Introduction
Reactive oxygen species (ROS) can damage DNA,
lipids, and proteins, and the accumulated oxidative damages play
key roles in the development of diseases, such as hypertension,
arteriosclerosis, arthritis, neurodegenerative disease and cancer
(1). In addition, ROS can serve
as messengers and can lead to the upregulation of several signaling
pathways such as the growth factor kinase (2,3),
Src/Abl kinase (4), and MAPK
(5) pathways. These specific
signaling pathways can activate several cancer-related
transcription factors, such as activator protein 1 and nuclear
factor-κB (6). NADPH oxidases
(NOXs) are a family of transmembrane proteins that generate ROS
through the transmission of electrons across membranes. Seven
isoforms of NOXs, including NOX1-5 and DUOX1-2, have been
identified to date (7). These
NOXs differ in organ expression, type of ROS produced, and the
activation mechanism. NOXs are activated by a cytosolic activator
or through the elevation of intracellular calcium ion levels. In
the present study, focus was placed on NOX5, which is activated by
the elevation of intracellular calcium ion levels (7). NOXs have physiological functions and
are also associated with several chronic diseases, such as
cardiovascular disease (8),
neurodegenerative disease (9),
inflammatory bowel disease (10),
and pulmonary fibrosis (11).
Additionally, NOXs have been reported to be associated with various
types of cancer, including colon (12), gastric (13), pancreas (14), lung (15), and prostate (16) cancer.
NOX5 has been linked to malignant diseases such as
prostate cancer (16), breast
cancer (17), melanoma (18), hairy cell leukemia (19), and esophageal cancer (20). It was previously reported that
patients with colon cancer with high NOX5 expression had
significantly poor prognosis (21). However, no study has examined the
physiological cellular functions of NOX5 in cancer progression. In
the present study, functional analyses were conducted using an
in vitro model to determine the relationship between NOX5
expression and cancer development, specifically for colon
cancer.
Materials and methods
Cell culture, antibodies and other
reagents
Two human colon cancer cell lines, HCT116 and SW48,
were purchased from RIKEN BioResource Center and Cell Resource
Center for Biochemical Research (KAC Co., Ltd.), respectively. Cell
authentication for these two cell lines was performed using short
tandem repeat profiling. Five human colon cancer cell lines (RKO,
SW620, SW480, DLD1, and HT29) were purchased from the American Type
Culture Collection (ATCC). HCT116 and RKO cells were cultured in
Dulbecco's modified Eagle's medium (DMEM) (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C. DLD-1 cells were cultured in RPMI-1640
medium (Gibco; Thermo Fisher Scientific, Inc.) at 37°C. HT29 cells
were cultured in McCoy's 5A and minimal essential medium (Gibco;
Thermo Fisher Scientific, Inc.) at 37°C, respectively. SW48, SW480,
and SW620 cells were cultured in Leibovitz's L-15 medium (Gibco;
Thermo Fisher Scientific, Inc.) in a CO2-free
environment at 37°C. All culture media were supplemented with 10%
fetal bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.)
and 1% penicillin/streptomycin.
A polyclonal NOX5 antibody (product code ab191010;
Abcam), a monoclonal integrin α2 antibody (product code ab133557;
Abcam) and β-actin antibody (product code ab8227; Abcam) were used
in the present study. A polyclonal NOX5 antibody (cat. no.
orb100974-CF405M; Biorbyt Ltd.) was used to confirm the results of
western blotting for NOX5 protein by the ab191010 antibody. β-actin
was used as a loading control in western blotting.
Immunohistochemistry
The detailed method of immunohistochemistry has been
previously described (21). Tumor
tissue and normal colonic mucosa were obtained from a 75-year-old
male patient who underwent curative surgery for colon (cecum)
cancer at the University of Yamanashi Hospital. The present study
was approved (approval no. of the institution: 1940) by the Ethics
Committee of the University of Yamanashi Hospital (Chuo, Japan) and
written informed consent was obtained from the patient. The tissue
sections were incubated overnight at 4°C with the NOX5 antibody
(1:100).
Western blotting
The expression levels of NOX5 were evaluated in 7
colon cancer cell lines using western blotting. Cells were lysed in
Pierce RIPA buffer (Thermo Fisher Scientific, Inc.) using a
protease/phosphatase inhibitor cocktail (Cell Signaling Technology,
Inc.). The protein concentration was assessed using NanoDrop 2000
(Thermo Fisher Scientific, Inc.) by Warburg-Christian method. Cell
lysates containing 4 mg/ml of total protein were separated using
4-12% sodium dodecyl sulphate-polyacrylamide gel electrophoresis
(Invitrogen; Thermo Fisher Scientific, Inc.) and
electrophoretically transferred to polyvinylidene difluoride
membranes (MilliporeSigma). The mass of protein loaded per lane was
40 µg. Nonspecific binding was blocked using the same amount
of Odyssey blocking buffer (LI-COR Biosciences) and
phosphate-buffered saline at room temperature for 90 min. The
membranes were probed with the indicated primary anti-bodies at 4°C
for 16 h. Each primary antibody was diluted as follows: Anti-NOX5
antibody (Abcam) (1:1,000), anti-NOX5 antibody (Biorbyt) (1:500),
anti-integrin α2 antibody (Abcam) (1:10,000), and anti-β-actin
antibody (Abcam) (1:5,000). Then, the membranes were incubated with
HRP conjugated anti-rabbit secondary antibody (1:2,000 dilution;
product code ab6721; Abcam) at room temperature for 1 h. The
proteins were visualized using Pierce™ ECL Plus western blotting
substrate (Thermo Fisher Scientific, Inc.), and the intensity of
the bands was assessed using ImageJ software version 1.8.0
(National Institutes of Health).
Small interfering RNA (siRNA)
transfection
HCT116 and SW48 cells were transfected with 10
nmol/l NOX5 small interfering (si)RNA (Stealth siRNA Assay ID
HSS128403; Invitrogen; Thermo Fisher Scientific, Inc.) using
Lipofectamine RNAi MAX Transfection Reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol.
In detail, 4 µl of Lipofectamine RNAi MAX Transfection
Reagent was diluted in 250 µl of Opti-MEM I Reduced-Serum
Medium (Gibco; Thermo Fisher Scientific, Inc.) and incubated at
room temperature for 5 min. The medium containing siRNA was
replaced with fresh medium 24 h after transfection. The Stealth
RNAiTM siRNA Negative Control Med GC Duplex #2 (cat. no. 12935112;
Invitrogen; Thermo Fisher Scientific, Inc.) was used as a negative
control siRNA (10 nmol/l). At 48 h after transfection, total RNA
was extracted from the cells and purified using the miRNeasy Mini
Kit (Qiagen GmbH) following the manufacturer's instructions.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was purified from the HCT116, SW48, RKO,
SW620, SW480, DLD1, and HT29 cells using the RNeasy Mini Kit
(Qiagen GmbH) following the manufacturer's instructions. The
concentration and purity of RNA were assessed using NanoDrop 2000.
Absorbance was measured at wavelengths of 280, 260, and 230 nm.
cDNA was synthesized using the high-capacity cDNA reverse
transcription kit (Applied Biosystems™; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Quantitative
PCR analysis was performed in 20-µl reaction systems
containing 2 µl of cDNA, 1 µl of appropriate primer,
10 µl of TaqMan Universal PCR Master Mix (Applied
Biosystems™; Thermo Fisher Scientific, Inc.), and 7 µl of
RNase-free water on a 96-well plate. The expression levels were
evaluated for the following human genes: NOX5 (Hs00225846_m1), Jun
proto-oncogene (JUN) (Hs01103582_s1), FBJ murine osteosarcoma viral
oncogene homolog (FOS) (Hs00170630_m1), plasminogen activator
urokinase (PLAU) (Hs01547054_m1), glycogen synthase kinase 3β
(GSK3B) (Hs01047719_m1), and integrin subunit α2 (ITGA2)
(Hs00158127_m1) (Thermo Fisher Scientific, Inc.). GAPDH (cat. no.
4310884E; Applied Biosystems™; Thermo Fisher Scientific, Inc.) was
used as an endogenous control for normalization. RT-qPCR was
performed using Applied Biosystems 7500 Real-Time PCR software
(7500 Software version 2.0.6; Applied Biosystems™; Thermo Fisher
Scientific, Inc.). The comparative cycle threshold (Cq) (ΔΔCq)
method was used to analyze the data (22). The PCR thermocycling conditions
were as follows: 2 min at 50°C, 10 min at 95°C, 40 cycles of 15 sec
at 95°C and 1 min at 60°C. For quantification, the threshold cycle
(Cq) was defined as the PCR cycle number. Each assay was performed
in triplicate.
Cell proliferation assay and cell cycle
analysis
Cells were seeded at a density of 0.5×105
cells/ml in a 6-well plate and incubated at 37°C with 5%
CO2. siRNA transfection was performed 24 h after
seeding. At 24, 48, and 72 h after transfection, the cells were
dislodged using 0.53 mmol/l trypsin-EDTA solution (Nacalai Tesque
Inc.), and the cells were counted using Countess Automated Cell
Counter (Invitrogen™; Thermo Fisher Scientific, Inc.). Each assay
was performed in triplicate. The cell cycle was evaluated using
fluorescence-activated cell sorting (FACS) (BD FACSDiva software
version 8.0; BD Biosciences) as previously described (23). Briefly, the cells were detached
using trypsin-EDTA solution and treated with Triton X-100 and
RNase. The nuclei were stained for 15 min at room temperature with
propidium iodide. Thereafter, the DNA content was assessed using
FACS with analysis of at least 2×104 cells.
Cell migration, invasion and wound
healing assays
A migration assay was conducted in a 24-well
Transwell system with control chambers of 8-µm pore size
(product no. 353097; Corning, Inc.) and an invasion assay in a
24-well Transwell system with Matrigel chambers of 8-µm pore
size (product no. 354480; Corning, Inc.). The Matrigel was
incubated to rehydrate at 37°C for 2 h. HCT116 and SW48 cells were
seeded at densities of 1.5×105 and 3.0×105
cells/ml in the upper chambers, respectively, at 24 h after siRNA
transfection. The upper chamber was filled with 0.75 ml of
serum-free medium, and the lower chamber was filled with 0.5 ml of
medium supplemented with 10% FBS. At 48 h for the migration assay
and 60 h for the invasion assay after incubation at 37°C with 5%
CO2, the cells that did not permeate the chamber were
removed using a cotton swab, and the cells that were able to
permeate the chamber were stained with Differential Quik III Stain
Kit (5% eosin Y and 5% azure-2) (Polysciences, Inc.) for each 30
sec at room temperature and counted with cell count software (ver.
2.2; Keyence Corporation) using a light microscope. Ibidi culture
inserts (Ibidi GmbH) were used for the wound healing assay. This
assay was started with 100% confluent HCT116 cells. HCT116 cells
were detached at 48 h after siRNA transfection, and
8.0×106 cells/70 µl were seeded in each area.
After 24 h, the insert was removed and each well was filled with
the medium with 10% FBS. Images of the migrated cells were captured
at 24 h after removing the insert using phase-contrast microscope,
and the rate of gap closure was analyzed with cell count software
(ver. 2.2). Each assay was performed in triplicate.
Gene microarray analysis
HCT116 cells were transfected with control siRNA
(n=1) and NOX5 siRNA (n=1). Total RNA was extracted from the cells
using a miRNeasy Mini Kit (Qiagen GmbH) according to the
manufacturer's protocol at 48 h after transfection. The RNA quality
was assessed using NanoDrop 2000 (Thermo Fisher Scientific, Inc.)
and Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.).
Microarray analyses were performed with a 3D-Gene Human Oligo chip
25k (Toray Industries, Inc.). This microarray adopted a columnar
structure to stabilize spot morphology and enable microbead
agitation for efficient hybridization. Total RNA was labeled with
Cy5 or Cy3 using the Amino Allyl MessageAMP II aRNA Amplification
Kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The Cy5-
or Cy3-labeled aRNA pools were mixed with hybridization buffer, and
subsequently hybridized for 16 h. The hybridization was performed
according to the manufacturer's protocols (www.3d-gene.com). The hybridization signals were
acquired using a 3D-Gene Scanner (Toray Industries, Inc.) and
processed using 3D-Gene Extraction software (ver.1.2.1.1; Toray
Industries, Inc.). The detected signals for each gene were
normalized using a global normalization method (Cy3/Cy5 ratio
median=1). The fold change of each molecule was calculated by the
normalized data of two samples, and a fold change cutoff value of
two or three times or more was set to identify significantly
downregulated molecular expressions. The network and pathway
analyses were performed using Ingenuity Pathway Analysis (IPA)
(ver. 62089861; Ingenuity Systems; Qiagen). Molecules were
represented by nodes, and the biological relationship between two
nodes was represented as an edge. The functional analysis was
identified along with biological function and diseases. The
microarray data used in the present study have been deposited in
the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nlh.gov/geo/) of NCBI and are
accessible through GEO Series accession number GSE175691.
Measurement of extracellular hydrogen
peroxide
The level of extracellular hydrogen peroxide
(H2O2) was assessed using the Amplex Red
Hydrogen Peroxidase Assay kit following the manufacturer's
instructions (Invitrogen™; Thermo Fisher Scientific, Inc.).
Briefly, the reaction mixture contained 50 µM Amplex Red
reagent, 0.1 U/ml HRP, and 5 µM ionomycin (Merck KGaA) in
Krebs-Ringer phosphate glucose (KRPG) and was incubated for 10 min
at 37°C. At 48 h after siRNA transfection, HCT116 and SW48 cells
were detached and 20 µl of 2.0×104 cells were
suspended in KRPG buffer and added to the prepared reaction
mixture. Fluorescence was assessed using a fluorescence microplate
reader at an excitation wavelength of 530 nm and fluorescence
detection at 590 nm. The absorbance was measured at 1 and 3 h after
the cells were added to the reaction mixture.
Statistical analysis
The results were expressed as the mean ± standard
error of the mean from at least three experiments. The data were
analyzed using JMP statistical software (version 14.2; SAS
Institute, Inc.). Unpaired Student's t-test was used to assess
statistical differences between mean values of control and treated
samples. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of NOX5 in human colon
cancer cell lines
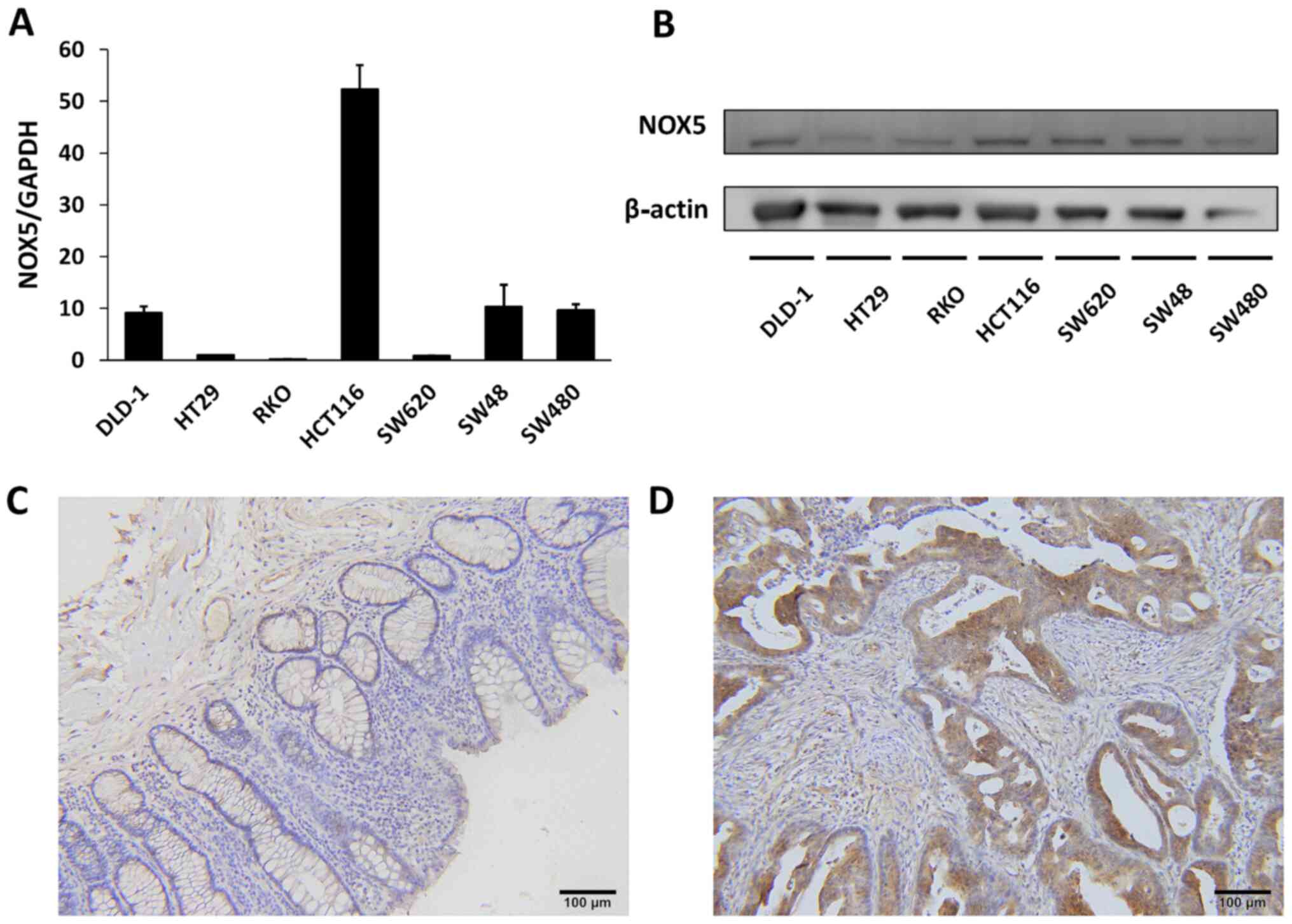
The expression levels of NOX5 mRNA and protein in 7
colon cancer cell lines were examined to investigate the role of
NOX5 in cancer development. NOX5 mRNA was highly expressed in
HCT116 cells, moderately expressed in DLD1, SW48, and SW480 cells,
and lowly expressed in HT29, RKO, and SW620 cells (Fig. 1A). NOX5 protein expression could
be detected at various levels in all cell lines (Fig. 1B). NOX5 protein expression could
be detected with similar levels using another anti-NOX5 antibody
(Fig. S1A and B). Using human
colon cancer tissues, immunohistochemical staining demonstrated
weaker NOX5 expression levels in the normal colonic mucosa than in
the tumor tissues (Fig. 1C and
D).
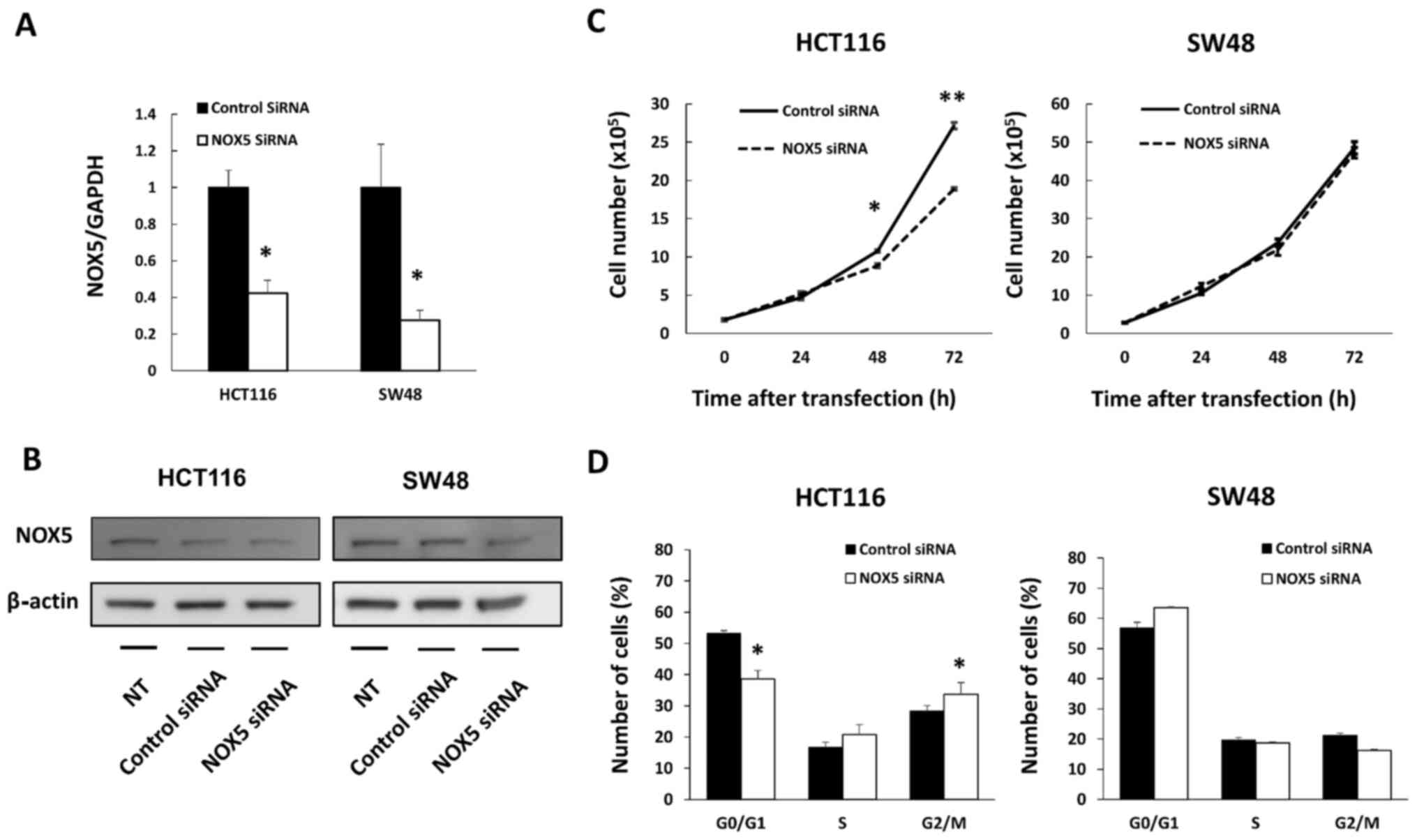
Limited effect of NOX5 on cell
proliferation in colon cancer cells
Further examination was conducted on HCT116 and SW48
cells. DLD1 cells, another cell line with a moderate level of NOX5
expression, was not selected due to the low efficiency of NOX5
knockdown according to the result of RT-qPCR (0.51±0.06; data not
shown). NOX5 siRNA transfection effectively downregulated NOX5 mRNA
expression in both cell lines (Fig.
2A) and NOX5 protein expression in HCT116 cells (Fig. 2B). In HCT116 cells, the
downregulation of NOX5 expression led to a significantly lower
number of viable cells at 48 (P=0.013) and 72 h (P<0.001) after
transfection, whereas there was no difference in the number of
cells at these time-points for SW48 cells (Fig. 2C). Cell cycle analysis also
revealed that downregulation of NOX5 expression led to
G2/M arrest only in HCT116 cells, which supported the
results of the cell proliferation assay (Fig. 2D).
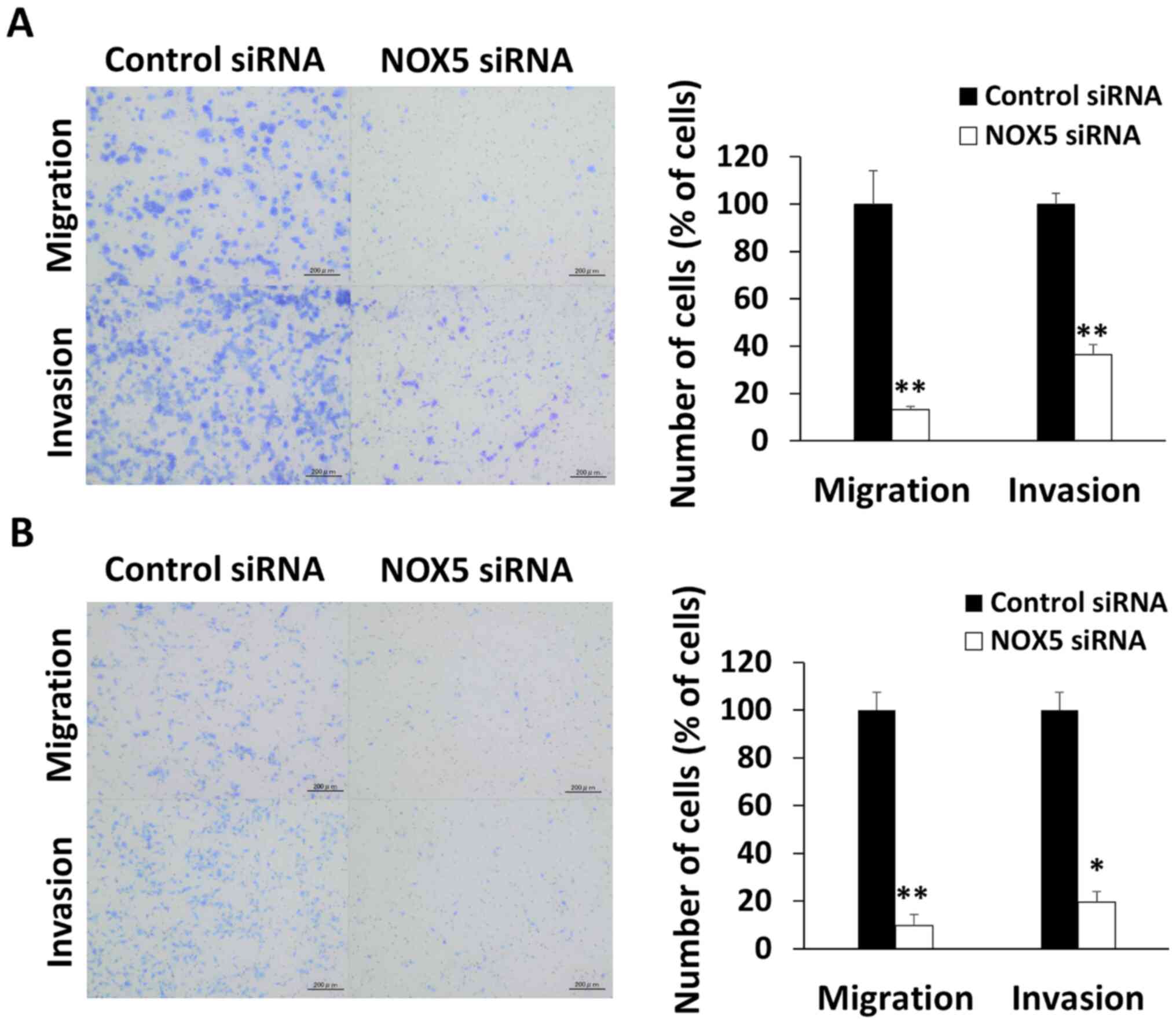
NOX5 regulates cell migration and
invasion in colon cancer cells
The role of NOX5 in cancer progression was further
explored. The migrated and invasive cell number ratio of
NOX5-knockdown cells was significantly reduced to 13.2±1.2 and
36.3±4.3% in HCT116 cells (P<0.01 and P<0.01, respectively)
(Fig. 3A). Similarly, the
migrated and invasive cell number ratio of NOX5-knockdown cells was
significantly reduced to 9.6±4.6 and 19.5±3.3% compared with that
in control SW48 cells (P<0.01 and P=0.010, respectively)
(Fig. 3B). The wound healing
assay revealed that the horizontal cellular motility of
NOX5-knockdown HCT116 cells was significantly inhibited (P<0.01)
(Fig. S2).
Level of extracellular
H2O2 in HCT116 and SW48 cells
To confirm the effect of NOX5 knockdown on ROS
production, the level of extracellular H2O2
in the culture medium of HCT116 and SW48 cells was assessed. The
H2O2 level was significantly increased by
NOX5 knockdown in both HCT116 and SW48 cells (Fig. S3).
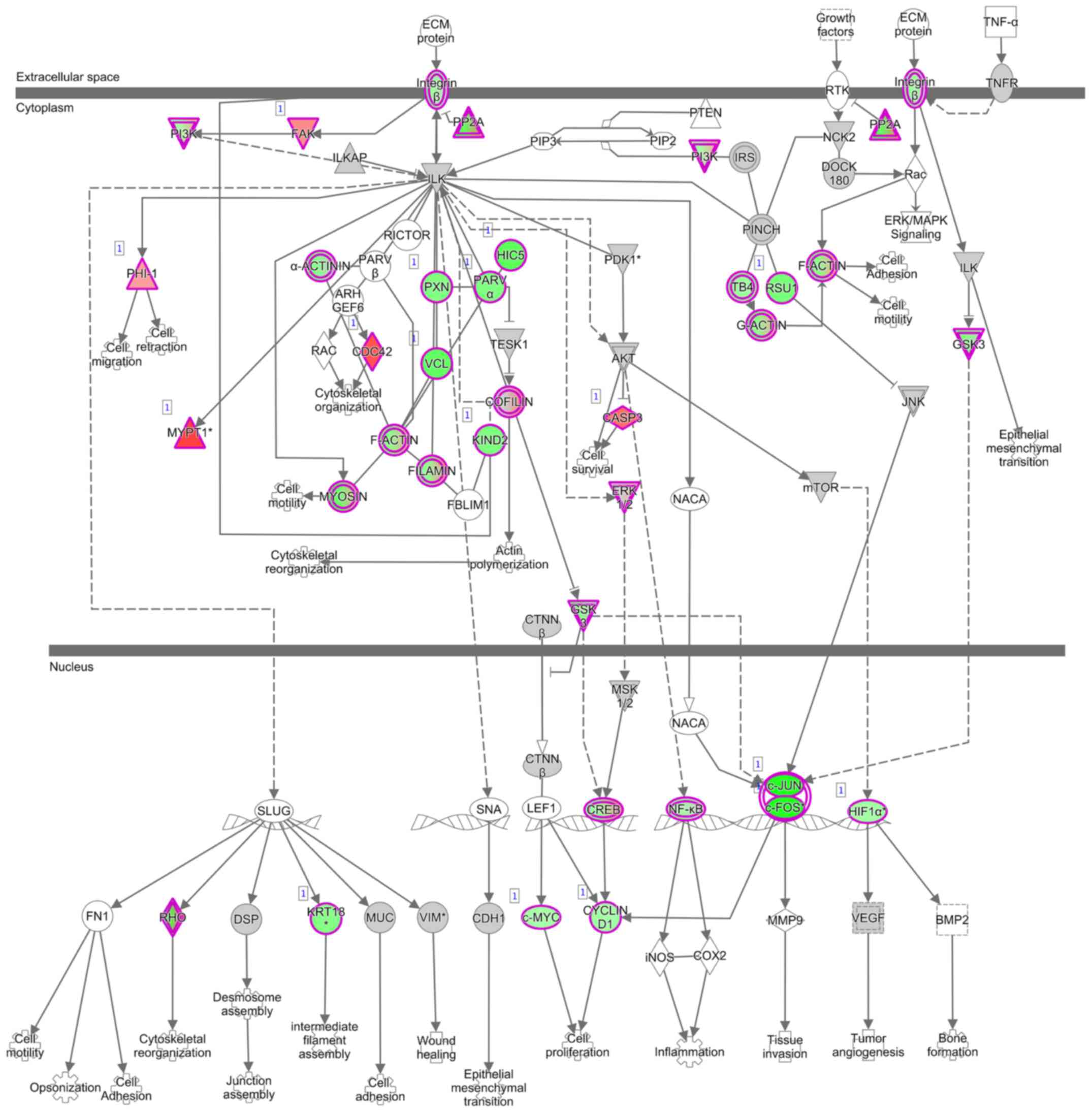
Gene expression profiling in NOX5
siRNA-transfected cells
A comprehensive analysis using mRNA microarray was
performed to examine changes in the gene expression profiles of
NOX5 siRNA-transfected HCT116 cells and control siRNA-transfected
HCT116 cells. The microarray analysis revealed a total of 4,043
genes with changes of ≥1.4-fold in HCT116 cells subjected to NOX5
knockdown. Of these genes, 2,323 were upregulated and 1,720 were
downregulated. IPA analysis revealed that cancer was one of the
top-ranked diseases and that integrin-linked kinase (ILK) signaling
was one of the top-ranked canonical pathways related to NOX5
knockdown (Table I; Fig. 4).
 | Table IMajor NADPH oxidase 5 canonical
pathways and disease/biological functions according to Ingenuity
Pathway Analysis. |
Table I
Major NADPH oxidase 5 canonical
pathways and disease/biological functions according to Ingenuity
Pathway Analysis.
A, Top canonical
pathways
|
|---|
| Name | P-value | Overlap |
|---|
| Protein
ubiquitination pathway |
1.58×10−9 | 30.8% |
| EIF2 signaling |
9.75×10−8 | 30.4% |
| Molecular
mechanisms of cancer |
2.19×10−7 | 26.3% |
| Integrin-linked
kinase signaling |
6.73×10−7 | 30.5% |
| Regulation of
actin-based motility by Rho |
7.21×10−7 | 37.2% |
|
B, Top Diseases and
Bio Functions (disease and disorders)
|
| Name | P-value range | Molecules |
| Cancer |
1.57×10−4-9.24×10−143 | 3,488 |
| Organismal injury
and abnormalities |
1.57×10−4-9.24×10−143 | 3,516 |
| Endocrine system
disorders |
1.57×10−4-1.75×10−98 | 2,721 |
| Gastrointestinal
disease |
1.57×10−4-1.99×10−67 | 3,002 |
| Reproductive system
disease |
1.57×10−4-7.16×10−16 | 1,974 |
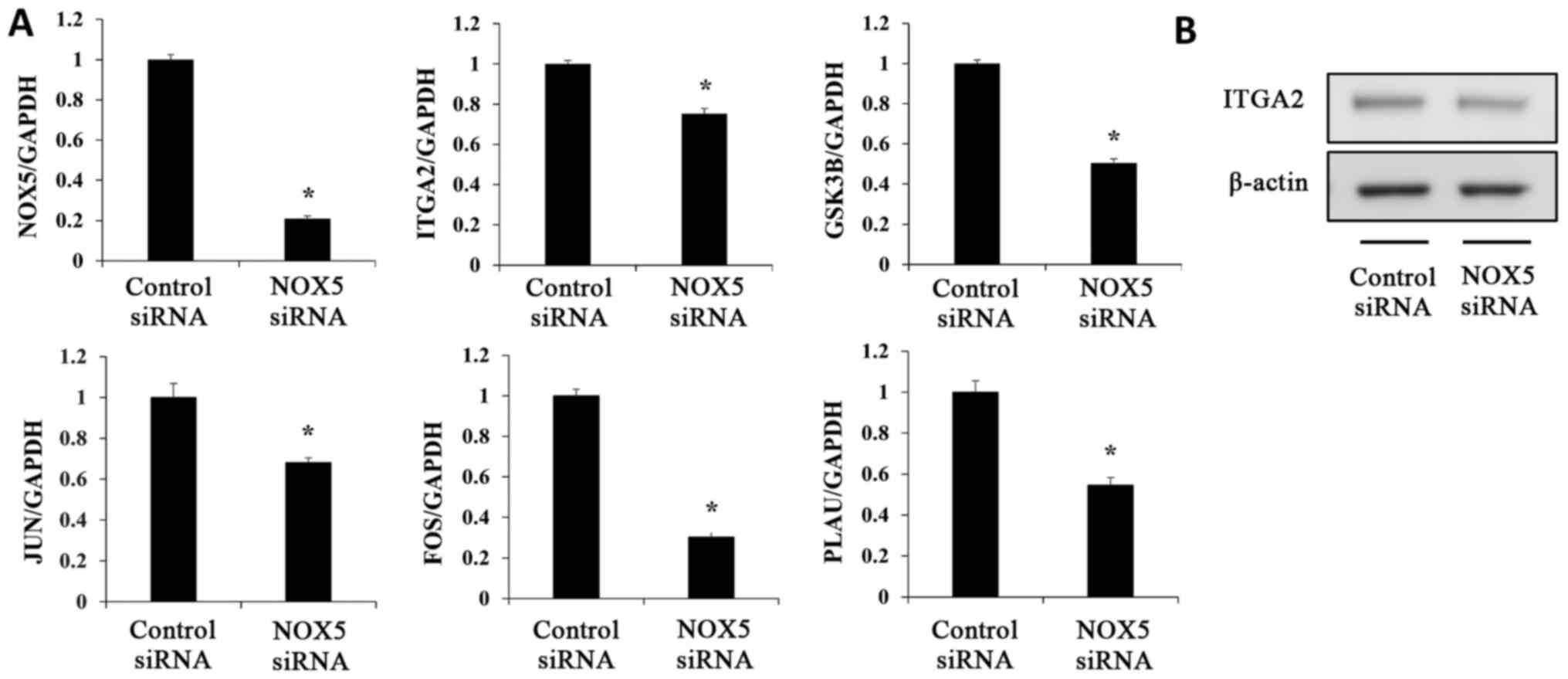
Verification of microarray results
A total of five genes related to the ILK signaling
pathway (ITGA2, GSK3B, JUN, FOS and
PLAU) were selected to confirm the results of the microarray
analysis (Table II; Fig. 4). The expression levels of these
genes were analyzed using RT-qPCR. The expression levels of each
gene were compared between NOX5 siRNA-transfected HCT116 cells and
control siRNA-transfected HCT116 cells. NOX5 siRNA-transfected
HCT116 cells revealed significantly decreased mRNA expression
levels of ITGA2, GSK3B, JUN, FOS, and
PLAU compared with the control cells (Fig. 5A). The ITGA2 protein level was
also decreased by NOX5 siRNA-transfection (Fig. 5B). These changes were consistent
with the results of the microarray analysis.
 | Table IIExpression level patterns of ILK
signaling pathway-related genes in HCT116 cells after NADPH oxidase
5 knockdown. |
Table II
Expression level patterns of ILK
signaling pathway-related genes in HCT116 cells after NADPH oxidase
5 knockdown.
| Gene symbol | UniGene ID | ILK signaling
pathway
| Fold change |
|---|
| Gene name |
|---|
| ITGA2 | H200016773 | Integrin subunit
α2 | −2.51 |
| GSK3B | H300019897 | Glycogen synthase
kinase 3β | −2.04 |
| JUN |
opHsV0400001031 | Jun
proto-oncogene | −5.69 |
| FOS | H200003548 | FBJ murine
osteosarcoma viral oncogene homolog | −4.50 |
| PLAU | H300019040 | Plasminogen
activator, urokinase | −4.61 |
Discussion
The role of NOX5 in tumor progression of colon
cancer was examined using human colon cancer cell lines. To the
best of our knowledge, this is the first study involving in
vitro experiments to determine the oncogenic role of NOX5 in
colon cancer.
The NOX family is composed of seven members
consisting of NOX1 to NOX5 and DUOX1 and 2, which generate ROS
(24). There have been numerous
studies concerning the roles of the NOX family in cancer. In colon
cancer, NOX1 has been well studied and it has been reported that
NOX1 is highly expressed in colon cancer and involved in cancer
progression (10,25,26). In addition, NOX4 was revealed to
be involved in protecting pancreatic cancer cells from apoptosis by
producing ROS (14). DUOX1 and
DUOX2 have been revealed to increase cell migration in lung cancer
(15). However, the functions of
NOX5 have not been investigated thoroughly as NOX5 was the latest
discovery among the members of the NOX family. It was only
discovered in 2001. The NOX5 gene is located on chromosome
15, with four splice variants (α, β, δ, and γ) of NOX5 and one
truncated variant (NOX5s or NOX5ε). NOX5α, NOX5β, NOX5δ, and NOX5γ
are known as NOX5-L (27). NOX5
is known to be expressed in a variety of organs such as lymphatic
tissues, testes (28), vascular
smooth muscles (29), and kidneys
(30). NOX5 has been revealed to
regulate monocyte differentiation into dendritic cells in the
immune system (31). NOX5 also
plays a role in vascular contraction and is involved in the
development of hypertension and atherosclerosis (32,33). In addition, NOX5 has been
associated with various types of cancer. Previous studies have
reported that NOX5 plays an important role in cancer growth,
migration, and invasion in prostate (16), breast (17), and esophageal cancer (34) through signaling pathways, such as
the MAPK pathway (35). However,
the role of NOX5 in colon cancer remains unclear unlike NOX1
(12). NOX5 reportedly promotes
cancer cell proliferation, migration, and invasion in breast cancer
cell lines (17). The present
study also demonstrated that NOX5 was involved in the tumor
motility of colon cancer. With regard to the cell growth rate and
cell cycle, different levels of NOX5 mRNA/protein expression in
colon cancer cells were associated with the results of the present
study. It was observed that the high levels of NOX5 expression in
HCT116 cells played a role in the regulation of cell cycle
progression and cell proliferation; however, SW48 cells only
moderately expressed NOX5 and were not involved in the same
mechanisms of cell cycle regulation and proliferation as those in
which HCT116 cells were involved. It was previously reported that
patients with colon cancer who had high NOX5 expression had
significantly poor prognosis with high rates of metastasis after
curative resection (21), which
corroborates with the results of the present study.
ILK is a protein kinase that is attached to the
integrin β-subunit, and the basic function of integrins is to
promote cell-to-cell and cell-to-extracellular matrix adhesion and
cell migration. ILK serves as an important regulator of signaling
cascades, such as P13K/AKT, AP-1, Hippo, NF-κB, and ERK (36), and plays important roles in cell
proliferation, invasion, and migration in various types of cancer.
Furthermore, ILK is overexpressed in colon cancer tissues and is
involved in epithelial-mesenchymal transition, metastasis and
chemoresistance (37). In the
present study, mRNA microarray experiments revealed that the
expression levels of several genes involved in the ILK signaling
pathway, such as ITGA2, GSK3B, JUN, and FOS, were significantly
downregulated by the depletion of NOX5.
Plasminogen activator (PA) is a serine protease that
comprises the plasminogen activator system, together with its
plasminogen activator receptor and its inhibitor, plasminogen
activator inhibitor (38). There
are two types of PA: Tissue type plasminogen activator and PLAU.
PLAU is mainly involved in cancer malignancies, including
colorectal cancer (39-41). It has been reported that PLAU
forms a complex with integrin and activates pathways such as the
PTK2 and ERK pathways and is involved in cell proliferation and
invasion (38,42). In the present study, both mRNA
microarray and its validation experiments revealed that the
depletion of NOX5 reduced PLAU expression. To the best of
our knowledge, there has been no study in the cancer-related field
on this topic, although there have been some studies that reported
the relationship between NOX and integrins or PLAU in the
noncancerous field (43,44). In addition, as the analysis of
microarray results revealed that the expression levels of some
Rho-related genes were also significantly altered (Table I), the Rho/ROCK signaling pathway
may be another potential target and further study is necessary.
As it has previously been demonstrated that an
increase in H2O2 levels is associated with
the triggering of cancer-related pathways in colorectal cancer
(45,46), changes in extracellular
H2O2 levels were also evaluated by
transfection with NOX5 siRNA. However, the extracellular
H2O2 levels did not decrease with NOX5
knockdown in both cell lines. These results indicated that other
mechanisms are involved, and further studies are needed to clarify
them. Although the lack of experiments in a normal colon cell line
exists as a potential limitation, the present study demonstrated
that changes in NOX5 expression could influence the ILK
signaling pathway and PLAU expression on colon cancer
development.
In conclusion, the present study revealed that NOX5
plays a role in cancer progression, especially on motility of
cancer cells. Microarray data revealed that NOX5 knockdown
influenced the expression of ILK signaling pathway-related genes.
Our observations indicated that NOX5 may be a useful biomarker and
a novel therapeutic target for the future treatment of colon
cancer, although further investigation of the underlying molecular
mechanism is necessary.
Supplementary Data
Availability of data and materials
All data generated or analyzed during this study are
included in this published article. The microarray data discussed
in this publication have been deposited in the Gene Expression
Omnibus (GEO, http://www.ncbi.nlm.nlh.gov/geo/) of NCBI and are
accessible through GEO Series accession number GSE175691.
Authors' contributions
NA performed all experiments and wrote the
manuscript. HS designed the project and wrote the manuscript. KS,
SF, HAk, NH, YK, HAm, HKa, MS, SI, and HKo participated in the
interpretation of the results and helped to edit the manuscript. KK
and AS helped to analyze the results of microarray analysis. DI
provided conceptual advice and edited manuscript. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved (approval no. of the
institution: 1940) by the Ethics Committee of the University of
Yamanashi Hospital (Kofu, Japan). Written informed consent was
obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by a Grant-in-Aid for Young
Scientists (grant no. 20K17609) from the Japan Society for the
Promotion of Science.
Abbreviations:
|
NOX
|
NADPH oxidase
|
|
ROS
|
reactive oxygen species
|
|
ILK
|
integrin-linked kinase
|
References
|
1
|
Valko M, Rhodes CJ, Moncol J, Izakovic M
and Mazur M: Free radicals, metals and antioxidants in oxidative
stress-induced cancer. Chem Biol Interact. 160:1–40. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bae YS, Kang SW, Seo MS, Baines IC, Tekle
E, Chock PB and Rhee SG: Epidermal growth factor (EGF)-induced
generation of hydrogen peroxide. Role in EGF receptor-mediated
tyrosine phosphorylation. J Biol Chem. 272:217–221. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Neufeld G, Cohen T, Gengrinovitch S and
Poltorak Z: Vascular endothelial growth factor (VEGF) and its
receptors. FASEB J. 13:9–22. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Esposito F, Chirico G, Montesano Gesualdi
N, Posadas I, Ammendola R, Russo T, Cirino G and Cimino F: Protein
kinase B activation by reactive oxygen species is independent of
tyrosine kinase receptor phosphorylation and requires SRC activity.
J Biol Chem. 278:20828–20834. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kyriakis JM and Avruch J: Mammalian
mitogen-activated protein kinase signal transduction pathways
activated by stress and inflammation. Physiol Rev. 81:807–869.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hsu TC, Young MR, Cmarik J and Colburn NH:
Activator protein 1 (AP-1)- and nuclear factor kappaB
(NF-kappaB)-dependent transcriptional events in carcinogenesis.
Free Radic Biol Med. 28:1338–1348. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brandes RP, Weissmann N and Schröder K:
Nox family NADPH oxidases: Molecular mechanisms of activation. Free
Radic Biol Med. 76:208–226. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cao J, Liu Z, Xu Q, Shi R and Zhang G:
Research progress in NADPH oxidase family in cardiovascular
diseases. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 44:1258–1267.
2019.In Chinese.
|
|
9
|
Sorce S, Stocker R, Seredenina T, Holmdahl
R, Aguzzi A, Chio A, Depaulis A, Heitz F, Olofsson P, Olsson T, et
al: NADPH oxidases as drug targets and biomarkers in
neurodegenerative diseases: What is the evidence? Free Radic Biol
Med. 112:387–396. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Makhezer N, Ben Khemis M, Liu D, Khichane
Y, Marzaioli V, Tlili A, Mojallali M, Pintard C, Letteron P,
Hurtado-Nedelec M, et al: NOX1-derived ROS drive the expression of
Lipocalin-2 in colonic epithelial cells in inflammatory conditions.
Mucosal Immunol. 12:117–131. 2019. View Article : Google Scholar
|
|
11
|
Ward PA and Hunninghake GW: Lung
inflammation and fibrosis. Am J Respir Crit Care Med.
157:S123–S129. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Juhasz A, Markel S, Gaur S, Liu H, Lu J,
Jiang G, Wu X, Antony S, Wu Y, Melillo G, et al: NADPH oxidase 1
supports proliferation of colon cancer cells by modulating reactive
oxygen species-dependent signal transduction. J Biol Chem.
292:7866–7887. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yamamoto T, Nakano H, Shiomi K, Wanibuchi
K, Masui H, Takahashi T, Urano Y and Kamata T: Identification and
characterization of a novel NADPH oxidase 1 (Nox1) inhibitor that
suppresses proliferation of colon and stomach cancer cells. Biol
Pharm Bull. 41:419–426. 2018. View Article : Google Scholar
|
|
14
|
Vaquero EC, Edderkaoui M, Pandol SJ,
Gukovsky I and Gukovskaya AS: Reactive oxygen species produced by
NAD(P) H oxidase inhibit apoptosis in pancreatic cancer cells. J
Biol Chem. 279:34643–34654. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luxen S, Belinsky SA and Knaus UG:
Silencing of DUOX NADPH oxidases by promoter hypermethylation in
lung cancer. Cancer Res. 68:1037–1045. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Höll M, Koziel R, Schäfer G, Pircher H,
Pauck A, Hermann M, Klocker H, Jansen-Dürr P and Sampson N: ROS
signaling by NADPH oxidase 5 modulates the proliferation and
survival of prostate carcinoma cells. Mol Carcinog. 55:27–39. 2016.
View Article : Google Scholar :
|
|
17
|
Dho SH, Kim JY, Lee KP, Kwon ES, Lim JC,
Kim CJ, Jeong D and Kwon KS: STAT5A-mediated NOX5-L expression
promotes the proliferation and metastasis of breast cancer cells.
Exp Cell Res. 351:51–58. 2017. View Article : Google Scholar
|
|
18
|
Antony S, Jiang G, Wu Y, Meitzler JL,
Makhlouf HR, Haines DC, Butcher D, Hoon DS, Ji J, Zhang Y, et al:
NADPH oxidase 5 (NOX5)-induced reactive oxygen signaling modulates
normoxic HIF-1α and p27Kip1 expression in malignant
melanoma and other human tumors. Mol Carcinog. 56:2643–2662. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kamiguti AS, Serrander L, Lin K, Harris
RJ, Cawley JC, Allsup DJ, Slupsky JR, Krause KH and Zuzel M:
Expression and activity of NOX5 in the circulating malignant B
cells of hairy cell leukemia. J Immunol. 175:8424–8430. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou X, Li D, Resnick MB, Wands J and Cao
W: NADPH oxidase NOX5-S and nuclear factor κB1 mediate acid-induced
microsomal prostaglandin E synthase-1 expression in Barrett's
esophageal adenocarcinoma cells. Mol Pharmacol. 83:978–990. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashizawa N, Shimizu H, Sudo M, Furuya S,
Akaike H, Hosomura N, Kawaguchi Y, Amemiya H, Kawaida H, Inoue S,
et al: Clinical significance of NADPH oxidase 5 in human colon
cancer. Anticancer Res. 39:4405–4410. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Maruyama S, Furuya S, Shiraishi K, Shimizu
H, Saito R, Akaike H, Hosomura N, Kawaguchi Y, Amemiya H, Kawaida
H, et al: Inhibition of apoptosis by miR-122-5p in
α-fetoprotein-producing gastric cancer. Oncol Rep. 41:2595–2600.
2019.PubMed/NCBI
|
|
24
|
Bedard K and Krause KH: The NOX family of
ROS-generating NADPH oxidases: Physiology and pathophysiology.
Physiol Rev. 87:245–313. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Laurent E, McCoy JW III, Macina RA, Liu W,
Cheng G, Robine S, Papkoff J and Lambeth JD: Nox1 is over-expressed
in human colon cancers and correlates with activating mutations in
K-Ras. Int J Cancer. 123:100–107. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang HP, Wang X, Gong LF, Chen WJ, Hao Z,
Feng SW, Wu YB, Ye T and Cai YK: Nox1 promotes colon cancer cell
metastasis via activation of the ADAM17 pathway. Eur Rev Med
Pharmacol Sci. 20:4474–4481. 2016.PubMed/NCBI
|
|
27
|
Fulton DJ: Nox5 and the regulation of
cellular function. Antioxid Redox Signal. 11:2443–2452. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bánfi B, Molnár G, Maturana A, Steger K,
Hegedûs B, Demaurex N and Krause KH: A Ca(2+)-activated NADPH
oxidase in testis, spleen, and lymph nodes. J Biol Chem.
276:37594–37601. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Montezano AC, Tsiropoulou S, Dulak-Lis M,
Harvey A, Camargo Lde L and Touyz RM: Redox signaling, Nox5 and
vascular remodeling in hypertension. Curr Opin Nephrol Hypertens.
24:425–433. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Holterman CE, Boisvert NC, Thibodeau JF,
Kamto E, Novakovic M, Abd-Elrahman KS, Ferguson SSG and Kennedy
CRJ: Podocyte NADPH oxidase 5 promotes renal inflammation regulated
by the toll-like receptor pathway. Antioxid Redox Signal.
30:1817–1830. 2019. View Article : Google Scholar
|
|
31
|
Marzaioli V, Hurtado-Nedelec M, Pintard C,
Tlili A, Marie JC, Monteiro RC, Gougerot-Pocidalo MA, Dang PM and
El-Benna J: NOX5 and p22phox are 2 novel regulators of human
monocytic differentiation into dendritic cells. Blood.
130:1734–1745. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Holterman CE, Thibodeau JF and Kennedy CR:
NADPH oxidase 5 and renal disease. Curr Opin Nephrol Hypertens.
24:81–87. 2015. View Article : Google Scholar
|
|
33
|
Montezano AC, De Lucca Camargo L, Persson
P, Rios FJ, Harvey AP, Anagnostopoulou A, Palacios R, Gandara ACP,
Alves-Lopes R, Neves KB, et al: NADPH oxidase 5 is a
pro-contractile nox isoform and a point of cross-talk for calcium
and redox signaling-implications in vascular function. J Am Heart
Assoc. 7:e0093882018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hong J, Li D and Cao W: Rho Kinase ROCK2
mediates acid-induced NADPH oxidase NOX5-S expression in human
esophageal adenocarcinoma cells. PLoS One. 11:e01497352016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pandey D and Fulton DJ: Molecular
regulation of NADPH oxidase 5 via the MAPK pathway. Am J Physiol
Heart Circ Physiol. 300:H1336–H1344. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zheng CC, Hu HF, Hong P, Zhang QH, Xu WW,
He QY and Li B: Significance of integrin-linked kinase (ILK) in
tumorigenesis and its potential implication as a biomarker and
therapeutic target for human cancer. Am J Cancer Res. 9:186–197.
2019.PubMed/NCBI
|
|
37
|
Tsoumas D, Nikou S, Giannopoulou E,
Champeris Tsaniras S, Sirinian C, Maroulis I, Taraviras S, Zolota
V, Kalofonos HP and Bravou V: ILK Expression in colorectal cancer
is associated with EMT, cancer stem cell markers and
chemoresistance. Cancer Genomics Proteomics. 15:127–141.
2018.PubMed/NCBI
|
|
38
|
Smith HW and Marshall CJ: Regulation of
cell signalling by uPAR. Nat Rev Mol Cell Biol. 11:23–36. 2010.
View Article : Google Scholar
|
|
39
|
Collen D and Lijnen HR: Basic and clinical
aspects of fibrinolysis and thrombolysis. Blood. 78:3114–3124.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Halamkova J, Kiss I, Pavlovsky Z, Tomasek
J, Jarkovsky J, Cech Z, Tucek S, Hanakova L, Moulis M, Zavrelova J,
et al: Clinical significance of the plasminogen activator system in
relation to grade of tumor and treatment response in colorectal
carcinoma patients. Neoplasma. 58:377–385. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Herszényi L, Farinati F, Cardin R, István
G, Molnár LD, Hritz I, De Paoli M, Plebani M and Tulassay Z: Tumor
marker utility and prognostic relevance of cathepsin B, cathepsin
L, urokinase-type plasminogen activator, plasminogen activator
inhibitor type-1, CEA and CA 19-9 in colorectal cancer. BMC Cancer.
8:1942008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ossowski L and Aguirre-Ghiso JA: Urokinase
receptor and integrin partnership: Coordination of signaling for
cell adhesion, migration and growth. Curr Opin Cell Biol.
12:613–620. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Terada LS and Nwariaku FE: Escaping
anoikis through ROS: ANGPTL4 controls integrin signaling through
Nox1. Cancer Cell. 19:297–299. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sánchez-Santos A, Martínez-Hernández MG,
Contreras-Ramos A, Ortega-Camarillo C and Baiza-Gutman LA:
Hyperglycemia-induced mouse trophoblast spreading is mediated by
reactive oxygen species. Mol Reprod Dev. 85:303–315. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kim SH, Kim KH, Yoo BC and Ku JL:
Induction of LGR5 by H2O2 treatment is associated with cell
proliferation via the JNK signaling pathway in colon cancer cells.
Int J Oncol. 41:1744–1750. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wu Y, Konaté MM, Lu J, Makhlouf H, Chuaqui
R, Antony S, Meitzler JL, Difilippantonio MJ, Liu H, Juhasz A, et
al: IL-4 and IL-17A cooperatively promote hydrogen peroxide
production, oxidative DNA damage, and upregulation of dual oxidase
2 in human colon and pancreatic cancer cells. J Immunol.
203:2532–2544. 2019. View Article : Google Scholar : PubMed/NCBI
|