ID: TCGA.
BRCA.sampleMap/HiSeqV2) (21), and
the associations of the DMTF1β/α and γ/α ratios with patient
survival were assessed using the log-rank and Gehan
Breslow-Wilcoxon tests. In general, the log-rank test tends to be
sensitive to distributional differences that are most evident later
in time, while the Wilcoxon test tends to be more powerful in
detecting differences early in time (22), and thus they can be used to
evaluate long- and short-term survivals of a patient cohort,
respectively. Consistent with the findings of a previous study
(12), the survival analyses
displayed significant associations between increased DMTF1β/α
ratios with the poor prognosis of breast cancer patients both at
long- and short-term follow-up (log-rank, P=0.0055; Breslow,
P=0.0071; Fig. 7A). Importantly,
it was also found that high DMTF1γ/α ratios were significantly
associated with the reduced long- and short-term survival rates of
the patients (log-rank, P=0.0041; Breslow, P=0.0023; Fig. 7B). These data suggested the
oncogenic activities of DMTF1β and γ in breast cancer.
The three DMTF1 mRNA isoforms originate from the
same pre-mRNA; thus, increased splicing of one isoform will
theoretically lead to reduced levels of the others. To prove this
notion, 1,095 patients from the aforementioned database were
divided into 8 groups with varying combinations of high (H-) or low
(L-) levels of the 3 isoforms, based on the DMTFα, β or γ
expression of each sample being higher or lower than its
corresponding mean value of all samples (Fig. 8A). In addition, H-α and L-α
expression was deliberately combined with 4 different β and γ
combinations; i.e., H-β,H-γ; H-β,L-γ; L-β,H-γ and L-β,L-γ (Fig. 8B). With H-α expression, only 9
patients had the H-β,H-γ status and the majority of the patients
(n=268) exhibited L-β,L-γ. On the other hand, at the L-α condition,
the majority of patients exhibited H-β,L-γ or H-β,H-γ levels (210
and 241 patients, respectively), but only 3 patients had the
L-β,L-γ status (Fig. 8A and B).
These data suggested the competitive splicing among the 3 DMTF1α
isoforms.
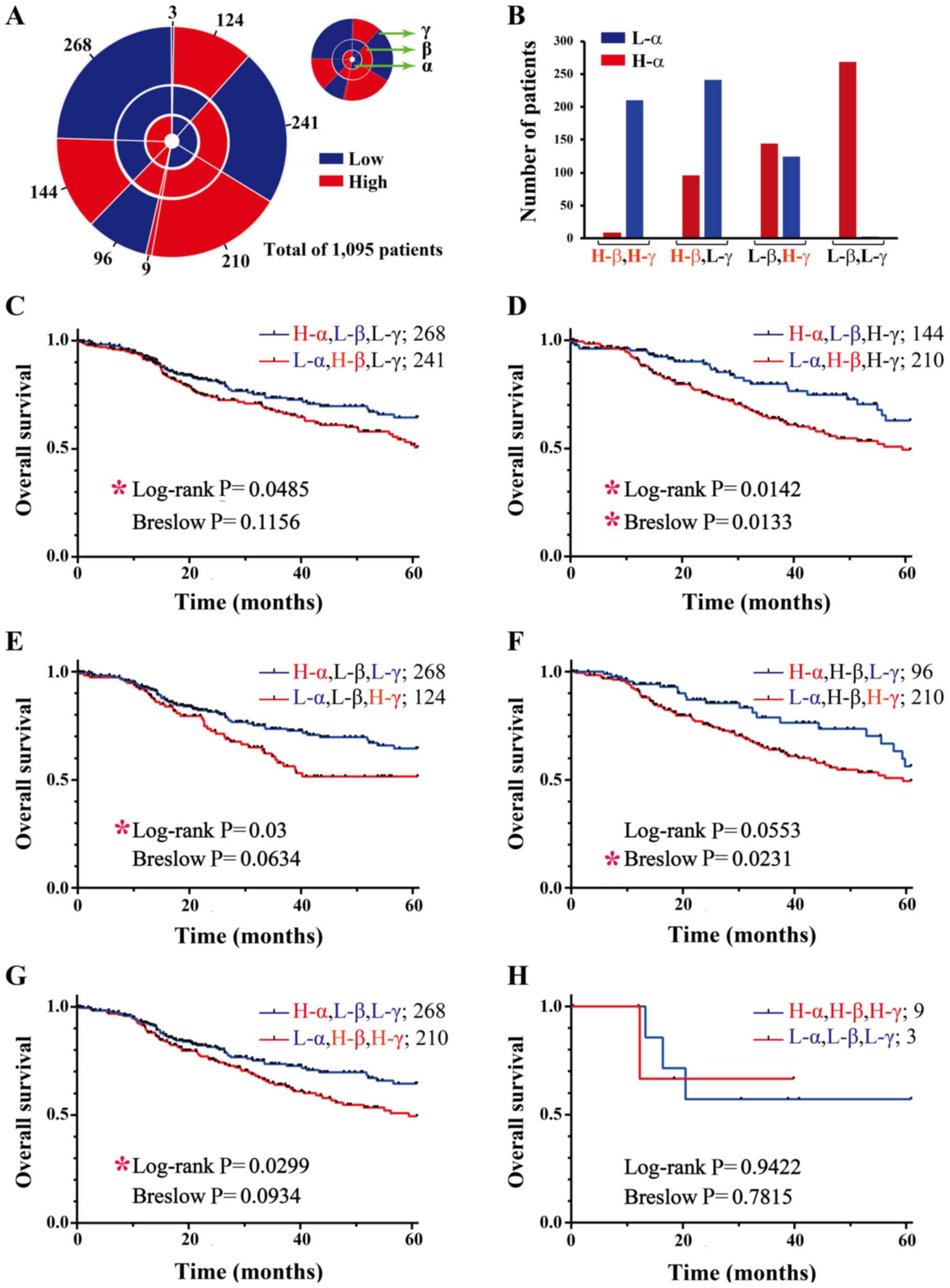 | Figure 8Survival curves of grouped breast
cancer patients based on combinations of high or low expression of
different DMTF1 isoforms. (A) Pie chart of grouped breast cancer
patients from the TCGA dataset (ID: TCGA.BRCA.sampleMap/HiSeqV2).
The 3 circles from inside to outside represent DMTF1α, β and γ. The
1,095 patients were divided into 8 groups with various combinations
of high (red) or low (blue) levels of the 3 isoforms, based on the
DMTFα, β or γ expression of each sample being higher or lower than
its corresponding mean value of all samples. The numbers of the 8
patient groups are labeled outside of the chart. (B) Using H and L
to denote high and low expression, respectively, the patients with
H-α and L-α were grouped and simultaneously expressing H or L
levels of β and γ to generate the bar chart, which allowed for the
intuitive visualization of the patient numbers of each group. (C-H)
The comparison of long- and short-term survival rates among the 8
groups of patients based on the log-rank p and Breslow P-values,
respectively. Patient numbers were labeled behind corresponding
group names. The red asterisk (*) denotes P<0.05, suggesting
statistically significant differences. DMTF1, cyclin D binding
myb-like transcription factor 1. |
To evaluate the contributions of DMTF1 isoforms to
breast cancer development, statistical analyses among were
performed for these 8 groups. First, patient survival according to
the varied DMTF1α and β expression at the L-γ or H-γ background was
assessed. At the L-γ condition, the patients in the L-α,H-β group
exhibited a statistically significant decrease in long-term
survival compared with the patients in the H-α,L-β group (log-rank,
P=0.0485); however, their short-term survival did not differ
significantly (Fig. 8C). However,
at the H-γ background, both the long- and short-term survival rates
of the L-α,H-β group were significantly decreased compared to the
H-α,L-β group (log-rank, P=0.0142; Breslow, P=0.0133; Fig. 8D). The data suggested that the
increased expression DMTF1γ enhanced DMTF1β-mediated mammary
oncogenesis. Second, the contribution of DMTFα and γ at the L-β or
H-β background to patient survival was evaluated. At the L-β
background, patients in the L-α,H-γ group exhibited a significantly
decreased long-term (log-rank, P=0.03), but not short-term
(Breslow, P=0.0634) survival comparted to the patients in the
H-α,L-γ group (Fig. 8E). However,
at the H-β background, significant difference were observed in the
short-term (Breslow, P=0.0231), but not the long-term survival
(log-rank, P=0.0553, Fig. 8F).
Third, two mixed conditions were analyzed. Surprisingly, the
patients with the L-α,H-β,H-γ status only exhibited a significantly
decreased long-term survival compared to those with H-α,L-β,L-γ
expression (log-rank, P=0.0299), while the difference of short-term
survival was not significant (Breslow, P=0.0934, Fig. 8G). The 2 patient groups with
L-α,L-β,L-γ and H-α,H-β,H-γ statuses did not exhibit significant
differences in survival due to very few numbers of patients
(Fig. 8H). Overall, the increased
splicing of DMTFβ and γ isoforms versus the α isoform was
associated with the poor prognosis of breast cancer patients. These
data also confirmed the tumor suppressive role of DMTF1α; its low
expression was always associated with the decreased survival of
patients.
Discussion
Alternative RNA splicing is a well-regulated process
in eukaryotic cells during development and contributes to
functional diversity of proteins and non-coding RNAs (ncRNAs) in
various cell signaling pathways. Under dysregulated conditions,
such as cancer cells, aberrantly alternative splicing can produce
RNAs or proteins with distinct or opposite functions compared to
cognate normal products. With the progress of the Ensembl Project,
a stunning amount of genomic data and their annotations are
publicly available, which include a huge number of new splicing
species for different genes. However, a number of transcripts in
the database only have partial sequences available and lack
information regarding their activity or cellular abundancy. For
example, the DMTF1 gene has 38 splicing variants and 20 of them can
possibly encode proteins based on the annotation in the Ensembl
Project Database. Currently, only 3 DMTF1 isoforms have been
reported (11). In a more recent
study, this group indicated that DMTF1β, but not γ could antagonize
DMTF1α-mediated ARF expression, and no immunoblot for the
expression of the two short protein isoforms was presented
(13). Thus, the present study
aimed to characterize the expression and function of DMTF1β and
γ.
Due to the relatively low expression of endogenous
DMTF1γ, and lack of antibody in discriminating DMTF1β and γ
isoforms, we designed two sets of vectors expressing 3 DMTF1
isoforms to indirectly evaluate the contributions of mRNA and
protein stability to their relative cellular levels. The results
may provide a conceptual base to understand differentially
regulated DMTF1 isoform expression in cancer cells. Many factors
can alter expression levels of protein coding genes. In the present
study, it was observed that vectors containing DMTF1β and γ coding
sequences could only express DMTF1 proteins when then were
immediately followed by a poly A sequence (Fig. 1C and D). The failure of DMTF1β and
γ expression when their stop codons are not adjacent to a poly A
sequence is reminiscent of nonsense-mediated decay (NMD) that
eukaryotes utilize to eliminates mRNA transcripts with premature
stop codons (23). This mechanism
can avoid the translation of aberrant mRNAs into proteins, which
potentially have deleterious gain-of-function or dominant negative
activity (24). Thus, it is
possible that the stop codon of DMTF1β and γ in intron 9 triggers
the NMD mechanism, which reduces their expression. The present
study also demonstrated markedly shorter half-lives of DMTF1β and γ
than α (Fig. 3A and B). In these
experiments, Flag tags were added to the DMTF1 N-terminals, which
shared 237 amino acids among the three isoforms (Fig. 1A). Thus, it was unlikely that Flag
tag could differentially affect protein expression in the plasmids.
These data suggest that DMTF1β and γ expression is antagonized
through both enhanced degradation of their transcripts and reduced
stability of their proteins to eliminate their potentially
deleterious effects (Figs. 2,
3 and S3). During the malignant transformation
or other biological deregulation, these mechanisms may be
attenuated, thereby leading to DMTF1β and γ accumulation.
Notably, semi-quantitative RT-PCR and RT-qPCR
analyses suggested that only DMTF1β- and γ-specific regions in the
transcripts were disrupted and the downstream coding sequence of
ZsGreen exhibited reduced levels, although the upstream regions
shared by the 3 isoforms remained apparently intact (Figs. 2 and S3). This observation indicates that the
5′ regions of DMTF1 coding sequences may possess a stable or
protective structure resistant to further RNA degradation.
The study by Tschan et al indicated that
DMTF1β was dominantly localized in the nucleus, but DMTF1γ was
located in both the nucleus and cytoplasm, when they were expressed
as fusion proteins with EGFP (13). They also observed that only DMTF1β,
but not DMTF1γ, could interfere with DMTF1α-mediated ARF
transcription. However, the data clearly demonstrated the nuclear
localization of all 3 DMTF1 isoforms (Figs. 4B and S5A). Importantly, we also discovered an
NLS sequence in a region shared by the three DMTF1 isoforms and a
putative NES sequence only present in DMTF1α (Figs. 4 and S5). The discrepancies between the
results in the aforementioned study and those of the present study
are likely due to the difference in DMTF1 expression strategies,
expression levels and employed cell lines. Tschan et al
expressed DMTF1 isoforms as fusion proteins with EGFP (13); however, the present study merely
added an N-terminal 3xFlag epitope; they expressed DMTF1γ with a
markedly reduced level compared to DMTF1β, but the present study
expressed all three isoforms at comparable levels; they used 293T
cells, but the present study transfected both MCF-7 and MDA-MB-231
cells with 3xFlag-DMTF1 expression vectors. Thus, it is unclear
whether EGFP can affect DMTF1γ localization and whether DMTF1γ
subcellular localization is cell type-specific. Importantly, it was
predicted the NLS location of DMTF1 in the region of
I47EPPHKRLCLSSE59 based on a published
algorithm and validated this prediction using mutagenesis
experiments, in which K52-R53 substitutions by alanines led to
cytoplasmic translocation of all 3 DMTF1 isoforms (Figs. 4C and S5B). To the best of our knowledge, the
present study is the first to map the NLS domain of DMTF proteins.
Notably, the KR-2A mutations caused the nuclear exclusion of
DMTF1α, but only rendered DMTFβ and γ present in both the cytoplasm
and nucleus. Whether the putative NES sequence
I351AELDV356 only presents in DMTF1α
(Fig. 4A) and contributes to its
nuclear exclusion warrants further investigation.
In reporter assays, luciferase expression driven by
the ARF promoter did not monotonically increase with the escalated
DMTF1α expression (Fig. 5B). It
was predicted that this phenomenon was likely caused by the
quenching effects (25) of
overexpressed DMTF1α that could bind and even saturate
transcription co-factors in a status unassociated with the ARF
promoter and thus interfere with the transcription mediated by the
target promoter. In addition, the observed basal level of the
ARF-promoter reporter was likely driven by endogenous DMTF1α. In
the EMSA experiments, the 2 short DMTF1 isoforms, particularly
DMTF1β, exhibited modest effects in attenuating DMTF1α binding to
its consensus element on the ARF promoter (Fig. 5E and F). Thus, the association of
DMTF1β and γ with α may attenuate DMTF1α-mediated transcription by
disrupting its recruited transcriptional machinery. In recent
years, phase separation caused by intrinsically disordered regions
of transcription factors has been demonstrated to play a key role
in regulating gene expression (26). It was noted that the N-terminal
region shared by the 3 DMTF1 proteins is highly disordered (data
not shown). Thus, the regulation of DMTF1α transcriptional activity
by DMTF1β and γ may also employ the mechanism of the phase
separation. In Fig. 5E and F, a
reduced complex intensity was observed when excessive DMTF1β and γ
proteins were added. It is possible that the incorporation of
DMTF1β and γ into DMTF1α-containing complex formed by the phase
separation mechanism could initially reduce its DNA binding
affinity. However, with the further increased expression of DMTF1β
and γ proteins, DMTF1α could be 'squeezed out' from the complex and
then bind to the probe, which may only occur in the in vitro
assays. Whether the interaction of DMTF1 isoforms is
physiologically significant and where the binding site(s) is
localized warrants further investigation (Fig. 6).
A number of studies have demonstrated positive
correlations between oncogene expression and the poor clinical
outcomes of breast cancer patients, such as PI3K, AKT, mTOR, MYC
and HER2 (27-30). When analyzing a breast cancer
database, the present study also discovered that both DMTF1β/α and
DMTF1γ/α ratios were inversely associated with the survival rates
of the patients (Fig. 7A and B),
supporting the oncogenic activities of DMTF1β and γ demonstrated in
the functional analyses. When the patients were divided into 8
groups based on their high or low DMTF1 isoform expression, it was
suggested that the 3 DMTF1 isoforms could competitively splice and
DMTF1a played a dominant role to suppress breast cancer development
(Fig. 8). Based on the analyses of
the log-rank and Breslow-Wilcoxon tests, DMTF1β and γ could
mutually enhance each other to reduce the long- or short-term
survival rates of breast cancer patients (Fig. 8C-H).
Overall, the data of the present study strongly
support that the oncogenic activity of DMTF1β in mammary
oncogenesis is through its antagonism of DMTF1α. Although DMTF1γ
can negatively affect DMTF1α-mediated ARF promoter activity, it
exerted marginal effects on the DNA binding affinity of DMTF1α. It
is possible that DMTF1β and γ can both contribute to oncogenesis in
a cell and tumor type-dependent manner, while the detailed
regulatory mechanisms warrant further exploration.
Supplementary Data
Acknowledgements
The authors would like to thank Dr Daniel B. Stovall
(College of Arts and Sciences, Winthrop University, Rock Hill, SC,
USA) for critically reading the manuscript.
Funding
The present study was supported by the Fundamental
Research Funds for the Central Universities (2572017AA14) to JL and
(2572020DY13) to GS, and the National Natural Science Foundation of
China (81872293 and 81672795) to GS.
Availability of data and materials
All data generated or analyzed during this study are
included either in this article or in the supplementary information
files.
Authors' contributions
JL, KS and GS conceived the study, wrote the
manuscript and generated the figures. JL, KS, TX, JH, TL and KC
conducted the experiments. JL and GL analyzed the bioinformatics
data. DL provided technical support and was involved in the
conceptual design in several key experiments. KI was involved in
conceiving the project and provided several important reagents and
suggestions to the research plan. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hirai H and Sherr CJ: Interaction of
D-type cyclins with a novel myb-like transcription factor, DMP1.
Mol Cell Biol. 16:6457–6467. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Inoue K and Sherr CJ: Gene expression and
cell cycle arrest mediated by transcription factor DMP1 is
antagonized by D-type cyclins through a
cyclin-dependent-kinase-independent mechanism. Mol Cell Biol.
18:1590–1600. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Inoue K, Roussel MF and Sherr CJ:
Induction of ARF tumor suppressor gene expression and cell cycle
arrest by transcription factor DMP1. Proc Natl Acad Sci USA.
96:3993–3998. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Inoue K, Wen R, Rehg JE, Adachi M,
Cleveland JL, Roussel MF and Sherr CJ: Disruption of the ARF
transcriptional activator DMP1 facilitates cell immortalization,
Ras transformation, and tumorigenesis. Genes Dev. 14:1797–1809.
2000.PubMed/NCBI
|
|
5
|
Inoue K, Zindy F, Randle DH, Rehg JE and
Sherr CJ: Dmp1 is haplo-insufficient for tumor suppression and
modifies the frequencies of Arf and p53 mutations in Myc-induced
lymphomas. Genes Dev. 15:2934–2939. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fry EA, Taneja P, Maglic D, Zhu S, Sui G
and Inoue K: Dmp1α inhibits HER2/neu-induced mammary tumorigenesis.
PLoS One. 8:e778702013. View Article : Google Scholar
|
|
7
|
Zhu S, Mott RT, Fry EA, Taneja P, Kulik G,
Sui G and Inoue K: Cooperation between Dmp1 loss and cyclin D1
overexpression in breast cancer. Am J Pathol. 183:1339–1350. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Peng Y, Dong W, Lin TX, Zhong GZ, Liao B,
Wang B, Gu P, Huang L, Xie Y, Lu FD, et al: MicroRNA-155 promotes
bladder cancer growth by repressing the tumor suppressor DMTF1.
Oncotarget. 6:16043–16058. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang X, Lou Y, Wang M, Liu C, Liu Y and
Huang W: miR-675 promotes colorectal cancer cell growth dependent
on tumor suppressor DMTF1. Mol Med Rep. 19:1481–1490. 2019.
|
|
10
|
Tian N, Li J, Shi J and Sui G: From
general aberrant alternative splicing in cancers and its
therapeutic application to the discovery of an oncogenic DMTF1
isoform. Int J Mol Sci. 18:1912017. View Article : Google Scholar :
|
|
11
|
Tschan MP, Fischer KM, Fung VS, Pirnia F,
Borner MM, Fey MF, Tobler A and Torbett BE: Alternative splicing of
the human cyclin D-binding Myb-like protein (hDMP1) yields a
truncated protein isoform that alters macrophage differentiation
patterns. J Biol Chem. 278:42750–42760. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Maglic D, Stovall DB, Cline JM, Fry EA,
Mallakin A, Taneja P, Caudell DL, Willingham MC, Sui G and Inoue K:
DMP1β, a splice isoform of the tumour suppressor DMP1 locus,
induces proliferation and progression of breast cancer. J Pathol.
236:90–102. 2015. View Article : Google Scholar :
|
|
13
|
Tschan MP, Federzoni EA, Haimovici A,
Britschgi C, Moser BA, Jin J, Reddy VA, Sheeter DA, Fischer KM, Sun
P and Torbett BE: Human DMTF1β antagonizes DMTF1α regulation of the
p14(ARF) tumor suppressor and promotes cellular proliferation.
Biochim Biophys Acta. 1849:1198–1208. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Niklaus NJ, Humbert M and Tschan MP:
Cisplatin sensitivity in breast cancer cells is associated with
particular DMTF1 splice variant expression. Biochem Biophys Res
Commun. 503:2800–2806. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mallakin A, Sugiyama T, Kai F, Kai F,
Taneja P, Kendig RD, Frazier DP, Maglic D, Matise LA, Willingham MC
and Inoue K: The Arf-inducing transcription factor Dmp1 encodes a
transcriptional activator of amphiregulin, thrombospondin-1, JunB
and Egr1. Int J Cancer. 126:1403–1416. 2010.
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Wan M, Huang W, Kute TE, Miller LD, Zhang
Q, Hatcher H, Wang J, Stovall DB, Russell GB, Cao PD, et al Yang
Yin: 1 plays an essential role in breast cancer and negatively
regulates p27. Am J Pathol. 180:2120–2133. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Deng Z, Wan M, Cao P, Rao A, Cramer SD and
Sui G: Yin Yang 1 regulates the transcriptional activity of
androgen receptor. Oncogene. 28:3746–3757. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kosugi S, Hasebe M, Tomita M and Yanagawa
H: Systematic identification of cell cycle-dependent yeast
nucleocytoplasmic shuttling proteins by prediction of composite
motifs. Proc Natl Acad Sci USA. 106:10171–10176. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
la Cour T, Kiemer L, Mølgaard A, Gupta R,
Skriver K and Brunak S: Analysis and prediction of leucine-rich
nuclear export signals. Protein Eng Des Sel. 17:527–536. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ryan M, Wong WC, Brown R, Akbani R, Su X,
Broom B, Melott J and Weinstein J: TCGASpliceSeq a compendium of
alternative mRNA splicing in cancer. Nucleic Acids Res. 44(D1):
D1018–D1022. 2016. View Article : Google Scholar :
|
|
22
|
Martinez RLMC and Naranjo JD: A pretest
for choosing between logrank and Wilcoxon tests in the two-sample
problem. METRON. 68:111–125. 2010. View Article : Google Scholar
|
|
23
|
Baker KE and Parker R: Nonsense-mediated
mRNA decay: Terminating erroneous gene expression. Curr Opin Cell
Biol. 16:293–299. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chang YF, Imam JS and Wilkinson MF: The
nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem.
76:51–74. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Latchman DS: Inhibitory transcription
factors. Int J Biochem Cell Biol. 28:965–974. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Soutourina J: Transcription regulation by
the Mediator complex. Nat Rev Mol Cell Biol. 19:262–274. 2018.
View Article : Google Scholar
|
|
27
|
Khan MA, Jain VK, Rizwanullah M, Ahmad J
and Jain K: PI3K/AKT/mTOR pathway inhibitors in triple-negative
breast cancer: A review on drug discovery and future challenges.
Drug Discov Today. 24:2181–2191. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Garte SJ: The c-myc oncogene in tumor
progression. Crit Rev Oncog. 4:435–449. 1993.PubMed/NCBI
|
|
29
|
Perrier A, Gligorov J, Lefèvre G and
Boissan M: The extracellular domain of Her2 in serum as a biomarker
of breast cancer. Lab Invest. 98:696–707. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Dittmer J: The role of the transcription
factor Ets1 in carcinoma. Semin Cancer Biol. 35:20–38. 2015.
View Article : Google Scholar : PubMed/NCBI
|