Introduction
Non-small cell lung cancer (NSCLC) is the world's
leading malignant tumor in terms of morbidity and mortality, of
which, lung adenocarcinoma is the most common subtype (1,2).
Surgical resection, chemotherapy and radiotherapy remain the
primary treatment options for lung adenocarcinoma. In recent
decades, despite improvements in diagnostics, surgical techniques
and radiochemotherapy regimens, as well as the application of
molecular targeted therapy, the long-term prognosis of patients
with lung adenocarcinoma remains poor due to the high metastasis
and recurrence rate, with a 5-year overall survival rate of <25%
(3). Platinum based chemotherapy
is the conventional treatment for lung adenocarcinoma (4). However, the effectiveness of
chemotherapy is limited due to the development of chemoresistance
(5), and the underlying molecular
mechanisms of chemotherapeutic drugs used for lung adenocarcinoma
are largely unknown, thus preventing resistance remains a
considerable challenge.
MicroRNAs (miRNAs/miRs) are a large class of
endogenous, small noncoding regulatory RNAs (~22 nucleotides in
length) that can suppress the expression of multiple target genes
by cleaving target mRNAs and/or inhibiting protein translation
(6,7). Studies have demonstrated that miRNAs
play critical roles in the initiation, progression and metastasis
of various types of cancer (8-12).
In addition, an increasing number of studies have suggested that
miRNAs are significantly associated with chemosensitivity in cancer
(13). For NSCLC cells, it has
been reported that miR-185-5p can overcome cisplatin sensitivity by
targeting ATP binding cassette subfamily C member 1 (14), and miR-106b-5p can reverse
cisplatin resistance by suppressing the expression of polycystin 2
(15). Moreover, overexpression
of miR-216b significantly increased the sensitivity of NSCLC cells
to cisplatin-induced apoptosis by targeting c-Jun (16).
miR-383 is a tumor suppressor that plays important
roles in multiple types of cancer, such as gastric cancer, prostate
cancer and cervical cancer (17,18). A previous study reported that in
the clinic, miR-383 was markedly downregulated in NSCLC carcinoma
tissues, in a stage dependent manner, and low tumoral miR-383
expression was negatively correlated with the overall survival of
patients with NSCLC (19).
However, it is not clear why miRNA-383 is downregulated in lung
adenocarcinoma tissues. Another study demonstrated that
overexpression of miR-383 in NSCLC cells significantly decreased
cell proliferation, migration and invasion through repression of
endothelial PAS domain-containing protein 1 (20). However, the role of miR-383 in the
survival of lung adenocarcinoma cells upon chemotherapy remains
unknown, to the best of our knowledge. The aim of the present study
was to determine the mechanism underlying miR-383 downregulation in
lung adenocarcinoma tissues, and how miR-383 regulated the
chemosensitivity of lung adenocarcinoma cells. The results
suggested a novel diagnostic and prognostic biomarker for lung
adenocarcinoma, which may highlight a potential therapeutic
strategy for the clinical reversal of chemotherapy resistance in
lung adenocarcinoma.
Materials and methods
Plasmids
Human miR-383-encoding DNA was subcloned into
pLKO.1-TRC cloning vector (plasmid #10878; Addgene, Inc.) between
AgeI and EcoRI restriction sites to construct the
miR-383 overexpression vector pLKO.1-miR-383. The empty pLKO.1-TRC
cloning vector served as a control. Human miR-383-encoding DNA was
amplified by PCR using genomic DNA extracted from A549 cells. Human
RBM24-encoding DNA was cloned into pLenti-CMV-GFP-Puro (plasmid
#17448, Addgene) between BamHI and SalI restriction
sites to form the overexpression vector pLenti-CMV-RBM24. Human
RBM24 DNA was also cloned into pCDNA3.1 between BamHI and
EcoRI to form the overexpression vector pCDNA3.1-RBM24.
Human RBM24-encoding DNA was amplified by PCR using cDNA prepared
from A549 cells. TargetScan (targetscan.org) and miRanda (microran.org) were used to predict the potential
target untranslated regions (UTRs) of miR-383. A 40-bp annealed DNA
fragment identical to part of the RBM24-3′UTR containing the
predicted miR-383-binding site (TCTGATC) was inserted into
PmeI and XbaI sites of pMIR-GLO, generating
pMIR-3′UTR/WT. pMIR-3′UTR/Mut, served as the control, and was
generated by introducing the same annealed DNA fragment with
mutations in the miR-383 seed region (CGCCTCG). Primers used for
PCR and cloning are listed in Table
I.
 | Table ISequences of the PCR primers,
annealing primers, siRNAs and miRNA inhibitors. |
Table I
Sequences of the PCR primers,
annealing primers, siRNAs and miRNA inhibitors.
| Gene | Sequence,
5′-3′ | Restriction
enzyme |
|---|
| miR-383 | | |
| Forward | AAACACCGGTCCTGCAATGTGTATTGTTTGAT | AgeI |
| Reverse | CCGGAATTCATGGATACCCAAGGTCTCATGA | EcoRI |
| RBM24 | | |
| Forward | CGCGGATCCATGCACACGACCCAGAAGGACA | BamHI |
| Reverse 1 | ACGCGTCGACGGTCTATTGCATTCGGTCTGTCTGC | SalI |
| Reverse 2 | CCGGAATTCGGTCTATTGCATTCGGTCTGTCTGC | EcoRI |
| RBM24-3′UTR | | |
| Forward |
AAACTAGCGGCCGCAGACCAGCCATCTGATCAAAGTTGAATTGTT | - |
| Reverse |
CTAGAACAATTCAACTTTGATCAGATGGCTGGTCTGCGGCCGCTAGTTT | - |
| Mut
RBM24-3′UTR | | |
| Forward |
AAACTAGCGGCCGCAGACCAGCCACGCCTCGAAAGTTGAATTGTT | - |
| Reverse |
CTAGAACAATTCAACTTTCGAGGCGTGGCTGGTCTGCGGCCGCTAGTTT | - |
| RBM24 siRNA |
GGAUCAUGCAACCAGGUUUDTDT | - |
| siRNA NC |
UUCUCCGAACGUGUCACGUDTDT | - |
| miR-383
inhibitor |
AGCCACAAUCACCUUCUGAUCU | - |
| miRNA NC |
CAGUACUUUUGUGUAGUACAA | - |
Cell culture and transfection
Human lung epithelial cells NuLi-1, human bronchial
epithelial cells HBE (CVCL_0287, Cellosaurus), Human lung cancer
cell lines A549, H1299, H1650, H460 and A549/CDDP, a cisplatin
resistant A549 cell line that was established in our previous study
(21), were cultured in RPMI-1640
medium (Cellgro; Corning, Inc.) supplemented with 10% FBS (HyClone;
Cytiva), 100 U/ml penicillin, and 100 mg/ml streptomycin (Cellgro;
Corning, Inc.) at 37°C with 5% CO2 in a humidified
incubator. For stable ectopic overexpression of miR-383 and RBM24,
a 2nd generation lentiviral system was used for lentivirus
packaging. Briefly, 5 μg of each pLKO.1-miR-383, pLKO.1-TRC
cloning, pLenti-CMV-GFP-Puro or pLenti-CMV-RBM24 vector as well as
lentiviral helper vectors pCMV-VSV-G (3.75 μg, Addgene
plasmid, cat. no. 8454) and pCMV-dR8.2 dvpr (1.25 μg,
Addgene plasmid, cat. no. 8455) were co-transfected into 293T cells
for 72 h, after which the lentiviral particles were harvested.
Cells were infected with lentiviral particles at a multiplicity of
infection of 10 for 24 h and selected with 2 μg/ml puromycin
(Sigma-Aldrich; Merck KGaA) for 1 week to establish
A549-Le/miR-383, A549-Le/RBM24 and A549/CDDP-Le/miR-383 cells.
Empty lentiviral particles infected and stable selected
A549-Le/control or A549/CDDP-Le/control cells served as controls.
Plasmids were transfected using Effectene Transfection Reagent
(Qiagen GmbH) according to the manufacturer's protocol. Human RBM24
small interfering RNA (RBM24 siRNA), nonsense control siRNA (siRNA
NC), miR-383 inhibitor and nonsense control miRNA (miRNA NC) were
synthesized by Shanghai GenePharma, Co., Ltd. The sequences are
listed in Table I. HiPerFect
Transfection Reagent (Qiagen GmbH) was used to transfect siRNA or
miRNA into cells according to the manufacturer instructions.
Briefly, 100 pmol of each siRNA or miRNA were transfected into one
well of the 6-well plate. Transfected cells were then incubated 2
days at 37°C before further analysis.
Patient samples
Lung adenocarcinoma tissues and their corresponding
non-cancerous tissues were randomly collected from 93 patients,
including 37 males and 56 females, (age range, 30-84; median age,
62 years) who underwent surgery at the Southwest Hospital of the
Army Medical University (Chongqing, China) between July 2008 and
June 2010. The subjects were diagnosed with lung adenocarcinoma
based on clinical manifestations, medical history and pathological
results, and had no history of other malignancies or relevant
antitumor treatments before the surgical resection. The
histopathological classification was based on the standard
formulated by World Health Organization in 1999 (22), and staging was based on the
criteria developed by the International Union Against Cancer in
2009 (23). Each pair of samples
were split into two halves. All samples were frozen in liquid
nitrogen within 10 min after surgery and stored at -80°C until
required for analysis. Procedures for the collection of human
samples and their use for tissue arrays and gene expression studies
were approved by the Ethical Committee of the Army Medical
University (Chongqing, China). Signed informed consent forms were
obtained from all patients prior to study participation.
Reverse transcription-quantitative
(RT-q)PCR
Total RNA from cells, tumor tissues and paired
adjacent non-tumor tissues were extracted using the QIAshredder and
RNeasy kit (Qiagen GmbH). Accordingly, 1 mg RNA was
reverse-transcribed to cDNA using SuperScript™ III Reverse
Transcriptase (Takara Bio, Inc.). PCR was performed using the
Phusion reaction system according to the manufacturer's protocol
(Thermo Fisher Scientific, Inc.). miR-383 cDNA was synthesized
using Hairpin-it™ miRNA RT-PCR Quantitation kit (Shanghai
GenePharma, Co., Ltd.) at 25°C for 30 min, 42°C for 30 min and 85°C
for 5 min. qPCR was performed on a RT-PCR system (Bio-Rad
Laboratories, Inc.). The thermocycling conditions were: 95°C for 3
min; followed by 40 cycles of at 95°C for 12 sec and 60°C for 40
sec. The expression levels of miR-383 were normalized to U6.
Detection of RBM24 mRNA was performed using a One Step SYBR
PrimeScript™ RT-PCR kit II (Takara Bio, Inc.) and GAPDH expression
was used as the loading control. The relative quantification of
gene expression was determined using the comparative CT method
(2−∆∆Cq) (24).
Primers used for RT-qPCR are listed in Table II.
 | Table IISequences of the primers used for
reverse transcription-quantitative PCR. |
Table II
Sequences of the primers used for
reverse transcription-quantitative PCR.
| Gene | Sequence,
5′-3′ |
|---|
| miR-383 | |
| Forward |
AGATCAGAAGGTGATTGTGGCT |
| Reverse |
TCTGACCAGGCAGTGCTGT |
| U6 snRNA | |
| Forward |
CTCGCTTCGGCAGCACATATACT |
| Reverse |
ACGCTTCACGAATTTGCGTGTC |
| RBM24 | |
| Forward |
CCAAGGATCATGCAACCAG |
| Reverse |
GCAGGTATCCCGAAAGGTCT |
| GAPDH | |
| Forward |
ATTCAACGGCACAGTCAAGG |
| Reverse |
GCAGAAGGGGCGGAGATGA |
Copy-number alteration (CNA) assays
The genomic DNA of each patient's tissue samples was
extracted using the GenElute kit (cat. no. G1N70; Sigma-Aldrich;
Merck KGaA) according to the manufacturer's protocol. The copy
number was analyzed by using TaqMan Copy Number assays (Applied
Biosystems; Thermo Fisher Scientific, Inc.) on the 7500 Fast Real
Time PCR system. Briefly, the test assay (FAM™ labeled), the
reference assay (VIC®-labeled) and the sample DNA were
combined with the TaqMan® MasterMix, and then the
standard TaqMan® Copy Number assay regimen was used to
calculate the relative changes in copy number.
Cell viability and invasion assay
Cell viability was assessed using a Cell Counting
Kit-8 (CCK-8) assay (cat. no. NQ646; Dojindo Molecular
Technologies, Inc.). Briefly, 5×103 cells/well were
seeded into a 96-well plate and cultured overnight. Then, cells
were treated with cisplatin for 48 h (for A549 cells the
concentration of cisplatin used were: 0, 0.5, 1, 1.5, 2 or 4
μg/ml; for A549/CDDP cells the concentrations of cisplatin
used were: 0, 2, 4, 8, 15 or 20 μg/ml). Next, 10 μl
CCK-8 solution was added to each well and incubated in the dark for
2 h. The absorbance at 450 nm was measured using a microplate
reader. The IC50 value was deduced according the cell
viability curve. The 24-well Transwell plates with 8.0 μm
pore size (cat. no. 3422, Corning) were used for invasion analysis.
Briefly, Transwell inserts in which the upper chambers were
precoated with Matrigel (cat. no. 356234; BD Biosciences) were
seeded with 3×104 cells in 200 μl serum-free
medium, and 500 μl medium supplemented with 10% FBS was
added to the lower chamber. Inserts were incubated at 37°C with 5%
CO2 for 24 h, and the cells which had invaded to
undersurface of the upper chamber were fixed with 4%
paraformaldehyde for 30 min at room temperature and stained with
0.05% crystal violet for 15 min at room temperature. A total of 5
random fields of view from each chamber were imaged and counted
under an inverted light microscope (magnification, ×200; Olympus
Corporation).
Tissue arrays
Arrays containing the 93 three tumor tissues and
adjacent non-tumor tissues were generated using a tissue
microarrayer (Leica Microsystems, Inc.). The streptavidin-biotin
peroxidase complex method was used to immunohistochemically stain
the paraffin embedded sections. The tissue arrays sections were
deparaffinized, blocked using normal goat serum at 37°C for 15 min
(cat. no. ab7481; Abcam), and then incubated with anti-human RBM24
antibody (1:250; cat. no. PA5-66881; Thermo Fisher Scientific,
Inc.) at 4°C overnight. Signals were visualized using
diaminobenzidine solution (Dako; Agilent Technologies, Inc.),
following staining with hematoxylin staining solution for 5 min at
room temperature (cat. no. C0107; Beyotime Institute of
Biotechnology). Then, a graded semiquantitative scoring system was
used to evaluate the expression of RBM24. The staining patterns
were classified based on the percentage of area stained and the
intensity of staining as follows: Negative, 0≤10%; sporadic,
11≤25%; focal, 26≤50%; or diffuse, ≥51%, and the staining intensity
was classified as none, 0; weak, 1; strong, 2; or very strong, 3.
An overall score was calculated by multiplying the intensity and
positivity scores and stratified as follows: 0, negative; 1-3, weak
staining; 4-6, moderate staining; and 7-9, strong staining
(25). To analyze the clinical
significance and prognosis, malignant samples with strong RBM24
staining were classified as high-expression, whereas low-expression
was indicated by moderate staining and weak staining.
Immunoblotting
Cells were lysed in ice cold RIPA lysis buffer
supplemented with a protease/phosphatase inhibitor cocktail (cat.
no. KGP2100; Nanjing KeyGen Biotech Co., Ltd.). The protein
concentration was determined using a Pierce BCA Protein Assay Kit
(cat. no. 23227; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. A total of 50 μg protein per
sample was loaded on an 12% SDS gel, resolved using and transferred
to a PVDF membrane (Invitrogen; Thermo Fisher Scientific, Inc.).
After blocking with 5% skim milk, the PVDF membrane was incubated
overnight at 4°C with the following primary antibodies: Anti-RBM24
(1:1,000; cat. no. PA5-66881; Thermo Fisher Scientific, Inc.),
anti-phosphorylated-(p-)P65 (1:1,000; cat. no. 3033S; Cell
Signaling Technology, Inc.), anti-P65 (1:1,000; cat. no. 8242S;
Cell Signaling Technology, Inc.), anti-p-IκBα (1:1,000; cat. no.
2859S; Cell Signaling Technology, Inc.), anti-IκBα (1:1,000; cat.
no. 4814S; Cell Signaling Technology, Inc.), anti-Bcl-2 (1:1,000;
cat. no. 15071S; Cell Signaling Technology, Inc.), anti-Bcl-xL
(1:1,000; cat. no. 2764S; Cell Signaling Technology, Inc.)
anti-cleaved caspase-3 (1:1,000; cat. no. 9661S; Cell Signaling
Technology, Inc.) and anti-GAPDH (1:2,500; cat. no. PA5-116420;
Thermo Fisher Scientific, Inc.). Then, the PVDF membranes were
incubated with the corresponding horseradish peroxidase-conjugated
Affinipure goat anti-rabbit IgG (1:5,000; cat. no. SA00001-1;
ProteinTech Group, Inc. Group, Inc.) or goat anti-mouse IgG
(1:5,000; cat. no. SA00001-2; ProteinTech Group, Inc. Group, Inc.)
secondary antibodies. at room temperature for 1 h, and visualized
using West Pico Super Signal chemiluminescent substrate (cat. no.
34095; Thermo Fisher Scientific, Inc.).
Dual-luciferase reporter assay
A549-Le/control and A549-Le/miR-383 cells were
transfected with pMIR-Report luciferase vector containing the
3′-UTR of RBM24, including WT or Mut fragments, using Effectene
Transfection Reagent. A total of 48 h after transfection, a
Dual-Luciferase Reporter kit (Promega Corporation) was used to
assay the luciferase activity according to the manufacturer's
protocol. Each transfection was repeated three times.
Alkaline comet assay
Alkaline comet assays were performed to assess
intra-strand and inter-strand crosslink formation using a Comet
assay kit (cat. no. ab238544; Abcam) according to the
manufacturer's protocol. Briefly, A549 cells stably overexpressing
RBM24 and A549/CDDP cells stably overexpressing miR-383 were
treated with their IC50 doses of cisplatin for 6 h.
Then, cells were trypsinized and resuspended at 1×105
cells/ml in ice-cold PBS (Mg2+ and Ca2+
free). Cell samples were combined with comet agarose and 75
μl of each sample was added onto the comet slide. After
solidification, the slide was immersed in the pre-chilled lysis
buffer for 1 h at 4°C and pre-chilled alkaline solution for a
further 1 h at 4°C. Next, the slides were electrophoresed in TBE
buffer for 30 min at 60 V at 4°C. After soaking in the pre-chilled
DI H2O three times (2 min/each), the slides were
immersed in cold 70% ethanol for 5 min. Subsequently, they were air
dried, and 100 μl/well diluted Vista Green DNA Dye was added
and incubated at room temperature for 15 min. Epifluorescence
microscopy (magnification, ×200; Olympus Corporation) with a FITC
filter was used to capture images of the cells, and the mean tail
moment was calculated using Comet Score version 1.5 (TriTeK
Corp.).
Generation of xenografts and drug
treatment
miR-control and miR-383 tumor xenografts were
established by subcutaneously inoculating 5×106 cells
(A549-Le/control and A549-Le/miR-383 cells resuspended in PBS at
5×107 cells/ml) into 4-week-old male BALB/c nude mice in
the right flank next to the hind limb (n=20 per group; 40 in total;
average weight, 16 g). A total of 28 days later, xenograft tumors
that were ~1.5 cm in length and diameter were selected for drug
treatment (n=9 per group). The volume of each tumor was calculated
according to the formula volume=A2B/2, where A
represents the length/diameter and B represents the short diameter.
Selected mice were subjected to peritoneal injection of 4 mg/kg
cisplatin once every week for 3 weeks. Every 6 days after
initiation of treatment, the tumor volumes were measured. Then all
mice were sacrificed to harvest the tumor tissues to perform
immunohistochemistry assays. All procedures for animal experiments
were approved by the Committee on the Use and Care on Animals [Army
Medical University (previously known as The Third Military Medical
University), Chongqing, China] and performed in accordance with
institutional guidelines. Mice were housed in a specific
pathogen-free facility with temperature at 24±1°C, relative
humidity at 40-60%, a 12:12 h light/dark cycle and free access to
food and water. All animals received humane care according to the
criteria outlined in the Guide for the Care and Use of Laboratory
Animals prepared by the National Academy (26). At the end of the experiment, all
animals were euthanized by CO2 inhalation with a chamber
volume displacement rate of 30% per minute (volume/min; performed
between March and September 2020).
Statistical analysis
Survival curves were plotted using the Kaplan-Meier
method, and the correlation between miR-383 or RBM24 expression and
the overall survival of patients with NSCLC was estimated using a
log-rank (Mantel-Cox) test. The comparison between two groups was
determined using a paired Student's t-test, and differences between
multiple groups were determined by ANOVA followed by a Tukey's
post-hoc test of pairwise differences (Tukey Honest Significant
Difference). The statistical analysis was performed using SPSS
version 19.0 (IBM, Corp.), and data are presented as the mean ± the
standard deviation. P<0.05 was considered to indicate a
statistically significant difference. The relationship between
RBM24 expression and miR-383 expression was evaluated using
Spearman's rank correlation analysis.
Results
miRNA-383 expression is downregulated in
patients with lung adenocarcinoma and this is associated with worse
survival outcomes
miR-383 expression profiling was performed on a
cohort of 93 patients with lung adenocarcinoma. Expression was
analyzed and compared with the matching non-cancerous tissues by
RT-qPCR. miR-383 expression was found to be significantly
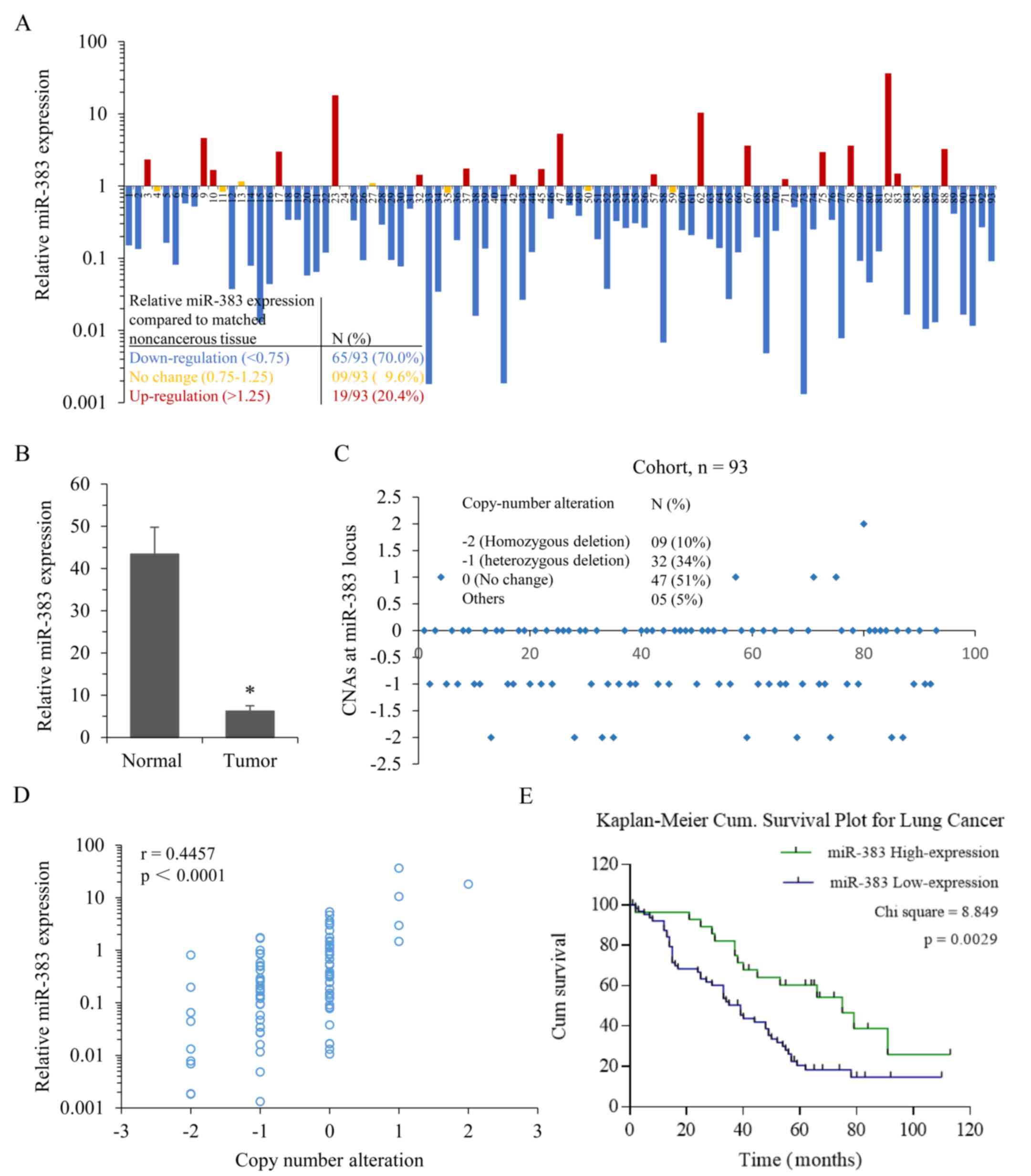
downregulated in 70% of lung adenocarcinoma cases (Fig. 1A). A total of 9.6% of lung
adenocarcinoma cases showed no change in expression, whereas 20.4%
of cases exhibited upregulated miR-383 expression. The average
miR-383 expression in lung adenocarcinoma tissues was ~6-fold lower
than that in non-cancerous tissues (P=0.017; Fig. 1B). miR-383 is located in a common
region of loss of heterozygosity (LOH) at the chr8p22 locus within
intron 3 of the Sarcoglycan ζ (SGCZ) gene (18). Next, the CNAs at this locus in
this cohort (n=93) were analyzed, and it was found that ~44% of
lung adenocarcinoma tissues exhibited heterozygous or homozygous
loss at the miR-383 locus compared with the adjacent non-cancerous
tissues (Fig. 1C). Additionally,
the correlation between CNAs and miR-383 expression within this
cohort was analyzed, and the results showed a positive correlation
(r=0.4457, P<0.0001; Fig. 1D).
In consideration of the universal downregulation of miR-383
expression in lung adenocarcinoma clinical specimens, analysis of
miR-383 expression with the clinicopathological parameters was
performed. The levels of miR-383 expression were stratified into
high (no change and upregulation, ≥0.75; n=28) and low
(downregulation, <0.75; n=65) expression groups relative to
matched noncancerous tissue. Kaplan-Meier survival analysis showed
that patients with low miR-383 expression had significantly poorer
survival outcomes (P=0.0029; Fig.
1E). Additionally, a low level of miR-383 expression in lung
adenocarcinoma was significantly associated with a higher mortality
rate (P=0.045), Tumor-Node-Metastasis stage (P=0.027) and
metastasis (P=0.044) (Table
III). These results suggested that low miR-383 expression may
be involved in the progression of lung adenocarcinoma.
 | Table IIIRelationship between miR-383
expression and of association with the clinicopathological
characteristics of the 93 patients used in the present study. |
Table III
Relationship between miR-383
expression and of association with the clinicopathological
characteristics of the 93 patients used in the present study.
| Characteristic | Number of
cases | miR-383 expression
| P-value |
|---|
| Low, n (%) | High (%) |
|---|
| Sex | | | | 0.121 |
| Male | 37 | 22 (59.5) | 15 (40.5) | |
| Female | 56 | 43 (17.1) | 13 (82.9) | |
| Age, years | | | | 0.247 |
| ≥60 | 50 | 38 (76.0) | 12 (24.0) | |
| <60 | 43 | 27 (62.8) | 16 (37.2) | |
| Death | | | | 0.045a |
| No | 65 | 50 (76.9) | 15 (23.1) | |
| Yes | 28 | 15 (53.6) | 13 (46.4) | |
| Tumor grading | | | | 0.381 |
| 1 | 19 | 11 (57.9) | 08 (42.1) | |
| 2 | 48 | 34 (70.8) | 14 (29.2) | |
| 3 | 26 | 20 (76.9) | 06 (23.1) | |
| Tumor-Node- | | | | |
| Metastasis
stage | | | | 0.027a |
| Early (I/II) | 42 | 24 (57.1) | 18 (42.9) | |
| Advanced
(III) | 51 | 41 (80.4) | 10 (19.6) | |
| Lymph node | | | | |
| metastasis | | | | 0.044a |
| No | 37 | 21 (56.8) | 16 (43.2) | |
| Yes | 56 | 44 (78.6) | 12 (21.4) | |
miRNA-383 sensitizes lung adenocarcinoma
cells to cisplatin
The expression patterns of miR-383 in lung
adenocarcinoma cell lines in vitro were assessed. Compared
with the NuLi-1 and HBE cells, endogenous miR-383 expression in
lung adenocarcinoma cell lines was significantly downregulated,
including H1299, H1650, H460, A549 and A549/CDDP cells (Fig. 2A). Compared with the parental A549
cells, the expression of miR-383 in cisplatin-resistant
counterparts (A549/CDDP cells) was further reduced. To study the
function of miR-383, a miR-383 overexpression plasmid was created
using the pLKO.1 vector (Fig.
2B). Using lentiviral infection and selection, miR-383 was
overexpressed in A549 and A549/CDDP cells (Fig. 2C and D). Overexpression of miR-383
inhibited the proliferation, migration and invasion of A549 cells
(Fig. S1). Cell viability
analysis showed that the high expression of miR-383 significantly
increased the sensitivity of cells to cisplatin; that is, the
IC50 of A549 cells decreased from 2 to 1.3 μg/ml,
whereas the IC50 of A549/CDDP decreased from 9 to 5.5
μg/ml (Fig. 2E and F).
These results suggested that miR-383 was involved in cisplatin
resistance in lung adenocarcinoma cells.
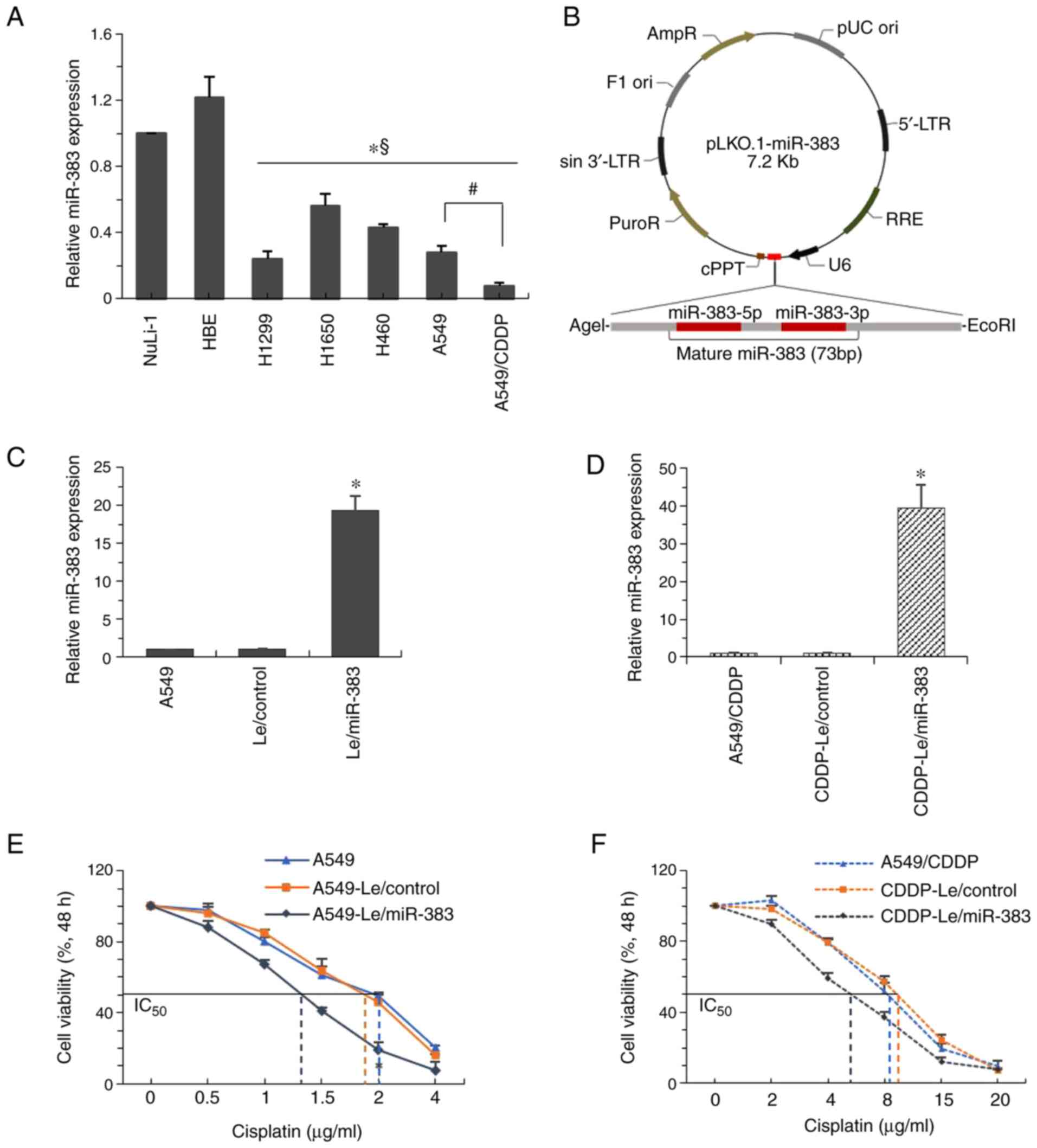 | Figure 2miR-383 sensitizes lung
adenocarcinoma cells to cisplatin. (A) RT-qPCR was used to quantify
the expression of miR-383 in the lung adenocarcinoma cell lines,
H1299, H1650, H460, A549 and A549/CDDP, and then normalized to
miR-383 expression in the normal human bronchial epithelium cell
line, NuLi-1. *P<0.05 vs. NuLi-1;
§P<0.05 vs. HBE cells; #P<0.05,
A549/CDDP vs. A549 cells. (B) The vector for overexpression of
miR-383. Mature miR-383-encoding DNA was directly inserted
downstream of the U6 promoter to create the pLKO.1-miR-383 vector.
(C) Expression of miR-383 was determined by RT-qPCR in
A549-Le/miR-383 cells and A549-Le/control cells, and normalized to
the expression in A549 cells. (D) miR-383 expression was determined
by RT-qPCR in A549/CDDP-Le/miR-383 cells and A549/CDDP-Le/control
cells, and normalized to the expression in A549/CDDP cells.
*P<0.05. The effect of miR-383 overexpression on the
viability of (E) A549 and (F) A549/CDDP cells after treatment with
different concentrations of cisplatin for 48 h. Data are presented
as the mean ± standard deviation of three sets of independent
experiments. miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; Le, lentiviral vector; A549/CDDP,
cisplatin-resistant A549 cells. |
miRNA-383 modulates RBM24 expression in
lung adenocarcinoma cells
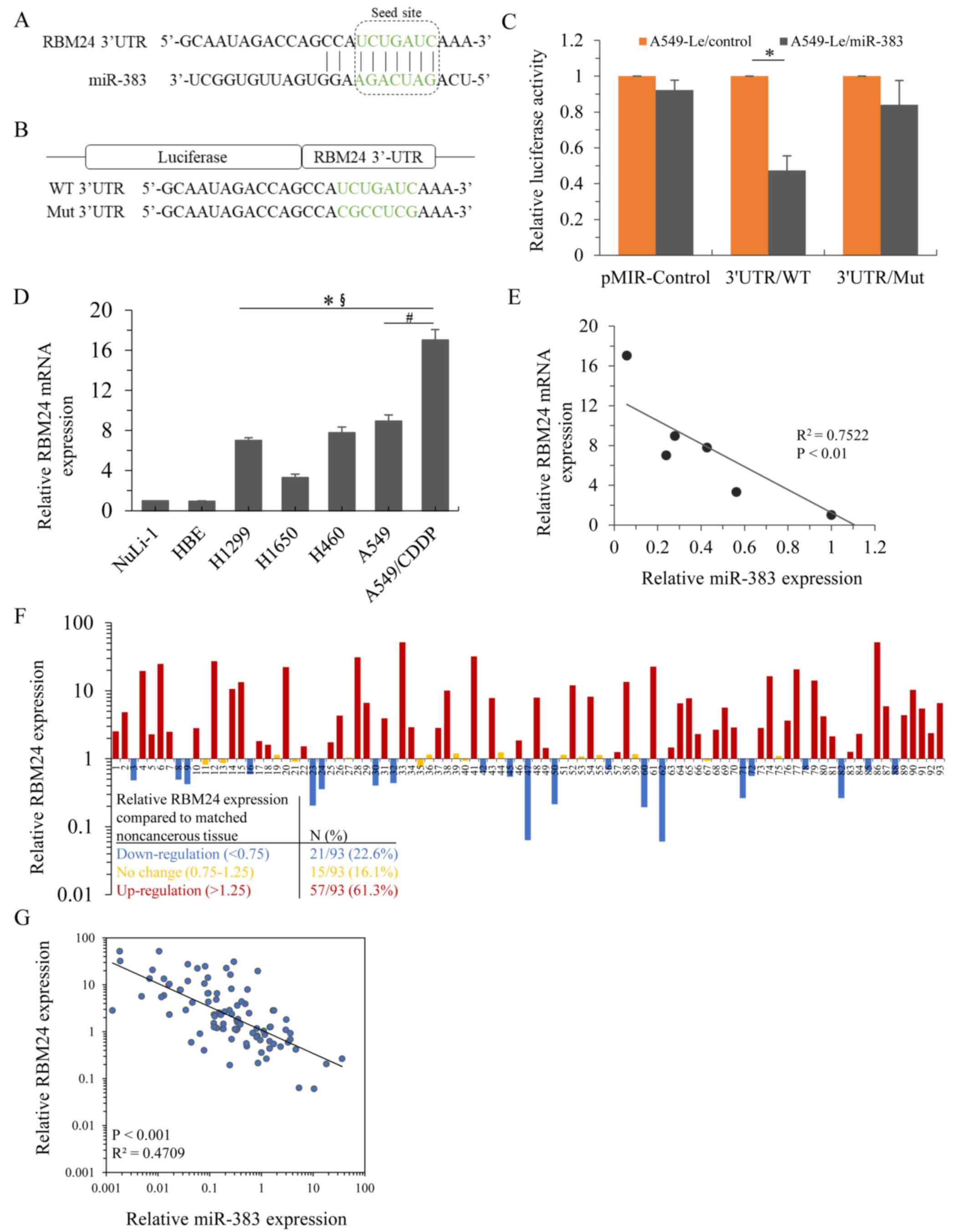
To investigate how miR-383 affected the sensitivity
of lung adenocarcinoma cells to cisplatin, TargetScan (targetscan.org) and miRanda (microran.org) were used, and they showed that that
RBM24 was a predicted target of miR-383. According to
bioinformatics analysis, the specific miR-383 binding site was
located within the 3′UTR of RBM24 mRNA (Fig. 3A). To investigate whether miR-383
directly targets the 3′UTR of RBM24 mRNA, the wild-type
(RBM24-3′UTR/WT) or mutant (RBM24-3′UTR/Mut) 3′UTR of RBM24 mRNA
was inserted into the pMIR-REPORT vector for luciferase activity
analysis (Fig. 3B). Using a dual
luciferase reporter assay, it was shown that miR-383 stably
overexpressing cells exhibited remarkably decreased luciferase
activity in the 3′UTR/WT group, whereas no significant reduction
was observed in the pMIR-control or 3′UTR/Mut group (Fig. 3C). Next, the mRNA expression
patterns of RBM24 in the abovementioned lung adenocarcinoma cell
lines was determined. Compared with the NuLi-1 and HBE cells,
endogenous RBM24 mRNA expression was significantly upregulated in
lung adenocarcinoma cell lines and further enhanced in
cisplatin-resistant A549 cells (Fig.
3D). Analysis of the expression patterns of miR-383 and RBM24
mRNA revealed that miR-383 and RBM24 were inversely associated in
these lung adenocarcinoma cell lines (Fig. 3E). Additionally, RBM24 mRNA
expression profiling was performed on the patient cohort samples,
and it was shown that RBM24 was significantly upregulated in 61.3%
of lung adenocarcinoma cases (Fig.
3F). A total of 16.1% of lung adenocarcinoma cases showed no
change, and 22.6% of cases exhibited reduced RBM24 mRNA expression.
Similar to what was observed in the lung adenocarcinoma cell lines,
the levels of miR-383 and RBM24 mRNA in the clinical specimens were
inversely associated (Fig. 3G).
Moreover, overexpression of miR-383 inhibited the expression of
RBM24 in both parental A549 cells and A549/CDDP cells (Fig. S2A and B). Together, these results
suggested an inverse association between RBM24 mRNA and miR-383
expression levels in lung adenocarcinoma cells.
RBM24 upregulation in lung cancer is
correlated with a poorer prognosis
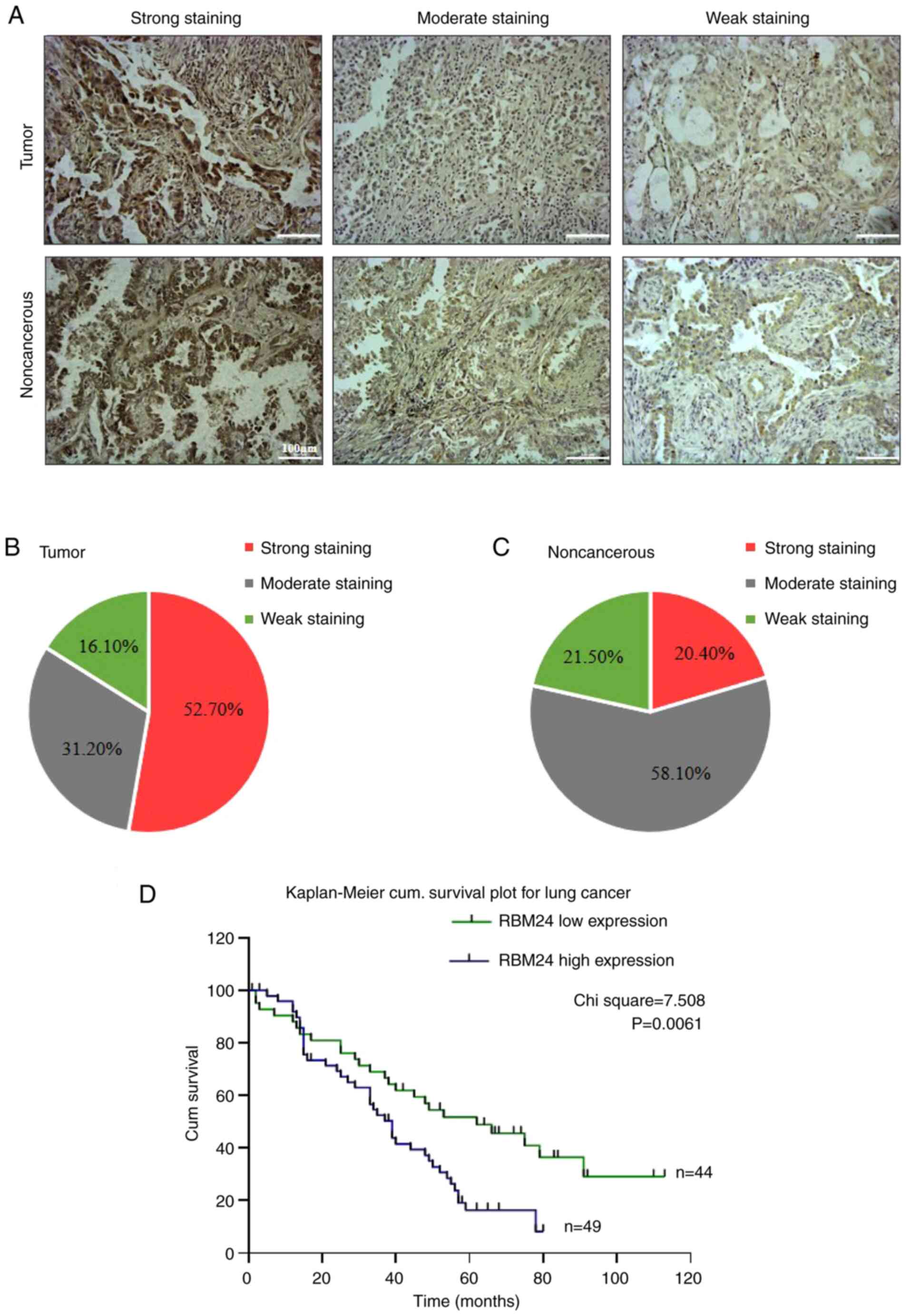
To further investigate whether aberrant RBM24
expression can be used to predict prognosis in patients with lung
adenocarcinoma, a tissue array was generated using the tissue
samples obtained from the cohort, and RBM24 expression was
determined using immunohistochemistry. Representative images with
strong, moderate and weak staining in the tumor tissue and
non-cancerous samples are shown in Fig. 4A. Strong staining was observed in
52.7% of tumor samples, whereas moderate and weak staining were
found in 31.2 and 16.1% of samples, respectively (Fig. 4B). In contrast, RBM24 expression
was relatively lower in non-cancerous lung tissues. Strong
positivity was found in only 20.4% of samples, whereas moderate and
weak RBM24 expression was observed in 58.1 and 21.5% of tissues,
respectively (Fig. 4C).
Importantly, RBM24 upregulation was associated with shorter overall
survival than RBM24 low expression (including moderate and weak
staining) in patients (P=0.0061; Fig.
4D).
miRNA-383 negatively regulates RBM24
expression, resulting in increased cisplatin sensitivity of lung
adenocarcinoma
Next, whether RBM24 could affect the sensitivity of
lung adenocarcinoma cells to cisplatin was assessed. First, the
expression of RBM24 in parental and cisplatin resistant A549 cells
was determined, and the results showed that A549/CDDP cells
demonstrated significantly increased expression of RBM24 (Fig. 5A). After knocking down RBM24
expression using siRNA (Fig.
S2C), the IC50 of A549/CDDP cells decreased from ~9
to ~3.5 μg/ml (Fig. 5B and
C). In contrast, with stable overexpression of RBM24 in A549
cells, the IC50 increased from 1.7 to ~3.0 μg/ml
(Fig. 5D and E). In addition,
neutralization of miR-383 using an inhibitor in A549 cells
(Fig. S2D) increased the
expression of RBM24 and enhanced tolerance to cisplatin, increasing
the IC50 from 1.6 to 2.8 μg/ml (Fig. 5F and G). Moreover, knockdown of
RBM24 overcame the upregulation of RBM24 expression induced by
neutralization of miR-383 and reduced the IC50 to ~0.8
μg/ml (Fig. 5H and I). In
addition, a comet assay demonstrated that overexpression of RBM24
inhibited the formation of DNA double-stranded breaks induced by
treatment with an IC50 dose cisplatin in A549 cells,
whereas overexpression of miR-383 in A549/CDDP cells promoted their
formation (Fig. 5J). Taken
together, these data demonstrated that miR-383 downregulation may
be responsible for the upregulation of RBM24, which partly
contributed to the cisplatin resistance of lung adenocarcinoma
cells.
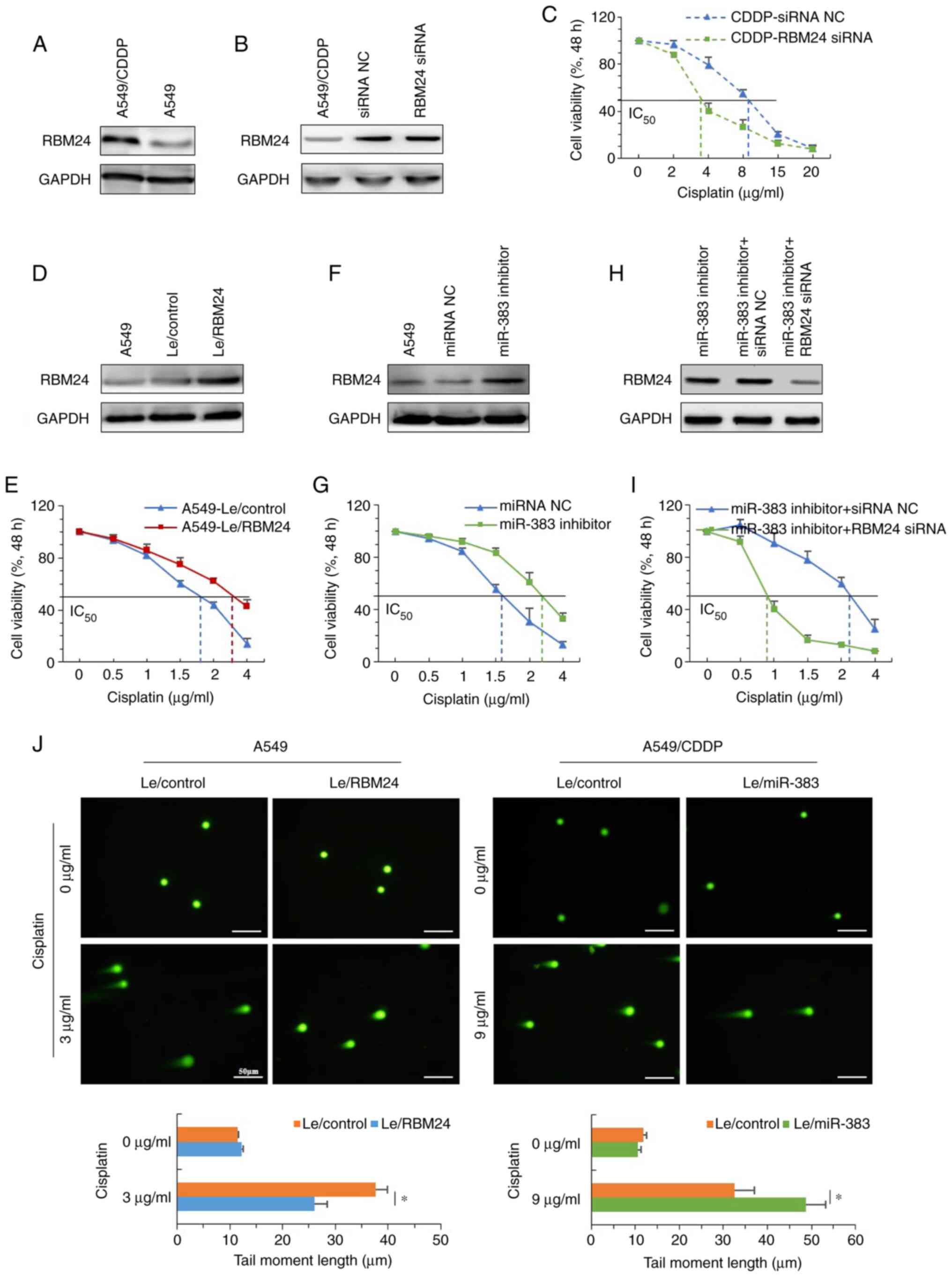 | Figure 5miR-383 enhances the sensitivity of
lung adenocarcinoma cells to cisplatin through negative regulation
of RBM24. (A) Protein expression levels of RBM24 were determined by
immunoblotting in A549/CDDP and parental A549 cells. (B) A549/CDDP
cells were transfected with siRNA NC or RBMA24 siRNA for 48 h, and
RBM24 expression as assessed using western blotting. (C) Effect of
RBM24 knockdown on the viability of A549/CDDP cells after treatment
with different concentrations of cisplatin for 48 h. (D) Western
blotting was used to determine the expression of RBM24 in A549
cells stably overexpressing RBM24 cells; uninfected and control
lentivirus-infected A549 cells served as controls. (E) Effect of
stable overexpression of RBM24 on the viability of A549 cells after
treatment with different concentrations of cisplatin for 48 h. (F)
A549 cells were transfected with miRNA NC or miR-383 inhibitor for
48 h, and RBM24 expression was assessed using western blotting. (G)
Effect of miR-383 knockdown on the viability of A549 cells after
treatment with different concentrations of cisplatin for 48 h. (H)
A549 cells were co-transfected with miR-383 inhibitor and either
RBM24 siRNA or siRNA NC for 48 h, and RBM24 expression was assessed
by western blotting. (I) Effect of miR-383 and RBM24 knockdown on
the viability of A549 cells after treatment with different
concentrations of cisplatin for 48 h. (J) Representative images
showing detectable comet tails visualized under a fluorescence
microscope. Magnification, ×200; scale bar, 50 μm. Data are
presented as the mean ± standard deviation of three independent
experiments. *P<0.05. miR, microRNA; RBM24, RNA
binding motif protein 24; A549/CDDP, cisplatin-resistant A549
cells; siRNA, small interfering RNA; NC, nonsense control;
miR/miRNA, microRNA. |
miRNA-383 suppresses the activation of
RBM24 mediated NF-κB signaling
Next, the effect of miR-383 on the activation of
NF-κB signaling was investigated. Western blot analysis
demonstrated that in both A549 and A549/CDDP cells, restoration of
miR-383 significantly decreased the Ser536-phosphorylation of p65
and Ser32-phosphorylation of IκBα. Moreover, consistent with the
expression of RBM24, the anti-apoptosis-related proteins Bcl-2 and
Bcl-xL were both downregulated (Fig.
6A). Using siRNA to knockdown RBM24 expression in A549/CDDP
cells, similar results were observed. The levels of phosphorylated
p65 and IκBα, as well as Bcl-2 and Bcl-xL expression were also
decreased (Fig. 6B). Given that
RBM24 was identified as the direct target of miR-383, whether
miR-383-mediated inactivation of NF-κB was associated with
downregulation of RBM24 was next assessed. Rescue experiments
showed that overexpression of RBM24 restored the phosphorylation of
p65 and IκBα by increasing the expression of Bcl-2 and Bcl-xL in
miR-383-overexpressing A549/CDDP cells (Fig. 6C). In addition, the results also
demonstrated that overexpression of RBM24 in A549 cells partially
inhibited the activation of caspase-3 (cleaved caspase-3) that were
induced by 3 μg/ml cisplatin. However, 3 μg/ml
cisplatin could not induce the activation of caspase-3 in A549/CDDP
Le-control cells, whereas the expression of RBM24 was significantly
inhibited in miR-383 overexpressing A549/CDDP cells, and the
quantity of active caspase-3 induced by 3 μg/ml cisplatin
was significantly increased (Fig.
S3). Taken together, these findings suggest that targeting
RBM24 was involved in the inhibition of NF-κB signaling by
miR-383.
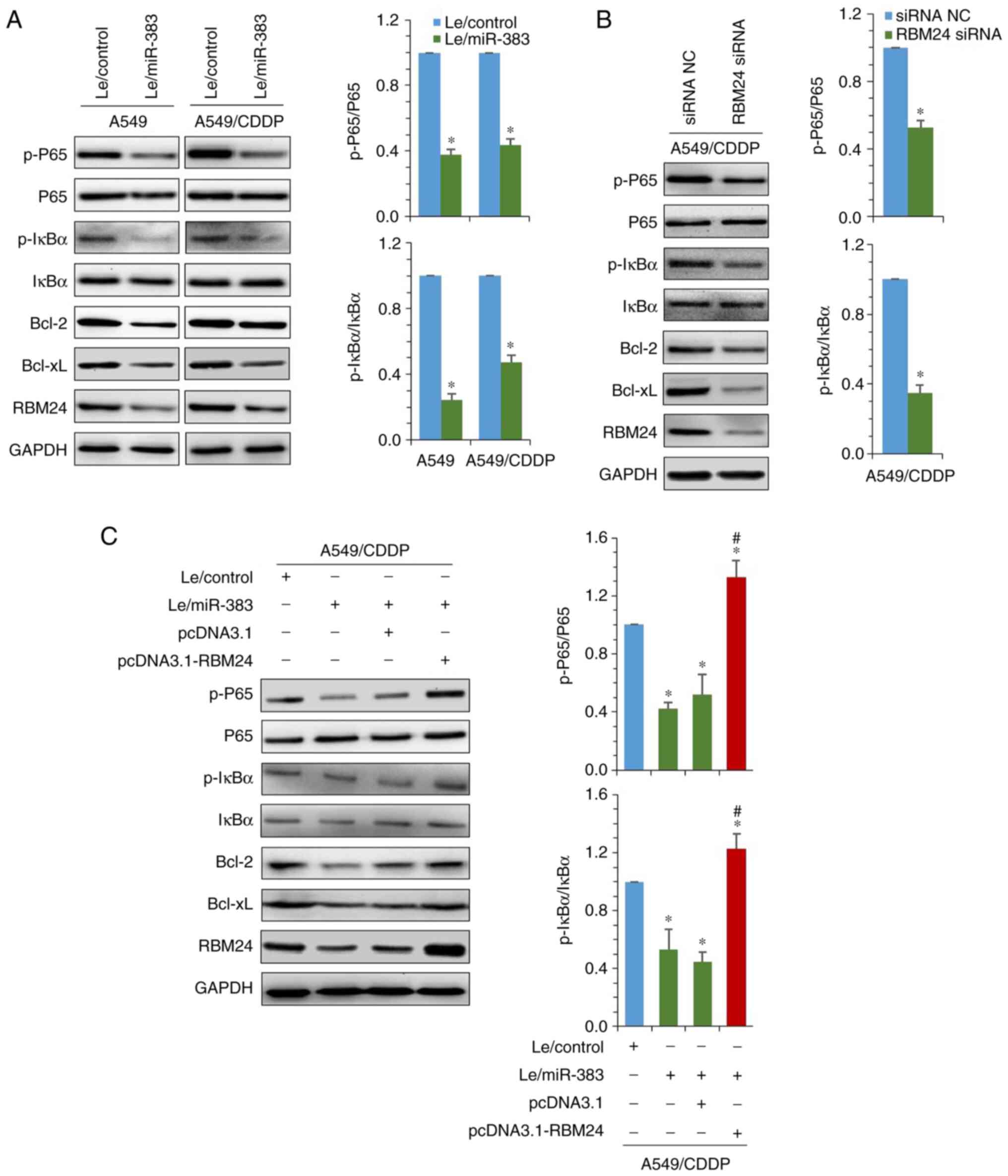 | Figure 6miR-383 inhibits RBM24-mediated NF-κB
signaling activation. (A) Immunoblotting was used to detect the
expression of NF-κB signaling regulatory factors and downstream
target genes in A549 and A549/CDPP cells stably overexpressing
miR-383 (left panel). Ratio of phosphorylated p65 and IκBα to total
expression of p65 and IκBα (right panels). (B) RBM24 expression was
knocked down using siRNA in A549/CDDP cells, and expression of the
indicated proteins was detected by western blotting (left panel).
Ratio of phosphorylated p65 and IκBα to total expression of p65 and
IκBα (right panels). (C) Immunoblotting was used to detect the
effect of transient overexpression of RBM24 on the expression of
the indicated proteins in A549/CDDP cells following restoration of
miR-383 expression (left panel). Ratio of phosphorylated p65 and
IκBα to total p65 and IκBα expression (right panels).
*P<0.05 vs. A549/CDDP-Le/control;
#P<0.05, pCDNA3.1-RBM24 group vs. pCDNA3.1 group.
Data are presented as the mean ± standard deviation of three
independent experiments. miR, microRNA; RBM24, RNA binding motif
protein 24; A549/CDDP, cisplatin-resistant A549 cells; NF-κB,
nuclear factor κB; IκBα, inhibitor α of NF-κB; p-, phosphorylated;
Le, lentiviral vector; Bcl-2, B-cell lymphoma-2; Bcl-xL, B-cell
lymphoma-xL. |
miR-383 enhances the cisplatin
sensitivity of lung adenocarcinoma cells in vivo
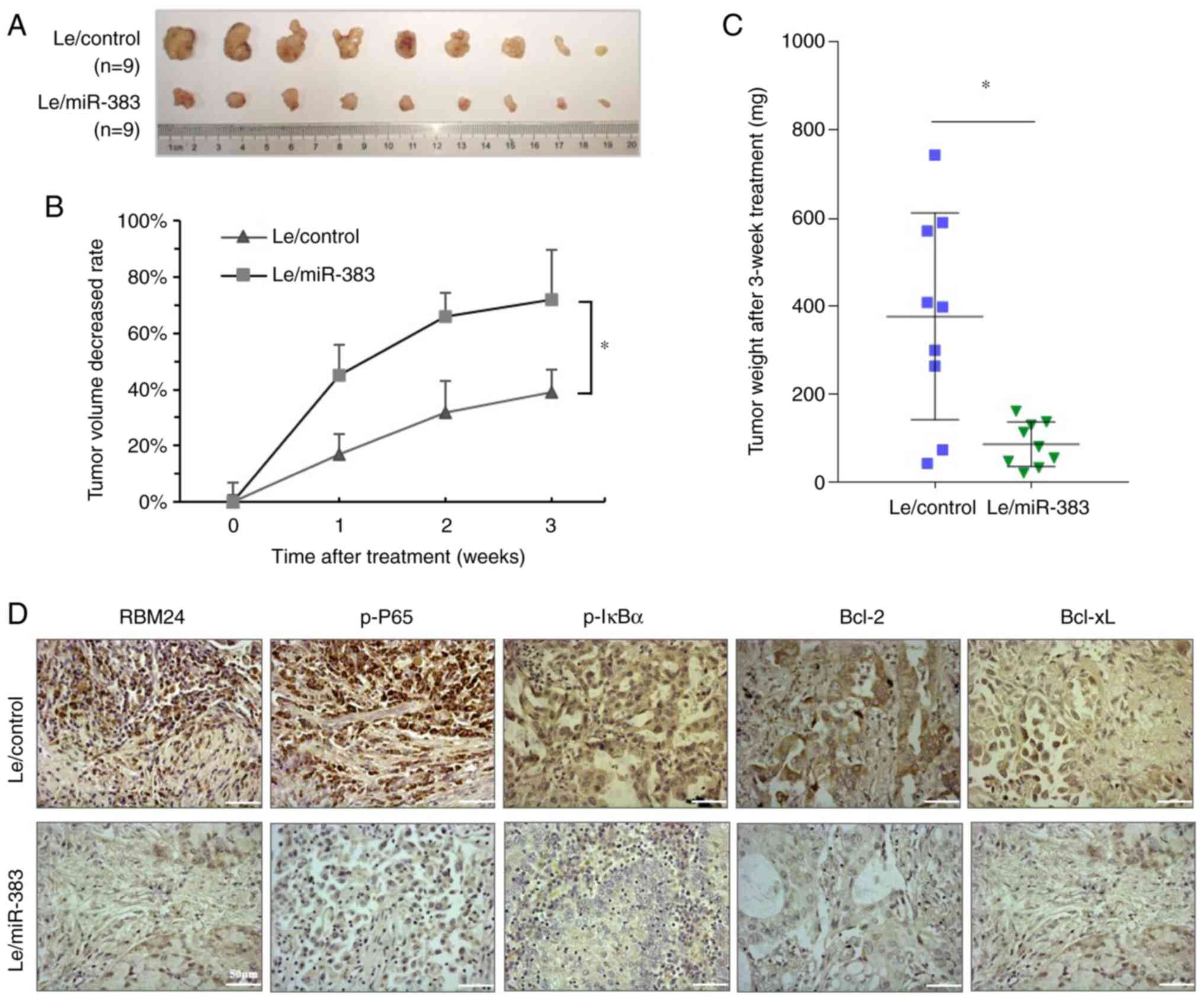
The role of miR-383 in the cisplatin sensitivity of
lung adenocarcinoma cells was also investigated in an animal model.
Xenograft tumors (n=9) of the A549/Le-miR-383 and A549/Le-control
groups ~1.5 cm in length and diameter were selected for cisplatin
treatment. After 3 weeks of treatment, tumors in the Le-miR-383
group were significantly smaller than those in the Le-control group
(Fig. 7A). The average tumor
volume gradually decreased after treatment in both groups, whereas
the average tumor volume reduction rates of the Le-miR-383 group
were significantly higher than those of the Le-control group
(Fig. 7B). Additionally, the
average weight of tumors was significantly lower in the Le-miR-383
group than in the Le-control group (Fig. 7C). Immunohistochemical staining
revealed that in the Le-miR-383 group, the expression of RBM24,
p-P65 and p-IκBα, as well as the downstream Bcl-2 and Bcl-xL was
suppressed compared with that in the Le-control group (Fig. 7D). These data suggested that
miR-383-targeted RBM24 enhanced the sensitivity of lung
adenocarcinoma cells to chemotherapeutics.
Discussion
Resistance to chemotherapeutic drugs is a difficult
problem in antitumor research worldwide. It has been reported that
>85% of patients with lung cancer treatment failure exhibit drug
resistance (27). Therefore,
research on the mechanism of lung cancer drug resistance, the
identification of new antitumor targets and the development of new
antitumor drugs have always been the focus of attention. miRNAs are
a relatively more recent and important discovery in the field of
RNA biology. There are >2,500 miRNAs expressed in the human
body, and 60% of human genes may be regulated by them (28). These target genes are involved in
a series of biological processes, such as individual development,
cell differentiation, proliferation and apoptosis (29). Therefore, miRNAs serve important
roles in the generation and development of human diseases, such as
tumors and metabolic disorders. However, the roles and mechanisms
in tumor drug resistance remain unclear.
miR-383 serves important roles in the progression of
multiple tumors (30,31). The present study found that the
expression of miR-383 in lung adenocarcinoma samples was reduced
and related to the poor prognosis of patients. This conclusion is
consistent with the results of previous studies assessing the same
miRNA and type of cancer (32,33). Restoration of miR-383 inhibited
the proliferation, migration and invasion of A549 cells. However,
for the first time, the present study demonstrated that restoration
of miR-383 expression promoted the cisplatin sensitivity of lung
adenocarcinoma cells. miR-383 promoted cisplatin sensitivity by
interacting with the 3′-UTR of RBM24 mRNA to negatively regulate
RBM24. Thus, these results highlight the potential of miR-383 and
RBM24 as biomarkers for the early diagnosis and prognosis of lung
adenocarcinoma.
The host gene SGCZ of miR-383 is located on chr8p22
(34). It has been reported that
a common region of LOH at the chr8p22 locus is related to breast
cancer and prostate cancer (18,35), and the present study also
confirmed that the low expression of miR-383 in lung adenocarcinoma
tissues was related to the deletion of this locus. In addition,
miR-383 was shown to inhibit the activation of the NF-κB signaling
pathway in the present study. Therefore, it was speculated that the
abnormal activation of the NF-κB signaling pathway caused by the
deletion of miR-383 may be partly responsible for the natural
resistance of certain lung adenocarcinoma cells to
chemotherapy.
Mechanistically, overexpression of miR-383 inhibited
the activation of NF-κB signaling in both A549 and A549/CDDP cells.
NF-κB is a key modulator of apoptosis in cancer cells, which
functions by regulating antiapoptotic-related genes, such as Bcl-2
and Bcl-xL (36). p65 is
phosphorylated at multiple sites, and Ser536-phosphorylation
enhances p65 transactivation potential (37). Chen et al (38) reported that IκBα-S32 and p65-S536
phosphorylation promoted the expression of Bcl-2. Bravo-Cuellar
et al (39) reported that
pentoxifylline and the proteasome inhibitor MG132 decreased the
p65-S536 phosphorylation and the expression of Bcl-2 and Bcl-xL,
inducing apoptosis in human leukemia U937 cells. The results of the
present study also showed that overexpression of miR-383 led to a
decrease in Bcl-2 and Bcl-xL expression in both A549 and A659/CDDP
cells, along with a reduction in Ser536-phosphorylation of p65 and
Ser32-phosphorylation of IκBα. However, the total expression of p65
and IκBα was not affected by miR-383. Further analysis showed that
miR-383-mediated inactivation of NF-κB was due to the
downregulation of RBM24. RBM24 is a member of the RNA binding
protein family, and has a conserved RNA recognition motif at its
N-terminus, which is mainly used as a post-transcriptional
regulator to regulate RNA metabolism, and plays an important role
in gene expression (40). A
previous study showed that RBM24 inhibits the progression of
nasopharyngeal carcinoma by upregulating miR-25 to target the
metastasis associated with lung adenocarcinoma transcript 1
(41). However, the present study
found that RBM24 is highly expressed in lung adenocarcinoma and is
positively correlated with the poor prognosis of patients. The
reason why the results of these studies are opposite may be due to
differences in the biological function of RBM24 in different
tissues/types of cancer. To the best of our knowledge, there are no
studies assessing the correlation between RBM24 and tumorigenesis.
It is necessary to further study the biological functions of RBM24
in different types of tumors. Studies have shown that RBM24
negatively regulates the expression of p53 and p63 by binding to
specific regions of their mRNA, suggesting that RBM24 may
negatively regulate the p53-mediated apoptosis pathway (42,43). The present study also showed that
RBM24 can activate the NF-κB signaling pathway to promote the
expression of downstream anti-apoptotic genes, including Bcl-2 and
Bcl-xL. Therefore, it is hypothesized that the abnormally high
expression of RBM24 mediates the cisplatin insensitivity of lung
adenocarcinoma through multiple pathways. It is worth mentioning
that RBM24 only changed the amount of phosphorylated p65 and IκBα,
not the total expression. It is suggested that part of the reason
is that RBM24 may affect the phosphorylation levels in cells
through post-transcriptional regulation of the expression of
certain kinases. Therefore, the exact mechanism by which RBM24
regulates the NF-κB signaling pathway needs further study.
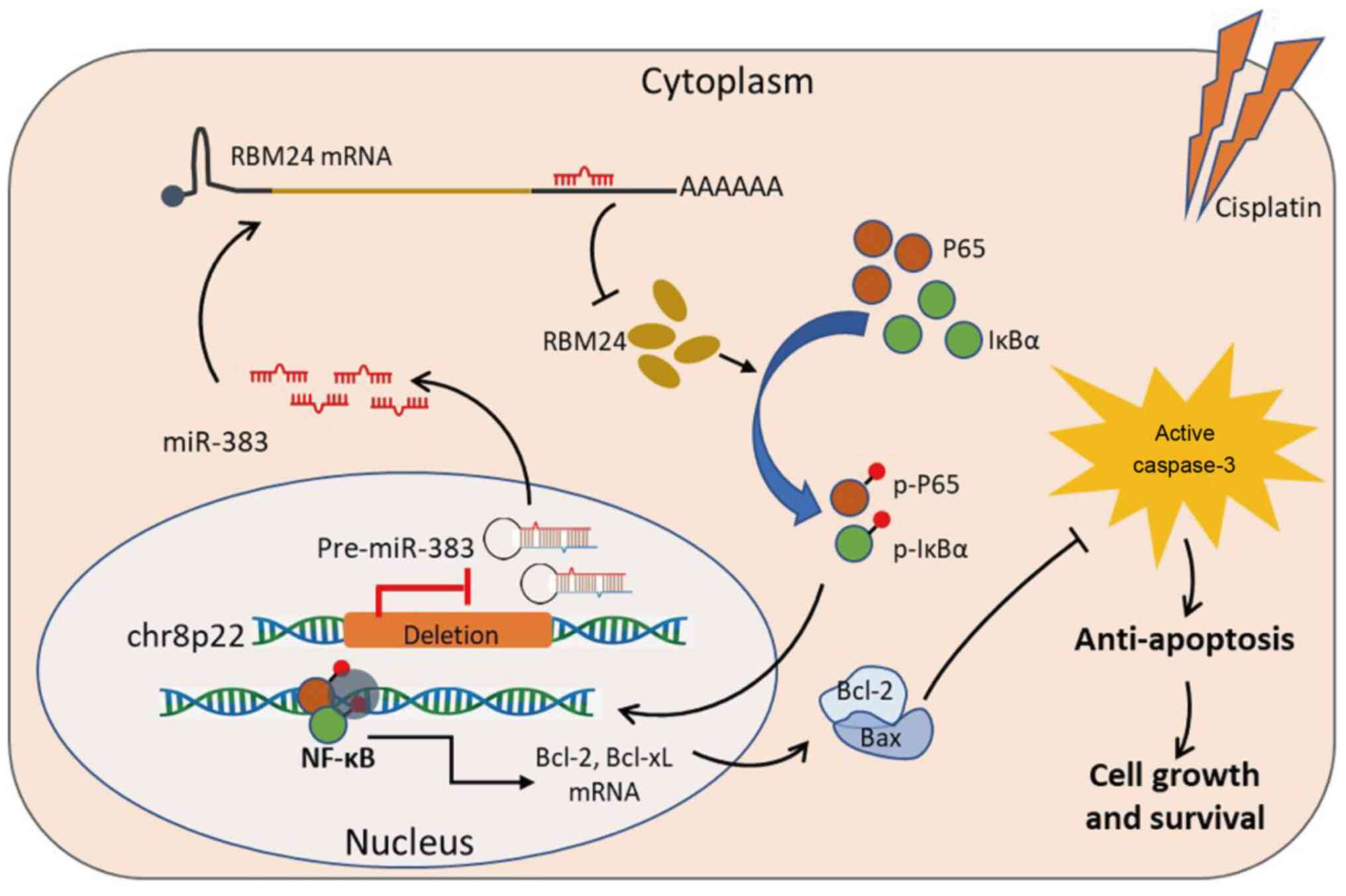
In conclusion, the results of the present study
demonstrated that chr8p22 loss resulted in defective expression of
miR-383 in lung adenocarcinoma cells, leading to the upregulation
of RBM24, which activates NF-κB signaling and causes the
upregulation of downstream anti-apoptosis-related proteins, thus
improving the tolerance of tumor cells to chemotherapy (Fig. 8). Restoration of miR-383 or
interference of RBM24 may provide potential therapeutic approaches
for reversing the chemotherapy resistance of lung
adenocarcinoma.
Supplementary Data
Availability of data and materials
All data generated and/or analyzed during the
present study are included in this published article.
Authors' contributions
YL and HW designed and conceived the study. BH and
CW performed the experiments. WS and YQ collated and analyzed the
clinicopathological data, and drafted the manuscript. JL, ZL and TJ
performed the statistical analysis and data interpretation. All
authors have read and approved the final manuscript. YL, BH and JL
confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
The study protocol was approved by the Ethics
Committee of the First Affiliated Hospital of the Army Medical
University (previously known as Third Military Medical University),
PLA (2015; Chongqing, China). Procedures for the collection of
human samples, and their use for tissue arrays and gene expression
studies were approved by the Ethical Committee of the Army Medical
University (Chongqing, China). All patients signed an informed
consent form and volunteered to participate in this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
We would like to thank Dr Xichao Xu (Key Laboratory
of Biorheological Science and Technology, College of
Bioengineering, Chongqing University, Chongqing, P.R. China) for
his assistance with image formatting.
Abbreviations:
|
A549/CDDP
|
cisplatin-resistant A549 cells
|
|
Bcl-2
|
B-cell lymphoma-2
|
|
Bcl-xL
|
B-cell lymphoma-xL
|
|
NF-κB
|
nuclear factor κB
|
|
IκBα
|
inhibitor α of NF-κB
|
|
LOH
|
loss of heterozygosity
|
|
miRNA/miR
|
microRNA
|
|
NSCLC
|
non-small cell lung cancer
|
|
RBM24
|
RNA binding motif protein 24
|
|
3′-UTR
|
3′-untranslated region
|
References
|
1
|
Schwartz AG and Cote ML: Epidemiology of
lung cancer. Adv Exp Med Biol. 893:21–41. 2016. View Article : Google Scholar
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2020. CA Cancer J Clin. 70:7–30. 2020. View Article : Google Scholar
|
|
3
|
Miller KD, Nogueira L, Mariotto AB,
Rowland JH, Yabroff KR, Alfano CM, Jemal A, Kramer JL and Siegel
RL: Cancer treatment and survivorship statistics, 2019. CA Cancer J
Clin. 69:363–385. 2019. View Article : Google Scholar
|
|
4
|
Xiong Y, Huang BY and Yin JY:
Pharmacogenomics of platinum- based chemotherapy in non- small cell
lung cancer: Focusing on DNA repair systems. Med Oncol. 34:482017.
View Article : Google Scholar
|
|
5
|
Denisenko TV, Budkevich IN and Zhivotovsky
B: Cell death- based treatment of lung adenocarcinoma. Cell Death
Dis. 9:1172018. View Article : Google Scholar
|
|
6
|
Zhao J, Xu T, Wang F, Cai W and Chen L:
miR- 493-5p suppresses hepatocellular carcinoma cell proliferation
through targeting GP73. Biomed Pharmacother. 90:744–751. 2017.
View Article : Google Scholar
|
|
7
|
Yu X and Li Z: MicroRNA expression and its
implications for diagnosis and therapy of tongue squamous cell
carcinoma. J Cell Mol Med. 20:10–16. 2016. View Article : Google Scholar
|
|
8
|
Gao Y, Luo LH, Li S and Yang C: miR- 17
inhibitor suppressed osteosarcoma tumor growth and metastasis via
increasing PTEN expression. Biochem Biophys Res Commun.
444:230–234. 2014. View Article : Google Scholar
|
|
9
|
Yue LU, Xiang JY, Sun P, Yao YS, Sun ZN,
Liu XP, Wang HB, Shen Z and Yao RY: Relationship between HSP70 and
ERBB2 expression in breast cancer cell lines regarding drug
resistance. Anticancer Res. 36:1243–1249. 2016.
|
|
10
|
Yu X, Li Z, Chan MT and Wu WK: microRNA
deregulation in keloids: An opportunity for clinical intervention?
Cell Prolif. 48:626–630. 2015. View Article : Google Scholar
|
|
11
|
Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong
FY, Bo T, He J, Hua RX, Hu WD, et al: miR- 134: A human cancer
suppressor? Mol Ther Nucleic Acids. 6:140–149. 2017. View Article : Google Scholar
|
|
12
|
Cao Q, Liu F, Ji K, Liu N, He Y, Zhang W
and Wang L: MicroRNA- 381 inhibits the metastasis of gastric cancer
by targeting TMEM16A expression. J Exp Clin Cancer Res. 36:292017.
View Article : Google Scholar
|
|
13
|
Yu X, Li Z, Yu J, Chan MTV and Wu WKK:
MicroRNAs predict and modulate responses to chemotherapy in
colorectal cancer. Cell Prolif. 48:503–510. 2015. View Article : Google Scholar
|
|
14
|
Pei K, Zhu JJ, Wang CE, Xie QL and Guo JY:
MicroRNA- 185-5p modulates chemosensitivity of human non- small
cell lung cancer to cisplatin via targeting ABCC1. Eur Rev Med
Pharmacol Sci. 20:4697–4704. 2016.
|
|
15
|
Yu S, Qin X, Chen T, Zhou L, Xu X and Feng
J: MicroRNA- 106b-5p regulates cisplatin chemosensitivity by
targeting polycystic kidney disease- 2 in non- small- cell lung
cancer. Anticancer Drugs. 28:852–860. 2017. View Article : Google Scholar
|
|
16
|
Gang H, Pan J, Ye Z, Fang B and Wei C:
Overexpression of miR- 216b sensitizes NSCLC cells to cisplatin-
induced apoptosis by targeting c- Jun. Oncotarget. 8:104206–104215.
2017. View Article : Google Scholar
|
|
17
|
Azarbarzin S, Feizi MAH, Safaralizadeh R,
Kazemzadeh M and Fateh A: The value of miR- 383, an intronic miRNA,
as a diagnostic and prognostic biomarker in intestinal- type
gastric cancer. Biochem Genet. 55:244–252. 2017. View Article : Google Scholar
|
|
18
|
Bucay N, Sekhon K, Yang T, Majid S,
Shahryari V, Hsieh C, Mitsui Y, Deng G, Tabatabai ZL, Yamamura S,
et al: MicroRNA- 383 located in frequently deleted chromosomal
locus 8p22 regulates CD44 in prostate cancer. Oncogene.
36:2667–2679. 2017. View Article : Google Scholar
|
|
19
|
Shang Y, Zang A, Li J, Jia Y, Li X, Zhang
L, Huo R, Yang J, Feng J, Ge K, et al: MicroRNA- 383 is a tumor
suppressor and potential prognostic biomarker in human non- small
cell lung caner. Biomed Pharmacother. 83:1175–1181. 2016.
View Article : Google Scholar
|
|
20
|
Ma H, Liu B, Wang S and Liu J: MicroRNA-
383 is a tumor suppressor in human lung cancer by targeting
endothelial PAS domain- containing protein 1. Cell Biochem Funct.
34:613–619. 2016. View Article : Google Scholar
|
|
21
|
Tan W, Liao Y, Qiu Y, Liu H, Tan D, Wu T,
Tang M, Zhang S and Wang H: miRNA 146a promotes chemotherapy
resistance in lung cancer cells by targeting DNA damage inducible
transcript 3 (CHOP). Cancer Lett. 428:55–68. 2018. View Article : Google Scholar
|
|
22
|
Wagenaar SS: New WHO- classification of
lung and pleural tumors. Ned Tijdschr Geneeskd. 143:984–990.
1999.In Dutch.
|
|
23
|
Tsim S, O'Dowd CA, Milroy R and Davidson
S: Staging of non- small cell lung cancer (NSCLC): A review. Respir
Med. 104:1767–1774. 2010. View Article : Google Scholar
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real- time quantitative PCR and
the 2(- Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering PUMILIO proteins. Cell.
164:69–80. 2016. View Article : Google Scholar
|
|
26
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals: Guide for the Care and Use of Laboratory Animals. 8th
edition. National Academies Press; Washington DC: 2011
|
|
27
|
Shanker M, Willcutts D, Roth JA and Ramesh
R: Drug resistance in lung cancer. Lung Cancer (Auckl). 1:23–36.
2010.
|
|
28
|
Panwar B, Omenn GS and Guan Y: miRmine: A
database of human miRNA expression profiles. Bioinformatics.
33:1554–1560. 2017.
|
|
29
|
Ludwig N, Leidinger P, Becker K, Backes C,
Fehlmann T, Pallasch C, Rheinheimer S, Meder B, Stähler C, Meese E
and Keller A: Distribution of miRNA expression across human
tissues. Nucleic Acids Res. 44:3865–3877. 2016. View Article : Google Scholar
|
|
30
|
Wan P, Chi X, Du Q, Luo J, Cui X, Dong K,
Bing Y, Heres C and Geller DA: miR-383 promotes cholangiocarcinoma
cell proliferation, migration, and invasion through targeting IRF1.
J Cell Biochem. 119:9720–9729. 2018. View Article : Google Scholar
|
|
31
|
Dong P, Xiong Y, Yu J, Chen L, Tao T, Yi
S, Hanley SJB, Yue J, Watari H and Sakuragi N: Correction: Control
of PD- L1 expression by miR- 140/142/340/383 and oncogenic
activation of the OCT4- miR- 18a pathway in cervical cancer.
Oncogene. 38:39722019. View Article : Google Scholar
|
|
32
|
Zhao S, Gao X, Zang S, Li Y, Feng X and
Yuan X: MicroRNA- 383-5p acts as a prognostic marker and inhibitor
of cell proliferation in lung adenocarcinoma by cancerous inhibitor
of protein phosphatase 2A. Oncol Lett. 14:3573–3579. 2017.
View Article : Google Scholar
|
|
33
|
Mu X, Wu H, Liu J, Hu X, Wu H, Chen L, Liu
W, Luo S and Zhao Y: Long noncoding RNA TMPO- AS1 promotes lung
adenocarcinoma progression and is negatively regulated by miR-
383-5p. Biomed Pharmacother. 125:1099892020. View Article : Google Scholar
|
|
34
|
Piovani G, Savio G, Traversa M, Pilotta A,
De Petro G, Barlati S and Magri C: De novo 1Mb interstitial
deletion of 8p22 in a patient with slight mental retardation and
speech delay. Mol Cytogenet. 7:252014. View Article : Google Scholar
|
|
35
|
Chi C, Murphy LC and Hu P: Recurrent copy
number alterations in young women with breast cancer. Oncotarget.
9:11541–11558. 2018. View Article : Google Scholar
|
|
36
|
Tsubaki M, Ogawa N, Takeda T, Sakamoto K,
Shimaoka H, Fujita A, Itoh T, Imano M, Satou T and Nishida S:
Dimethyl fumarate induces apoptosis of hematopoietic tumor cells
via inhibition of NF-κB nuclear translocation and down- regulation
of Bcl- xL and XIAP. Biomed Pharmacother. 68:999–1005. 2014.
View Article : Google Scholar
|
|
37
|
Viatour P, Merville MP, Bours V and
Chariot A: Phosphorylation of NF- kappaB and IkappaB proteins:
Implications in cancer and inflammation. Trends Biochem Sci.
30:43–52. 2005. View Article : Google Scholar
|
|
38
|
Chen Y, Wang D, Peng H, Chen X, Han X, Yu
J, Wang W, Liang L, Liu Z, Zheng Y, et al: Epigenetically
upregulated oncoprotein PLCE1 drives esophageal carcinoma
angiogenesis and proliferation via activating the PI- PLCε- NF-κB
signaling pathway and VEGF- C/Bcl- 2 expression. Mol Cancer.
18:12019. View Article : Google Scholar
|
|
39
|
Bravo-Cuellar A, Hernández- Flores G,
Lerma- Díaz JM, Domínguez- Rodríguez JR, Jave-Suárez LF, De Célis-
Carrillo R, Aguilar- Lemarroy A, Gómez-Lomeli P and Ortiz-Lazareno
PC: Pentoxifylline and the proteasome inhibitor MG132 induce
apoptosis in human leukemia U937 cells through a decrease in the
expression of Bcl- 2 and Bcl- XL and phosphorylation of p65. J
Biomed Sci. 20:132013. View Article : Google Scholar
|
|
40
|
Yao Y, Yang B, Cao H, Zhao K and Chen X:
RBM24 stabilizes hepatitis B virus pregenomic RNA but inhibits core
protein translation by targeting the terminal redundancy sequence.
Emerg Microbes Infect. 7:862018. View Article : Google Scholar
|
|
41
|
Hua WF, Zhong Q, Xia TL, Chen Q, Zhang MY,
Zhou AJ, Tu ZW, Qu C, Li MZ, Xia YF, et al: RBM24 suppresses cancer
progression by upregulating miR- 25 to target MALAT1 in
nasopharyngeal carcinoma. Cell Death Dis. 7:e23522016. View Article : Google Scholar
|
|
42
|
Xu E, Zhang J, Zhang M, Jiang Y, Cho SJ
and Chen X: RNA- binding protein RBM24 regulates p63 expression via
mRNA stability. Mol Cancer Res. 12:359–369. 2014. View Article : Google Scholar
|
|
43
|
Zhang Q, Wang YZ, Zhang W, Chen X, Wang J,
Chen J and Luo W: Involvement of cold inducible RNA- binding
protein in severe hypoxia- induced growth arrest of neural stem
cells in vitro. Mol Neurobiol. 54:2143–2153. 2016. View Article : Google Scholar
|