Introduction
In 2018, hepatocellular carcinoma (HCC) was
estimated to be the sixth most common cancer and the third most
common cause of cancer-related mortality, resulting in ~841,000 new
cases and 781,000 deaths worldwide (1,2).
Although advances in therapeutic strategies have benefited patients
who are diagnosed at an early stage, the majority of patients with
HCC are diagnosed at an advanced stage and their overall survival
remains poor, which is primarily attributed to the recurrence and
metastasis of the disease (3).
Therefore, identifying novel causative genes and molecular
mechanisms underlying HCC progression is important for the
development of therapeutic targets with improved efficacy.
Recent studies revealed that the Hippo signaling
pathway is implicated in tumorigenesis and may display tumor
suppressor effects (4-6). The core of Hippo signaling consists
of macrophage stimulating 1/2, which regulates activation of large
tumor suppressor kinase 1/2 (LATS1/2). Active LATS1/2
phosphorylates the downstream transcriptional co-activator
Yes-associated protein (YAP)/tafazzin (TAZ). In the cytoplasm, the
proteasome mediates ubiquitination and degradation of
phosphorylated YAP/TAZ, which suppresses the transcription of
proliferation- and survival-associated genes (7). Increasing evidence has indicated
that the Hippo signaling pathway is crucial for HCC initiation and
progression (8-10). Several factors have been reported
to regulate Hippo signaling, including actin cytoskeleton, cell
polarity and cell contact (11).
Moreover, recent studies have demonstrated that cytoskeletal
proteins regulate HCC progression by activating the Hippo signaling
pathway (6,12,13).
Septins are highly conserved GTP-binding proteins
family incorporating 13 members, which are ubiquitously expressed
in the majority of eukaryotes (14). Recently, septins were categorized
as the fourth cytoskeletal component, which interacts with cellular
membranes, actin filaments and microtubules, and regulates various
cellular processes (15). Among
the 13 members, septin 6 (SEPT6) primarily regulates filament
dynamics, cytokinesis, proliferation, cell cycle transition,
survival and chemotaxis (14,16-18). In prostate cancer tissues, SEPT6
expression was decreased, and SEPT6 knockdown contributed to
prostate cancer survival and invasion (18). Recently, it was reported that
SEPT6 expression was upregulated in liver fibrosis, which promoted
hepatic stellate cell activation, proliferation and migration
(19). A previous study
demonstrated that Hepatitis B surface antigen (HBsAg) knockdown
blocked HCC growth, whereas HBsAg knockdown decreased SEPT6
expression in HepG2.2.15 cells (20), indicating that SEPT6 may be
involved in HCC pathogenesis. However, the functional importance of
SEPT6 in HCC development and the regulation of the Hippo signaling
pathway is not completely understood.
The present study aimed to investigate whether SEPT6
expression was upregulated in HCC tissues and to determine its
association with prognosis. The effects of SEPT6 overexpression on
HCC cell proliferation, cell cycle transition, migration and
invasion, and the role of the Hippo signaling pathway and YAP
activation were investigated. The results of the present study may
indicate a novel therapeutic strategy for HCC.
Materials and methods
HCC samples and cell lines
A total of 64 patients (51 male patients and 13
female patients; age range, 26-78 years; average age, 52.58±12.73)
were enrolled in the present study at Tongji Hospital (Wuhan,
China) between January 2011 and December 2014. The inclusion
criteria were as follows: i) Patients were pathologically diagnosed
with HCC; ii) patients underwent surgical excision; and iii)
patients were aged >18 years. The exclusion criteria were as
follows: i) Patients received preoperative therapy; and ii)
patients with more than one primary tumor. The tumor and
corresponding adjacent non-tumor (distance from tumor margin, >2
cm) tissues were collected. Tissues were fixed with 4%
paraformaldehyde at room temperature for 48 h, embedded in paraffin
and sectioned to 5-µm thick sections for
immunohistochemistry staining. Alternatively, tissues were
immediately preserved at −80°C for RNA and protein extraction. The
clinicopathological characteristics of the patients were recorded,
including sex, age, hepatitis B virus infection, α-fetoprotein
levels, tumor size, tumor number and metastasis (Table I). Written informed consent was
obtained from all patients. The present study was approved by the
Ethics Committee of Tongji Hospital (approval no. TJ-IRB20180404)
and was conducted according to the principles outlined in the
Declaration of Helsinki. Two normal hepatocyte cell lines (THLE-2
and THLE-3), Hep3B and Huh7 were purchased from American Type
Culture Collection. MHCC-97L and MHCC-97H were purchased from The
Cell Bank of Type Culture Collection of The Chinese Academy of
Sciences. HCC-LM3 was obtained from the Liver Cancer Institute,
Zhongshan Hospital, Fudan University (Shanghai, China). Cells were
cultured in DMEM supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin and 100 µg/ml
streptomycin at 37°C with 5% CO2.
 | Table IAssociation between SEPT6 expression
and clinicopathological variables in human HCC tissues. |
Table I
Association between SEPT6 expression
and clinicopathological variables in human HCC tissues.
| Variable | SEPT6 expression
| P-value |
|---|
| Low (n=18) | High (n=46) |
|---|
| Sex | | | 0.530 |
| Female | 4 | 9 | |
| Male | 14 | 37 | |
| Age (years) | | | 0.578 |
| ≤45 | 6 | 16 | |
| >45 | 12 | 30 | |
| HBsAg | | | 0.291 |
| Negative | 5 | 18 | |
| Positive | 13 | 28 | |
| AFP (ng/ml) | | | 0.172 |
| ≤400 | 8 | 13 | |
| >400 | 10 | 33 | |
| Tumor diameter
(cm) | | | 0.010a |
| ≤5 | 11 | 12 | |
| >5 | 7 | 34 | |
| Tumor number | | | 0.490 |
| Single | 13 | 35 | |
| Multiple | 5 | 11 | |
| Metastasis | | | 0.016a |
| No | 11 | 13 | |
| Yes | 7 | 33 | |
Reagent
The F-actin inhibitor latrunculin B (Lat. B) was
purchased from Abcam (cat. no. ab144291) and was used following the
standard protocol. Briefly, latrunculin B (1 mg) was dissolved in
40 µl DMSO to make a 25 mg/ml stock solution. The stock
solution was stored at −20°C until subsequent use. Before use, Lat.
B was thawed and added to DMEM to a final concentration of 10
µM. Cells were treated with Lat. B for 2 h at 37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA extraction, reverse transcription, and
RT-qPCR were performed following a standard protocol, as previously
described (19). Total RNA was
extracted from liver tissues and cell lines using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA concentrations were determined using a NanoDrop 2000
Spectrophotometer (Thermo Fisher Scientific, Inc.). Total RNA was
reverse transcribed into cDNA using the PrimeScript reagent kit
(Takara Biotechnology Co., Ltd.) according to the manufacturer's
protocol. The following temperature protocol was used for reverse
transcription: 37°C for 15 min and 85°C for 5 sec. Subsequently,
qPCR was performed using SYBR Premix ExTaq (Takara Biotechnology
Co., Ltd.) and an ABI StepOne Real-Time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. The following thermocycling conditions
were used for qPCR: Pre-denaturation at 95°C for 30 sec; followed
by 40 cycles of 95°C for 5 sec and 60°C for 30 sec. mRNA expression
levels were quantified using the 2−∆∆Cq method (21) and normalized to the internal
reference gene β-actin. The sequences of the primers used for qPCR
are presented in Table SI.
Western blotting and
co-immunoprecipitation (co-IP)
Total protein was extracted from liver tissues and
cell lines using RIPA buffer containing protease inhibitors PMSF
and cocktail (Servicebio Technology Co., Ltd.). Nuclear proteins
were extracted using NE-PER (Thermo Fisher Scientific, Inc.).
Protein concentrations were determined using a BCA kit (Boster
Biological Technology). Western blotting was performed as
previously described (19).
Briefly, proteins (30 µg per lane) were separated via 10%
SDS-PAGE and transferred onto PVDF membranes (EMD Millipore), which
were then blocked with 5% BSA (cat. no. 4240GR100; Guangzhou Saiguo
Biotech Co., Ltd.) at room temperature for 1 h. Subsequently, the
membranes were incubated with primary antibodies at 4°C overnight.
After washing three times in TBST, the membranes were incubated
with HRP-conjugated secondary antibodies (Beyotime Institute of
Biotechnology) at room temperature for 1 h. After washing three
times, protein bands were detected using an ECL assay kit
(Advansta, Inc.). Protein expression was semi-quantified using
ImageJ software (version 1.44p; National Institutes of Health) with
β-actin as the loading control. The primary and secondary
antibodies used for western blotting are listed in Table SII.
For the co-IP assay, MHCC-97H cells
(2-5×107) were washed twice with cold PBS twice and
lysed using 1% NP-40 buffer (cat. no. P0013F; Beyotime Institute of
Biotechnology) containing protease inhibitors at 4°C for 30 min.
After centrifugation at 12,000 × g for 15 min at 4°C, the
supernatant was collected. Protein A/G PLUS-Agarose beads (cat. no.
sc-2003; Santa Cruz Biotechnology, Inc.) were washed three times
with PBS and diluted in PBS to 50% concentration. Subsequently, the
Agarose beads (100 µl/ml) were added to the supernatant
(containing 200-600 µg protein). The mixture was incubated
for 30 min at 4°C on a horizontal shaker. After centrifugation at
1,000 × g for 5 min at 4°C, the supernatant was collected and
divided into two parts. SEPT6 antibody (2 µg/500 µg
cell lysate) or isotype normal IgG antibody (2 µg/500
µg cell lysate; cat. no. sc-2026; Santa Cruz Biotechnology,
Inc.) was added to the supernatant (~500 µl total volume)
and incubated for 1 h at 4°C. Subsequently, additional Agarose
beads (100 µl/ml) were added and incubated at 4°C overnight.
After centrifugation at 1,000 × g for 5 min at 4°C, the supernatant
was discarded and the pellets were washed four times with 1.0 ml
NP-40 buffer. The samples were boiled with sample loading buffer
for 10 min, and the Agarose beads were discarded. The supernatant
was collected and analyzed via western blotting using anti-bodies
targeted against SEPT6, LATS1 and LATS2 according to the
aforementioned protocol. The antibodies used for co-IP are listed
in Table SII.
Immunohistochemistry (IHC) staining
The expression of SEPT6 in HCC samples and
corresponding adjacent non-tumor tissues were analyzed by IHC, as
previously described (19).
Briefly, paraffin-embedded slides were de-paraffinized in xylene
and rehydrated using an alcohol gradient. Antigen retrieval was
performed by heating samples in 0.01 mol/l citrate buffer (pH 6.0)
for 15 min in a microwave. Subsequently, the slides were immersed
in 3% H2O2 at room temperature for 15 min to
eliminate the endogenous peroxidase. After washing three times with
PBS, the sections were blocked using 10% goat serum (Boster
Biological Technology) at room temperature for 30 min.
Subsequently, the sections were incubated with an anti-SEPT6 (cat.
no. 12805-1-AP; 1:100; ProteinTech Group, Inc.) at 4°C overnight.
After washing three times with PBS, the sections were incubated
with a biotinylated secondary antibody (cat. no. SP-9000; OriGene
Technologies, Inc.) at 37°C for 1 h. After washing three times with
PBS, peroxidase activity was visualized using DAB (OriGene
Technologies, Inc.) at room temperature for ~10 sec. Then, the
sections were counterstained with hematoxylin (OriGene
Technologies, Inc.) at room temperature for ~1 min. Stained samples
were visualized using an IX71 light microscope (Olympus
Corporation; magnification, ×100).
Immunofluorescence staining
Huh7 cells (5×104 each well) were seeded
onto glass cover slides in 24-well plates overnight. Subsequently,
cells were fixed with 4% formaldehyde at room temperature for 20
min, permeabilized using 0.3% Triton X-100 and blocked with 5% BSA
(cat. no. 4240GR100; BioFroxx; Saiguo Biological Technology Co.,
Ltd.) at room temperature for 30 min. Subsequently, the slides were
incubated with ActinRed (cat. no. KGMP0012; Nanjing KeyGen Biotech
Co., Ltd.) at room temperature for 20 min. The nuclei were
counter-stained with DAPI solution (cat. no. G1012; Wuhan
Servicebio Technology Co., Ltd.) at room temperature for 10 min.
Stained cells were observed using an IX71 fluorescence microscope
(Olympus Corporation; magnification, ×400).
Plasmid transfection and stable cell line
selection
The plasmids used for SEPT6 and YAP knockdown and
overexpression were purchased from Shanghai GeneChem Co., Ltd. At
80-90% confluence, cells were transfected with 2 µg plasmid
using Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) in Opti-MEM (Gibco; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. At 6-8 h
post-transfection, the cell culture medium was replaced with DMEM
supplemented with 10% FBS. The control shRNA was a non-targeting
shRNA and the overexpression control was an empty vector. The shRNA
sequences are presented in Table
SI. At 48 h post-transfection, transfected cell lines were
treated with G418 (400 µg/ml) for 2 weeks to select stably
transfected cells. Transfection efficiencies were assessed and the
stable cell lines were used for subsequent experiments. The
following cell lines were established: MHCC-97H-shcontrol,
MHCC-97H-shSEPT6, Huh7-Vector, Huh7-SEPT6, MHCC-97H-shSEPT6+YAP,
Huh7-SEPT6+shYAP, HCC-LM3-shcontrol, HCC-LM3-shSEPT6, Hep3B-Vector
and Hep-3B-SEPT6.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation was detected using the CCK-8 kit
(cat. no. C0037; Beyotime Institute of Biotechnology) according to
the manufacturer's protocol, as previously described (19). Briefly, cells were seeded
(1×103 cells/well) into 96-well plates and cultured for
24, 48, 72 or 96 h. Subsequently, the culture medium was replaced
with 100 µl DMEM and 10 µl CCK-8. After incubation
for 2 h, the absorbance of each well was measured at a wavelength
of 450 nm using an ELISA reader.
Flow cytometry analysis of the cell
cycle
Cell cycle distribution was assessed by flow
cytometry, as previously described (22). Briefly, cells (1×106)
were harvested, washed with cold PBS and fixed using 75% ethanol
overnight at 4°C. After washing twice with PBS, cells were
incubated with PI staining solution containing RNase (cat. no.
KGA511-KGA512; Nanjing KeyGen Biotech Co., Ltd.) at 37°C for 30 min
in the dark. Subsequently, cell cycle distribution was analyzed
using a BD FACSVerse flow cytometer (BD Biosciences) and
CELLQuestPro software (version 5.1; BD Biosciences).
Transwell assays
Transwell insert chambers (pore size, 8 µm;
Corning, Inc.) were used to examine cell invasion and migration,
respectively. For the invasion assay, the upper chamber inserts
were precoated with Matrigel (BD Biosciences) at 37°C for 1 h.
Briefly, 200 µl serum-free DMEM containing cells
(2×104) was plated into the upper chamber and the lower
chamber was filled with 600 µl DMEM supplemented with 20%
FBS. Following incubation at 37°C for 24 h, migratory/invading
cells were fixed using absolute methanol at room temperature for 10
min, and stained using 0.2% crystal violet solution at room
temperature for 1 h. Cells were observed using an IX71 microscope
(Olympus Corporation; magnification, ×100) in at least three fields
of view.
Database
The Gene Expression Profiling Interactive Analysis
(GEPIA) database (gepia.cancer-pku.cn) was used to determine SEPT6
mRNA expression levels in human liver HCC specimens and
corresponding adjacent non-tumor specimens.
Statistical analysis
Each experiment was performed in triplicate. Data
are presented as the mean ± SD. Statistical analyses were performed
using GraphPad Prism (version 5.0; GraphPad Software, Inc.) or SPSS
(version 19.0; IBM Corp.) software. Comparisons between two groups
were analyzed using the paired or unpaired Student's t-test.
Comparisons among multiple groups were analyzed using one-way ANOVA
followed by Tukey's post hoc test. Categorical data were analyzed
using Fisher's exact test. Patient survival was analyzed via
Kaplan-Meier analysis and log-rank tests. P<0.05 was considered
to indicate a statistically significant difference.
Results
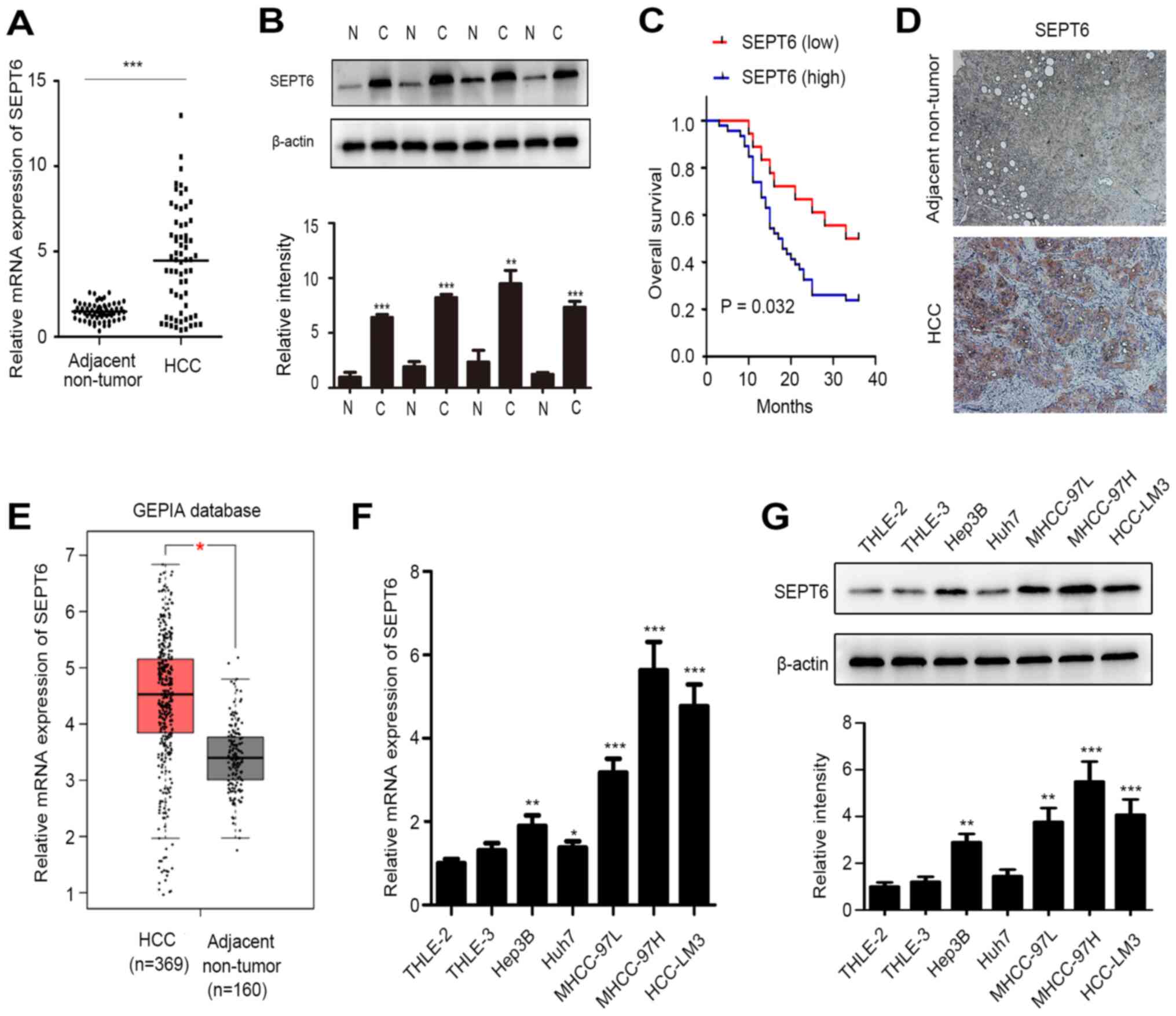
SEPT6 is upregulated in human HCC and
predicts poor prognosis
To examine SEPT6 expression in HCC, SEPT6 mRNA
expression levels were assessed in 64 paired HCC samples. Compared
with corresponding adjacent non-tumor samples, SEPT6 mRNA
expression levels were significantly higher in 46 paired HCC
samples (71.88%; Fig. 1A).
Subsequently, we selected 20 paired tissues, including 16 paired
tissues with higher SEPT6 mRNA expression in the HCC tisues
compared with the adjacent non-tumor tissues, and 4 paired tissues
with lower SEPT6 mRNA expression in HCC tissues compared with the
adjacent non-tumor tissues. The protein expression levels of SEPT6
were determined via western blotting. The results demonstrated that
the protein expression levels of SEPT6 were significantly higher in
16 HCC tissues compared with the adjacent non-tumor tissues
(Fig. 1B and S1A, C and D), whereas the protein
expression levels of SEPT6 were significantly lower in 4 HCC
tissues compared with the adjacent non-tumor tissues (Fig. S1B), indicating a positive
association between mRNA and protein expression levels of SEPT6 in
human patients with HCC. The association between SEPT6 expression
levels and clinicopathological characteristics was investigated.
SEPT6 expression levels were significantly associated with tumor
size and metastasis, but not significantly associated with sex,
age, hepatitis B virus infection, tumor number or α-fetoprotein
levels (Table I). Furthermore,
the association between SEPT6 expression and overall survival was
assessed. SEPT6 high/low expression represented that SEPT6
expression in the HCC tissues was higher/lower compared with the
corresponding adjacent non-tumor tissues (fold change >1.5),
respectively. The results demonstrated that high SEPT6 expression
levels indicated significantly worse overall survival in patients
with HCC compared with low SEPT6 expression levels (Fig. 1C). IHC staining demonstrated that
SEPT6 expression levels were notably higher in HCC samples compared
with corresponding adjacent non-tumor samples, and SEPT6 protein
expression was primarily localized in the cytoplasm (Fig. 1D). In addition, analysis of the
GEPIA database demonstrated significantly upregulated SEPT6
expression in HCC compared with adjacent non-tumor tissues
(Fig. 1E). Subsequently, SEPT6
expression levels were examined in two normal hepatocyte cell lines
(THLE-2 and THLE-3) and several HCC cell lines. Among HCC cell
lines, MHCC-97H and HCC-LM3 cells display the highest metastatic
potential (23-25). The results suggested that SEPT6
expression was significantly higher in the majority of the HCC cell
lines, particularly in those with high metastatic potential
(MHCC-97H and HCC-LM3), compared with normal hepatocytes (Fig. 1F and G). Collectively, the results
indicated that SEPT6 expression was upregulated in human HCC and
may serve as a predictor of poor prognosis.
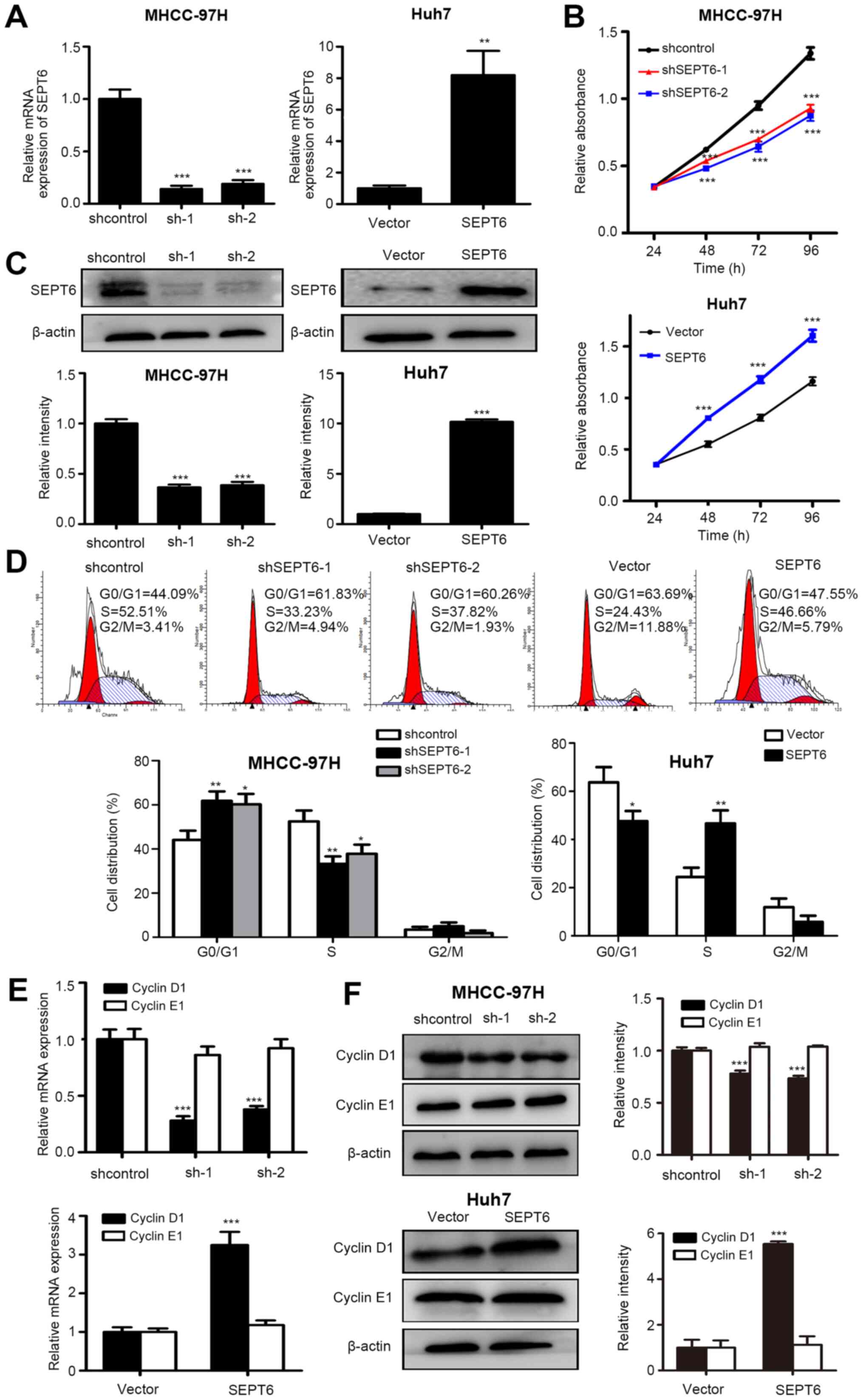
SEPT6 promotes HCC cell
proliferation
Subsequently, gain- and loss-of-function assays were
performed to assess the effect of SEPT6 on HCC cell function.
Following assessment of SEPT6 endogenous expression levels in
different HCC cells, MHCC-97H and Huh7 cells were selected for
SEPT6 knockdown or overexpression, respectively, and stably
transfected cells were established. Transfection efficiencies were
determined by measuring SEPT6 mRNA and protein expression levels
(Fig. 2A and C). The CCK-8 assay
results indicated that SEPT6 knockdown significantly inhibited
MHCC-97H cell proliferation compared with the control group,
whereas SEPT6 overexpression significantly increased Huh7 cell
proliferation compared with the Vector group (Fig. 2B). Subsequently, flow cytometry
was performed to investigate whether SEPT6 regulated the cell
cycle. Compared with the control group, SEPT6 knockdown resulted in
significantly increased cell cycle arrest at the G1/S
phase in MHCC-97H cells, but displayed no significant effect on the
G2/M transition (Fig.
2D). By contrast, compared with the Vector group, SEPT6
overexpression significantly promoted G1/S transition in
Huh7 cells, but had no significant effect on G2/M
transition. The results suggested that SEPT6 primarily regulated
the G1/S transition, whereas its effect on the
G2/M transition was not significant. Furthermore, cyclin
D1 and cyclin E1 expression levels are significantly associated
with G1/S cell cycle transition (15). The RT-qPCR results indicated that
SEPT6 knockdown significantly decreased cyclin D1 expression in
MHCC-97H cells compared with the control group, whereas SEPT6
overexpression significantly increased cyclin D1 expression in Huh7
cells compared with the Vector group (Fig. 2E). However, cyclin E1 mRNA
expression levels were not significantly altered by SEPT6 knockdown
or overexpression compared with the control and Vector groups,
respectively. Consistent results were obtained for protein
expression levels (Fig. 2F).
Collectively, the results indicated that SEPT6 promoted HCC cell
proliferation and G1/S transition in vitro.
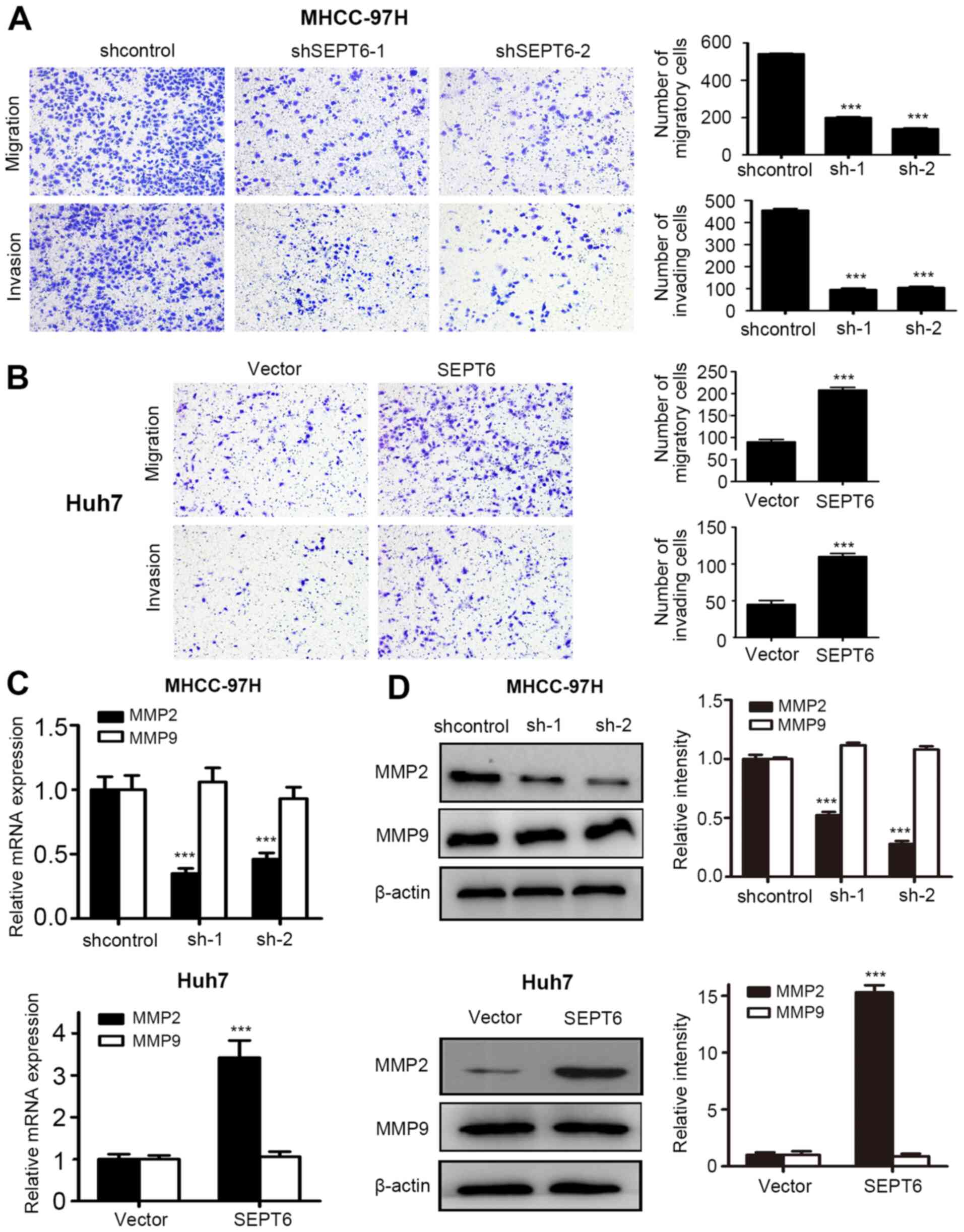
SEPT6 promotes HCC cell migration and
invasion
Metastasis is the leading cause of HCC-related
mortality (26). The Transwell
assay results demonstrated that SEPT6 knockdown significantly
decreased MHCC-97H cell migration and invasion compared with the
control group, whereas SEPT6 overexpression significantly increased
Huh7 cell migration and invasion compared with the Vector group
(Fig. 3A and B). Moreover, matrix
metallopeptidase (MMP)2 expression levels were significantly
decreased by SEPT6 knockdown in MHCC-97H cells compared with the
control group, whereas SEPT6 overexpression significantly increased
MMP2 expression levels in Huh7 cells compared with the Vector group
(Fig. 3C). However, MMP9 mRNA
expression levels were not significantly altered in response to
SEPT6 knockdown or overexpression compared with the control and
Vector groups, respectively (Fig.
3C). Similar results were obtained via western blotting
(Fig. 3D). Collectively, the
in vitro results demonstrated that SEPT6 enhanced HCC cell
migration and invasion.
SEPT6 regulates the Hippo/YAP signaling
pathway in HCC
As aforementioned, SEPT6 promoted HCC cell
proliferation and migration; therefore, the mechanism underlying
its action was investigated. Increasing evidence has demonstrated
that Hippo signaling is crucial for HCC tumorigenesis and
metastasis (8-10). Furthermore, Hippo signaling is
primarily regulated by the actin cytoskeleton (11). Septin proteins are considered as
the fourth cytoskeletal component and SEPT6 regulates actin
cytoskeleton dynamics (15);
therefore, the present study further investigated whether SEPT6
promoted HCC cell progression by regulating Hippo signaling. The
transfection efficiency of SEPT6 knockdown and overexpression in
HCC-LM3 and Hep3B cells, respectively, was verified via RT-qPCR and
western blotting (Fig. S2). The
western blotting results indicated that compared with the control
group, SEPT6 knockdown significantly promoted the phosphorylation
of LATS1 and YAP in MHCC-97H and HCC-LM3 cells, but notably
decreased the overall expression of YAP. By contrast, compared with
the vector group, SEPT6 overexpression significantly decreased the
phosphorylation of LATS1 and YAP, and markedly increased the
overall expression of YAP both in Huh7 and Hep3B cells (Fig. 4A). The results indicated that
SEPT6 may regulate the activity of Hippo signaling, while
modulating the activity, stability and overall expression of YAP.
Subsequently, the present study examined whether SEPT6 regulated
YAP nuclear translocation. The western blotting results
demonstrated that SEPT6 knockdown significantly decreased nuclear
YAP protein expression levels in MHCC-97H and HCC-LM3 cells
compared with the control group, whereas SEPT6 overexpression
significantly upregulated nuclear YAP protein expression levels in
Huh7 and Hep3B cells compared with the Vector group (Fig. 4B). The results suggested that
SEPT6 upregulation may inactivate Hippo signaling by inhibiting the
phosphorylation of LATS1, which resulted in inhibition of YAP
phosphorylation, as well as proteasome-induced YAP ubiquitination
and degradation. Therefore, higher protein expression levels of YAP
were translocated to the nucleus, resulting in enhanced gene
transcription.
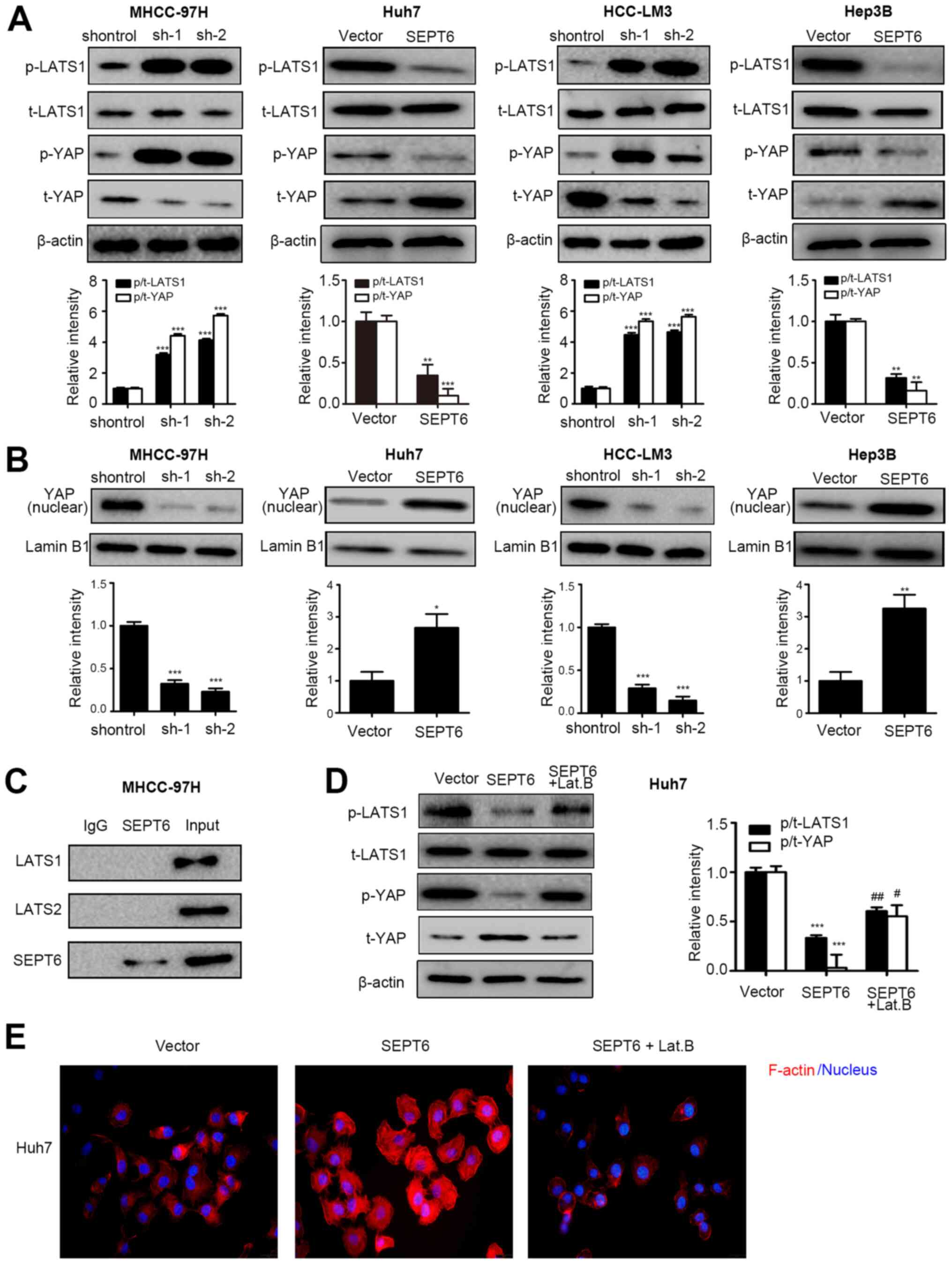 | Figure 4SEPT6 regulates the Hippo/YAP
signaling pathway in HCC. (A) p-LATS1, LATS1, p-YAP and YAP protein
expression levels were measured via western blotting. (B) Nuclear
YAP protein expression levels were measured via western blotting
using Lamin B1 as the loading control. (C) Co-immunoprecipitation
assays were performed to determine the interaction between SEPT6
and LATS1 or LATS2. SEPT6-overexpression Huh7 cells were treated
with 10 µM Lat. B for 2 h to disrupt the cytoskeleton. (D)
Protein expression levels of p-LATS1, LATS1, p-YAP and YAP were
measured via western blotting. (E) F-actin formation was determined
by performing immunofluorescence assays (magnification, ×400).
*P<0.05, **P<0.01 and
***P<0.001 vs. shcontrol or Vector;
#P<0.05 and ##P<0.01 vs. SEPT6. SEPT6,
septin 6; YAP, yes-associated protein; HCC, hepatocellular
carcinoma; p, phosphorylated; LATS1, large tumor suppressor kinase
1; Lat. B, Latrunculin B; t, total; sh, short hairpin RNA. |
Furthermore, an endogenous co-IP assay was performed
to investigate whether SEPT6 interacted with LATS1 and LATS2. The
results indicated that SEPT6 did not directly interact with LATS
(Fig. 4C). Septins, the fourth
component of the cytoskeleton, lack the kinase activity domain
(6); therefore, SEPT6 may
regulate LATS1 phosphorylation indirectly, which may be associated
with the cytoskeleton-regulating function of the septin proteins.
To verify this hypothesis, SEPT6-overexpression Huh7 cells were
treated with the F-actin inhibitor Lat. B to disrupt the
cytoskeleton. Subsequently, F-actin levels and the phosphorylated
and overall expression levels of LATS1 and YAP were assessed. The
results indicated that compared with the vector group, SEPT6
overexpression notably facilitated F-actin formation, which was
markedly disrupted by Lat. B (Fig.
4E). Furthermore, Lat. B treatment increased LATS1 and YAP
phosphorylation, thus notably decreasing YAP overall expression in
SEPT6-overexpression Huh7 cells (Fig.
4D). Collectively, the results indicated that SEPT6 may
regulate Hippo/YAP signaling in HCC by modulating F-actin
formation.
SEPT6 regulates HCC cell proliferation,
cell cycle progression, migration and invasion via the Hippo/YAP
signaling pathway
The present study further investigated whether SEPT6
exerted its effects via regulating the Hippo/YAP signaling pathway.
Firstly, the transfection efficiencies of YAP-overexpression
plasmids in MHCC-97H cells and YAP-knockdown plasmids in Huh7 cells
were verified by measuring mRNA and protein expression levels,
which suggested that these plasmids were appropriate for YAP
overexpression and knockdown (Fig.
S3). Subsequently, YAP was overexpressed by plasmid
transfection in stable SEPT6-knockdown MHCC-97H cells and stable
cells were selected by G418. YAP upregulation in MHCC-97H-shSEPT6
cells was validated by protein expression analysis (Fig. 5A). Compared with the control
group, SEPT6 knockdown significantly decreased MHCC-97H cell
proliferation, G1/S transition, migration and invasion
(Fig. 5B-D). SEPT6
knockdown-induced effects were significantly reversed by YAP
overexpression. YAP downregulation was achieved by shRNA
transfection in stable SEPT6-overexpression Huh7 cells, and stable
cells were selected for further experiments. YAP knockdown was
verified in Huh7-SEPT6 cells (Fig.
5E). Compared with the Vector group, SEPT6 overexpression
significantly enhanced Huh7 cell proliferation, migration, invasion
and G1/S phase transition (Fig. 5F-H). SEPT6 overexpression-mediated
effects were significantly reversed by YAP knockdown. Furthermore,
the present study assessed whether YAP was involved in
SEPT6-regulated cyclin D1 and MMP2 expression. YAP overexpression
significantly upregulated cyclin D1 and MMP2 expression levels in
SEPT6-knockdown MHCC-97H cells, whereas YAP knockdown significantly
decreased cyclin D1 and MMP2 expression levels in
SEPT6-overexpression Huh7 cells (Fig.
5I). Furthermore, to analyze SEPT6-independent effects of YAP
on the regulation of cyclin D1 and MMP2 expression, four stable
cell lines were established by plasmid transfection and G418
selection using Huh7 cells, namely Huh7-Vector, Huh7-SEPT6,
Huh7-shYAP and Huh7-shYAP-SEPT6. Compared with the Vector group,
SEPT6 overexpression significantly upregulated cyclin D1 and MMP2
expression levels, whereas YAP knockdown not only inhibited
SEPT6-mediated effects on cyclin D1 and MMP2 expression, but also
significantly decreased cyclin D1 and MMP2 expression levels
(Fig. S4). Collectively, the
results demonstrated that the Hippo/YAP signaling axis may serve a
key role in SEPT6-induced HCC cell proliferation, cell cycle
progression, migration and invasion.
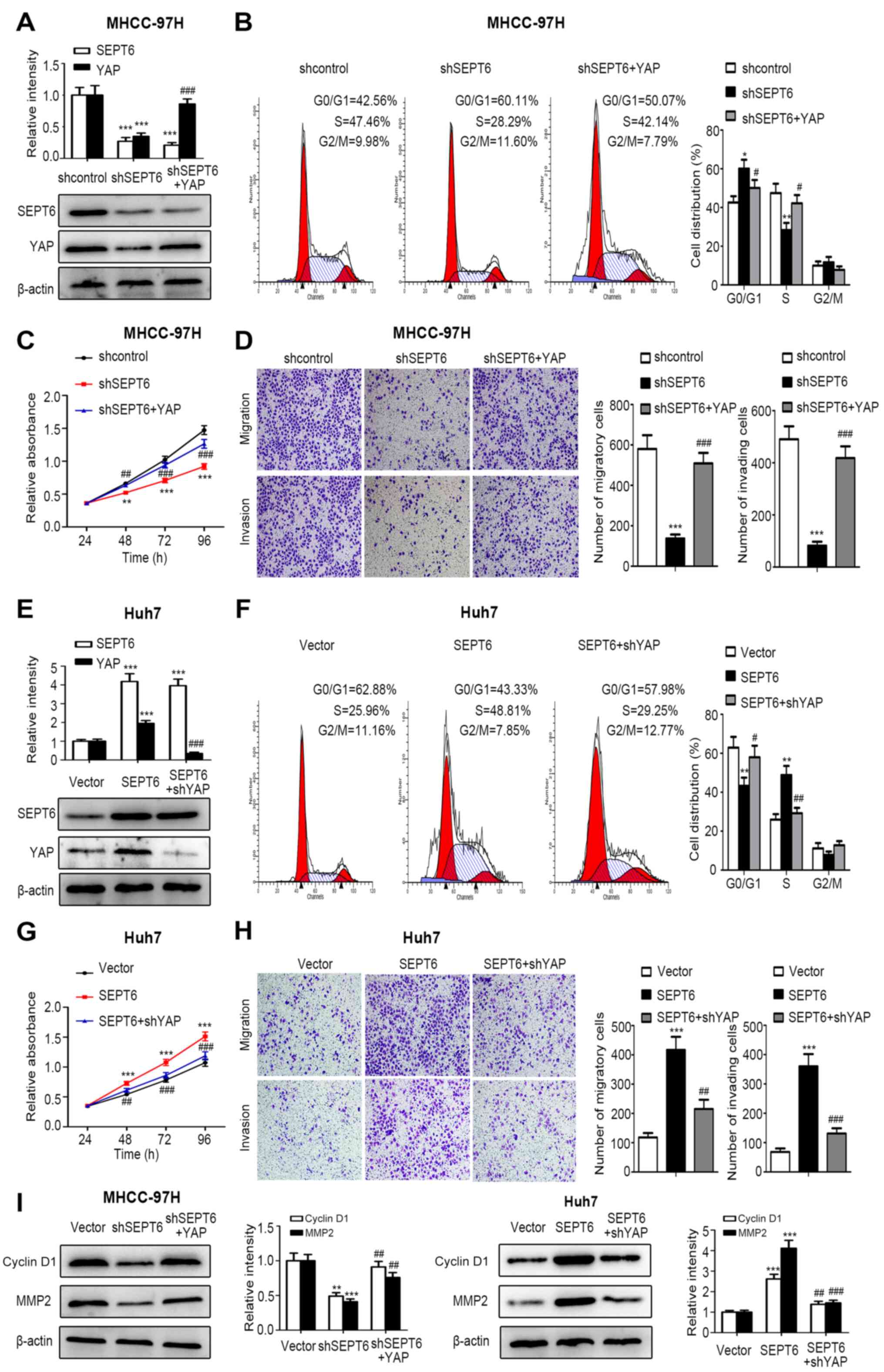 | Figure 5SEPT6 regulates HCC cell
proliferation, cell cycle progression, migration and invasion via
the Hippo/YAP signaling pathway. MHCC-97H-shSEPT6 cells were
transfected with YAP and stable Huh7-SEPT6 cells were transfected
with shYAP. Subsequently, G418 was used for stable cell selection.
(A) Transfection efficiencies of shSEPT6 and YAP were determined
via western blotting. MHCC-97H cell (B) cycle distribution, (C)
proliferation, (D) migration and invasion (magnification, ×100)
were assessed by performing CCK-8, flow cytometry and Transwell
assays, respectively. (E) Transfection efficiencies of SEPT6 and
shYAP were determined via western blotting. Huh7 cell (F) cycle
distribution, (G) proliferation, (H) migration and invasion
(magnification, ×100) were assessed by performing CCK-8, flow
cytometry and Transwell assays, respectively. (I) Cyclin D1 and
MMP2 protein expression levels were determined via western
blotting. *P<0.05, **P<0.01 and
***P<0.001 vs. Vector; #P<0.05,
##P<0.01 and ###P<0.001 vs. shSEPT6 or
SEPT6. SEPT6, septin 6; HCC, hepatocellular carcinoma; YAP,
yes-associated protein; sh, short hairpin RNA; CCK-8, Cell Counting
Kit-8; MMP2, matrix metallopeptidase 2. |
Discussion
HCC is the third leading cause of cancer-related
mortality, and its prognosis is extremely poor due to early relapse
and metastasis following curative resection (3). Although various therapeutic targets
have been identified, the prognosis of patients with HCC remains
poor (3). To the best of our
knowledge, the present study was the first to demonstrate that
SEPT6 inhibited Hippo signaling, activated the downstream effector
YAP, and enhanced cyclin D1 and MMP2 expression levels, which
promoted HCC growth and metastasis.
SEPT6 is primarily implicated in hematological
malignancies (27), nervous
system development (16) and
tumor progression (18). In
prostate cancer, SEPT6 expression is downregulated, and SEPT6
knockdown promotes cancer cell survival and invasion, suggesting a
tumor suppressor role (18).
However, the present study demonstrated that SEPT6 expression was
significantly increased in HCC tissues compared with corresponding
adjacent non-tumor tissues, which was associated with poor
prognosis. The results prompted investigation into why SEPT6
expression was upregulated in HCC tissues. It was hypothesized that
the malignant transformation of tumor cells and the complicated
tumor microenvironment, which involves hypoxia, inflammatory
cytokines stimulation, metabolic reprogramming and epigenetic
regulation, might be the leading causes for SEPT6 upregulation in
HCC. Moreover, the leading cause of HCC and whether the causes work
synergistically requires further investigation. SEPT6
overexpression significantly enhanced HCC cell proliferation, cell
cycle transition, migration and invasion compared with the Vector
group, whereas SEPT6 knockdown displayed the opposite effects
compared with the control group. Therefore, SEPT6 was identified as
an oncogene in HCC, which contrasted to its role in prostate
cancer. It was previously reported that SEPT6 promoted liver
fibrogenesis (19). Since liver
fibrosis and liver cirrhosis are considered as the precancerous
states of HCC, it is reasonable to hypothesize that SEPT6 may
promote HCC progression. The results of the present study indicated
that the expression patterns and effects of SEPT6 in prostate
cancer and HCC were opposite, which may be due to different genetic
backgrounds, including gene mutation, or the tumor
microenvironment. The roles of certain proteins are
context-dependent; therefore, the difference in the tumor
microenvironment between HCC and prostate cancer may result in
different expression patterns and functional roles of SEPT6.
Subsequently, the mechanism underlying the oncogenic
action of SEPT6 was examined. The present study focused on the
Hippo signaling pathway, which is crucial for HCC tumorigenesis and
progression (8-10). Hippo signaling is primarily
regulated by the actin cytoskeleton (11). For example, the cytoskeletal
protein PDZ and LIM domain 1 inhibited HCC metastasis by activating
Hippo signaling (6). SEPT6 has
been reported to regulate actin and microtubule remodeling
(17). Based on the
aforementioned studies, the potential role of SEPT6 in promoting
HCC progression in a Hippo/YAP-dependent manner was assessed. The
results indicated that compared with the Vector group, SEPT6
overexpression inactivated Hippo signaling, and dephosphorylated
and stabilized the downstream effector YAP, leading to the
translocation of active YAP into the nucleus and the
transactivation of cyclin D1 and MMP2, which resulted in HCC cell
proliferation and metastasis (Fig.
6). Furthermore, YAP knockdown significantly reversed the
oncogenic effects of SEPT6 overexpression on HCC progression,
whereas YAP overexpression significantly reversed SEPT6
knockdown-mediated inhibitory effects. As previously reported, YAP
regulates cyclin D1 and MMP2 expression independently, regardless
of SEPT6 expression (26,28-30). The results of the present study
also demonstrated that YAP knockdown inhibited SEPT6-mediated
upregulation of cyclin D1 and MMP2 expression. The results
demonstrated that SEPT6 regulated cyclin D1 and MMP2 expression via
YAP, and YAP independently regulated cyclin D1 and MMP2 expression
to a certain extent.
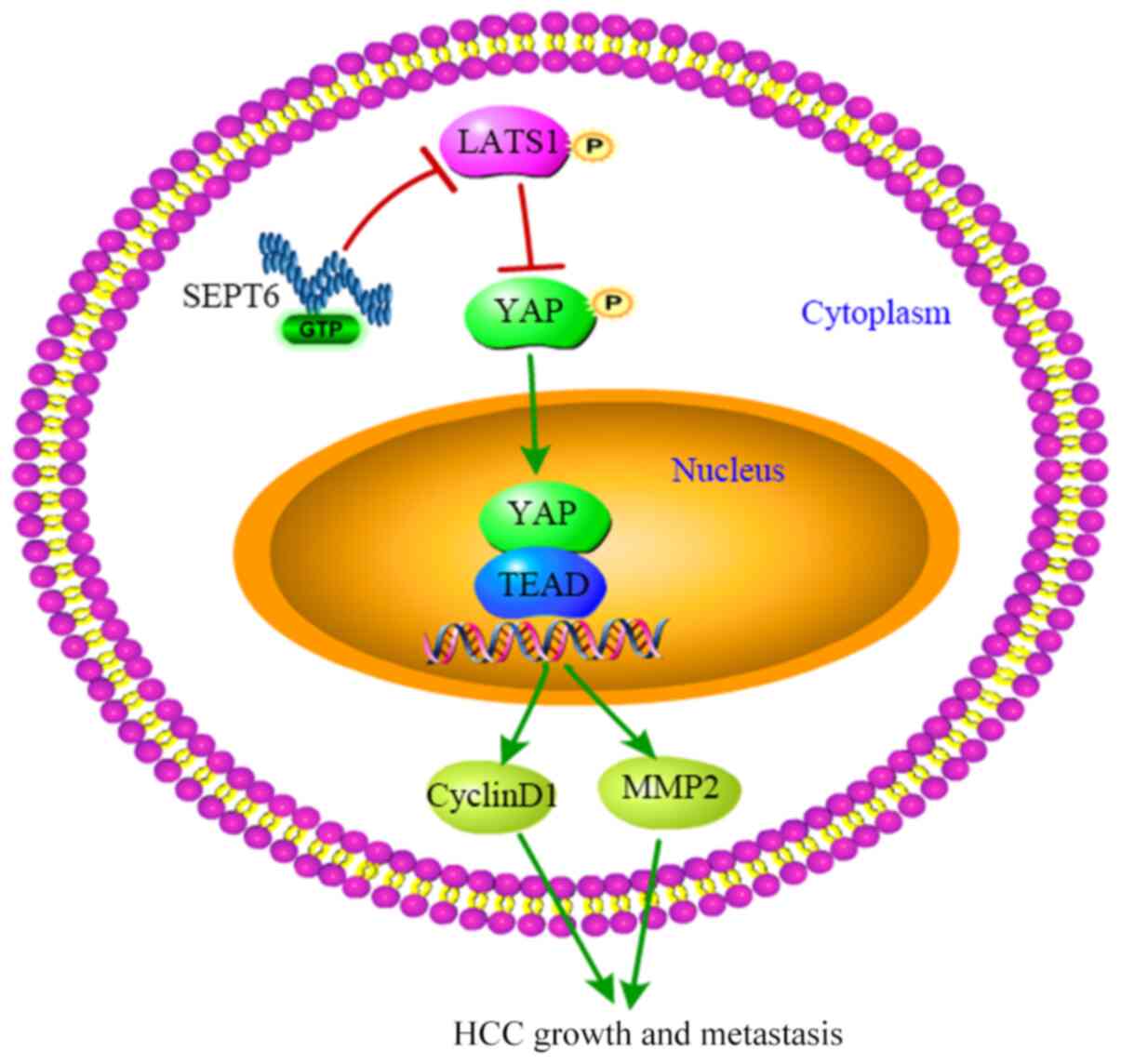 | Figure 6Summary of SEPT6-regulated Hippo/YAP
signaling pathway in HCC. SEPT6 expression was upregulated in HCC,
which inactivated Hippo signaling, dephosphorylated and stabilized
the downstream effector YAP and upregulated YAP expression.
Subsequently, active YAP was translocated to the nucleus and
promoted transactivation of cyclin D1 and MMP2, resulting in HCC
cell proliferation and metastasis. SEPT6, septin 6; YAP,
yes-associated protein; HCC, hepatocellular carcinoma; MMP2, matrix
metallopeptidase 2; TEAD, TEA domain transcription factor; p,
phosphorylated |
The regulatory mechanism underlying Hippo signaling
has received increasing attention. Hippo signaling can be regulated
by mechanical force, the extracellular matrix, cell-cell contact
and cytoskeletal interactions (7,11,31,32). With regard to cytoskeletal
interactions, Hippo signaling can be regulated by F-actin levels,
F-actin activity and cytoskeletal tension (11). In HCC cell lines, F-actin was
confirmed to bind to LATS1, resulting in the dephosphorylation and
inactivation of Hippo signaling (6). Furthermore, the Rho GTPase serves an
important role in regulating Hippo signaling activity by the
cytoskeleton (31). Septins
belong to a family of GTP-binding proteins and are considered as
the fourth cytoskeletal component. In addition, SEPT6 was reported
to regulate actin cytoskeleton dynamics (14,15). Based on previous studies, it was
hypothesized that SEPT6 may regulate Hippo via three possible
mechanisms, one of which involves the regulation of F-actin
formation by SEPT6 in order to affect cytoskeleton dynamics.
Subsequently, F-actin binds to LATS1 and causes Hippo inactivation.
The hypothesis was confirmed by the present study, since compared
with the Vector group, SEPT6 overexpression notably facilitated
F-actin formation, whereas disruption of F-actin by Lat. B
abrogated SEPT6-induced LATS1 dephosphorylation and Hippo
inactivation. Furthermore, whether SEPT6 regulates other proteins
associated with cytoskeleton dynamics, including Ezrin and
neurofibromin 2 (NF2), requires further investigation. Previous
studies reported that Ezrin and NF2 were involved in the regulation
of Hippo signaling (33,34). Secondly, as a GTP-binding protein,
SEPT6 may regulate GTPase activity and thus, Hippo signaling
activity. Finally, it may be possible that SEPT6 mediates the
recruitment of certain phosphatases to repress LATS1
phosphorylation. Collectively, the results of the present study
indicated a possible mechanism by which SEPT6 regulated Hippo
signaling via upregulation of F-actin formation. However, further
investigation of the underlying mechanism is required.
In conclusion, the present study demonstrated that
SEPT6 was upregulated in HCC and displayed an oncogenic function in
HCC progression. SEPT6 promoted HCC cell proliferation, cell cycle
progression, migration and invasion, which was mediated at least
partly via the SEPT6/Hippo/YAP axis. Therefore, the results of the
present study may provide novel insight into HCC treatment.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
YF performed the cytologic and mechanistic
experiments. ZD and QD analyzed the data. JZ analyzed the clinical
data. QD and JZ confirm the authenticity of all the raw data. MODW
and ALG made substantial contributions to the conception of the
study and drafted the manuscript. ML and CJS designed the study and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Tongji
Hospital Ethics Committee (approval no. TJ-IRB20180404). Written
informed consent was obtained from each patient in accordance with
the ethical standards of the World Medical Association Declaration
of Helsinki.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Deutsche
Forschungsgemeinschaft (grant nos. DFG STE 1022/2-3 and DFG STE
1022/4-1), the National Natural Science Foundation of China (grant
nos. 81272657 and 81572422) and the China Scholarship Council
(grant nos. 201908080017 and 201606230249).
References
|
1
|
Yang JD, Hainaut P, Gores GJ, Amadou A,
Plymoth A and Roberts LR: A global view of hepatocellular
carcinoma: Trends, risk, prevention and management. Nat Rev
Gastroenterol Hepatol. 16:589–604. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kanwal F and Singal AG: Surveillance for
hepatocellular carcinoma: Current best practice and future
direction. Gastroenterology. 157:54–64. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ji S, Liu Q, Zhang S, Chen Q, Wang C,
Zhang W, Xiao C, Li Y, Nian C, Li J, et al: FGF15 activates Hippo
signaling to suppress bile acid metabolism and liver tumorigenesis.
Dev Cell. 48:460–474.e9. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dent P, Booth L, Roberts JL, Liu J,
Poklepovic A, Lalani AS, Tuveson D, Martinez J and Hancock JF:
Neratinib inhibits Hippo/YAP signaling, reduces mutant K-RAS
expression, and kills pancreatic and blood cancer cells. Oncogene.
38:5890–5904. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang Z, Zhou JK, Wang K, Chen H, Qin S,
Liu J, Luo M, Chen Y, Jiang J, Zhou L, et al: PDLIM1 inhibits tumor
metastasis through activating Hippo signaling in hepatocellular
carcinoma. Hepatology. 71:1643–1659. 2020. View Article : Google Scholar
|
|
7
|
Meng Z, Moroishi T and Guan KL: Mechanisms
of Hippo pathway regulation. Genes Dev. 30:1–17. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhou D, Conrad C, Xia F, Park JS, Payer B,
Yin Y, Lauwers GY, Thasler W, Lee JT, Avruch J, et al: Mst1 and
Mst2 maintain hepatocyte quiescence and suppress hepatocellular
carcinoma development through inactivation of the Yap1 oncogene.
Cancer Cell. 16:425–438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng X, Lu T, Li J, Yang R, Hu L, Ye Y,
Mao F, He L, Xu J, Wang Z, et al: The tumor suppressor interferon
regulatory factor 2 binding protein 2 regulates Hippo pathway in
liver cancer by a feedback loop in mice. Hepatology. 71:1988–2004.
2020. View Article : Google Scholar
|
|
10
|
Li Y, Lu J, Chen Q, Han S, Shao H, Chen P,
Jin Q, Yang M, Shangguan F, Fei M, et al: Artemisinin suppresses
hepatocellular carcinoma cell growth, migration and invasion by
targeting cellular bioenergetics and Hippo-YAP signaling. Arch
Toxicol. 93:3367–3383. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun S and Irvine KD: Cellular organization
and cytoskeletal regulation of the Hippo signaling network. Trends
Cell Biol. 26:694–704. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen Q, Zhou XW, Zhang AJ and He K: ACTN1
supports tumor growth by inhibiting Hippo signaling in
hepatocellular carcinoma. J Exp Clin Cancer Res. 40:232021.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang XM, Cao XY, He P, Li J, Feng MX,
Zhang YL, Zhang XL, Wang YH, Yang Q, Zhu L, et al: Overexpression
of Rac GTPase activating protein 1 contributes to proliferation of
cancer cells by reducing Hippo signaling to promote cytokinesis.
Gastroenterology. 155:1233–1249.e22. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Longtine MS, DeMarini DJ, Valencik ML,
Al-Awar OS, Fares H, De Virgilio C and Pringle JR: The septins:
Roles in cytokinesis and other processes. Curr Opin Cell Biol.
8:106–119. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Mostowy S and Cossart P: Septins: The
fourth component of the cytoskeleton. Nat Rev Mol Cell Biol.
13:183–194. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hu J, Bai X, Bowen JR, Dolat L, Korobova
F, Yu W, Baas PW, Svitkina T, Gallo G and Spiliotis ET:
Septin-driven coordination of actin and microtubule remodeling
regulates the collateral branching of axons. Curr Biol.
22:1109–1115. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kremer BE, Adang LA and Macara IG: Septins
regulate actin organization and cell-cycle arrest through nuclear
accumulation of NCK mediated by SOCS7. Cell. 130:837–850. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wei Y, Yang J, Yi L, Wang Y, Dong Z, Liu
Z, Ou-yang S, Wu H, Zhong Z, Yin Z, et al: MiR-223-3p targeting
SEPT6 promotes the biological behavior of prostate cancer. Sci Rep.
4:75462014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan Y, Du Z, Steib CJ, Ding Q, Lu P, Tian
D and Liu M: Effect of SEPT6 on the biological behavior of hepatic
stellate cells and liver fibrosis in rats and its mechanism. Lab
Invest. 99:17–36. 2019. View Article : Google Scholar
|
|
20
|
Xiangji L, Feng X, Qingbao C, Weifeng T,
Xiaoqing J, Baihe Z, Feng S, Hongyang W and Mengchao W: Knockdown
of HBV surface antigen gene expression by a lentiviral
microRNA-based system inhibits HBV replication and HCC growth. J
Viral Hepat. 18:653–660. 2011. View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Kondo R, Ishino K, Wada R, Takata H, Peng
WX, Kudo M, Kure S, Kaneya Y, Taniai N, Yoshida H, et al:
Downregulation of protein disulfide isomerase A3 expression
inhibits cell proliferation and induces apoptosis through STAT3
signaling in hepatocellular carcinoma. Int J Oncol. 54:1409–1421.
2019.PubMed/NCBI
|
|
23
|
Li Y, Tang ZY and Hou JX: Hepatocellular
carcinoma: Insight from animal models. Nat Rev Gastroenterol
Hepatol. 9:32–43. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sun F, Wang J, Sun Q, Li F, Gao H, Xu L,
Zhang J, Sun X, Tian Y, Zhao Q, et al: Interleukin-8 promotes
integrin β3 upregulation and cell invasion through PI3K/Akt pathway
in hepatocellular carcinoma. J Exp Clin Cancer Res. 38:4492019.
View Article : Google Scholar
|
|
25
|
Ding ZB, Shi YH, Zhou J, Shi GM, Ke AW,
Qiu SJ, Wang XY, Dai Z, Xu Y and Fan J: Liver-intestine cadherin
predicts microvascular invasion and poor prognosis of hepatitis B
virus-positive hepatocellular carcinoma. Cancer. 115:4753–4765.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xia H, Dai X, Yu H, Zhou S, Fan Z, Wei G,
Tang Q, Gong Q and Bi F: EGFR-PI3K-PDK1 pathway regulates YAP
signaling in hepatocellular carcinoma: The mechanism and its
implications in targeted therapy. Cell Death Dis. 9:2692018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cerveira N, Bizarro S and Teixeira MR:
MLL-SEPTIN gene fusions in hematological malignancies. Biol Chem.
392:713–724. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Pan Y, Tong JH, Lung RW, Kang W, Kwan JS,
Chak WP, Tin KY, Chung LY, Wu F, Ng SS, et al: RASAL2 promotes
tumor progression through LATS2/YAP1 axis of hippo signaling
pathway in colorectal cancer. Mol Cancer. 17:1022018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang J, Xu ZP, Yang YC, Zhu JS, Zhou Z
and Chen WX: Expression of Yes-associated protein in gastric
adenocarcinoma and inhibitory effects of its knockdown on gastric
cancer cell proliferation and metastasis. Int J Immunopathol
Pharmacol. 25:583–590. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xie K, Xu C, Zhang M, Wang M, Min L, Qian
C, Wang Q, Ni Z, Mou S, Dai H, et al: Yes-associated protein
regulates podocyte cell cycle re-entry and dedifferentiation in
adriamycin-induced nephropathy. Cell Death Dis. 10:9152019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang C, Wang F, Gao Z, Zhang P, Gao J and
Wu X: Regulation of Hippo signaling by mechanical signals and the
cytoskeleton. DNA Cell Biol. 39:159–166. 2020. View Article : Google Scholar
|
|
32
|
Matsui Y and Lai ZC: Mutual regulation
between Hippo signaling and actin cytoskeleton. Protein Cell.
4:904–910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xue Y, Bhushan B, Mars WM, Bowen W, Tao J,
Orr A, Stoops J, Yu Y, Luo J, Duncan AW, et al: Phosphorylated
Ezrin (Thr567) regulates Hippo pathway and yes-associated protein
(Yap) in liver. Am J Pathol. 190:1427–1437. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Matsuda T, Zhai P, Sciarretta S, Zhang Y,
Jeong JI, Ikeda S, Park J, Hsu CP, Tian B, Pan D, et al: NF2
activates Hippo signaling and promotes ischemia/reperfusion injury
in the heart. Circ Res. 119:596–606. 2016. View Article : Google Scholar : PubMed/NCBI
|