Introduction
Colorectal cancer (CRC) is the third most commonly
diagnosed cancer type worldwide, with ~2.000.000 new cases in 2020
(1). It is the third most common
cancer type in males, and the second most common cancer type in
females after breast cancer (2).
CRC is the second most common cause of cancer-associated
mortalities worldwide (2). Thanks
to the advances in treatments, including endoscopic and surgical
excision, radiotherapy, targeted therapy, immunotherapy and
chemotherapy, CRC-based therapies have improved, and promising
results have been reported (1,3).
5-Fluorouracil (5-FU) is the first-line
chemotherapeutic agent for CRC treatment (4). As an anti-metabolite drug that has
been widely used in cancer treatments and clinical studies, 5-FU
induces cytotoxicity, mainly by interfering with essential
biosynthetic processes through the inhibition of thymidylate
synthase or by leading to errors in base pairing during RNA and DNA
synthesis (5). Furthermore, 5-FU
exposure induces DNA damage, thereby promoting apoptosis in cancer
cells (6). However, despite its
several advantages in cancer management, a critical limitation to
the clinical application of 5-FU is the development of
chemotherapeutic drug resistance in cancer cells. Resistance to
5-FU has been proposed to develop through drug uptake inhibition,
target alterations, elevated DNA repair abilities and resistance to
apoptosis (5). Numerous genes have
been detected to be involved in the development of resistance to
5-FU in cancer cells (7). Thus,
targeting key genes or signaling pathways involved in drug
resistance has the potential as a therapeutic strategy to overcome
5-FU resistance during cancer treatment (5,7).
Methyltransferase-like 3 (METTL3) is a key
N6-methyladenosine (m6A)
methyltransferase enzyme that catalyzes the m6A
modification of its target transcripts. Due to its role in
m6A modification, METTL3 regulates various biological
processes, including the cell cycle, cell proliferation, apoptosis,
migration invasion and differentiation, and inflammatory response
(8). Previous studies have
demonstrated the functions of METTL3 and m6A
modification in tumorigenesis and cancer progression in different
cancer types, including CRC (9,10).
METTL3 was consistently overexpressed in CRC and facilitated CRC
progression (11). Additionally,
higher METTL3 expression in CRC was associated with poor prognosis.
However, the function and mechanism of METTL3 chemotherapeutic
efficacy and drug resistance in CRC remain unexplored.
In the present study, a series molecular,
biochemical and cellular experiments were performed to explore the
function of METTL3 with regard to chemotherapeutic response and
drug resistance in CRC. The present study demonstrated the function
of METTL3 in regulating cell proliferation, chemotherapeutic
response and 5-FU resistance in CRC cells. It was observed that
silencing METTL3 suppressed cell proliferation, enhanced
chemotherapeutic response and overcame 5-FU resistance in CRC cells
through the regulation of RAD51 associated Protein 1 (RAD51AP1),
which is a key homologous recombination (HR) repair protein.
Therefore, the present results suggest the potential use of METTL3
as a therapeutic target for CRC treatment, either alone or as a
combined treatment strategy.
Materials and methods
Plasmid construction
For the knockdown of METTL3 and RAD51AP1, silencing
plasmids containing small hairpin RNA (shRNA) METTL3, 5′-CTC AGT
GGA TCT GTT GTG ATA-3′ (12); and
RAD51AP1, 5′-GCA CTA GCT TTA TCA GTG A-3′ sequences (13) were constructed based on the
psilencer3.0-H1. The sequence of the shRNA negative control (sh-CN)
was 5′-GTC AGG CTA TCG CGT ATC G-3′. The METTL3-overexpressing
plasmid was obtained from WZ Biosciences, Inc. ORF of human METTL3
was constructed in pENTER vector, with C terminal Flag and His tag.
All plasmids were verified through sequencing. For plasmid
transfection, cells were seeded at a density of 3×105
cells/well in 6-well plates. Subsequently, 24 h later, the cells
were transfected with sh-CN, sh-METTL3, or shRAD51AP1 plasmid (2.0
µg per well) using HighGene transfection reagent (cat. no.
RM09014; Abclonal Biotech Co., Ltd.) at 37°C, following the
manufacturer's instructions. Finally, 48 h after transfection, the
cells were collected for reverse transcription-quantitative PCR
(RT-qPCR) analysis, western blotting assay, or subsequent
experiments assay.
METTL3-knockdown (KD) and METTL3-overexpressing
lentiviruses were constructed by Corues Biotechnology Company,
using the oligonucleotide METTL3-shRNA and METTL3-overexpressing
vector (pPENTER), respectively. For lentiviral particle production,
293T cells (cat. no. GNHu17; Shanghai Institute of Cell Biology,
Chinese Academy of Sciences) were seeded 24 h before lentiviral
infection. Lentiviruses were packaged by transfecting the
aforementioned vectors (sh-METTL3, sh-CN transfer vector
(psilencer3.0-H1), control pENTER or METTL3-overexpressing vector),
psPAX2, and pMD2.G under a 4.5:2:1 ratio into 293T cells. The
collected viral mix was purified by ultracentrifugation at 30,000 ×
g for 2 h at 4°C. Purified viral particles were used to infect
target cells with polybrene (Sigma-Aldrich-Merck KGaA). After a
48-h incubation at 37°C, infected cells were screened with 1 mg/ml
of puromycin (InvivoGen, Inc.) for two weeks.
Cell lines and cell culture
The human CRC HCT-8 cell line was obtained from the
Shanghai Institute of Cell Biology, Chinese Academy of Sciences
(cat. no. TCHu 18). In addition, human normal epithelial cell line,
NCM460, was obtained from INCELL Corporation LLC, and cultured in
M3 media supplemented with 10% FBS, 100 U/ml penicillin and 100
mg/ml streptomycin at 37°C in a 95% humidified atmosphere with 5%
CO2. HCT-8 cells were cultured in the recommended
RMPI-1640 medium (cat. no. KGM31800N-500; Nanjing KeyGEN BioTECH
Co, Ltd.) and F12 medium (cat. no. KGM21700-500; Nanjing KeyGEN
BioTECH Co., Ltd.), supplemented with 10% fetal bovine serum (FBS;
Invigentech, Inc.), 100 U/ml penicillin, and 100 U/ml streptomycin,
following incubation at 5% CO2 and a temperature of
37°C. Subsequently, 5-FU-resistant HCT-8 cells (HCT-8R) were
selected and established from HCT-8 cells as previously described
(7). For METTL3-overexpressing or
METTL3-KD HCT-8 and HCT-8R stable cells, the cells were infected
with specific lentivirus vectors (MOI, 10) for 48 h and then
selected with puromycin (1 mg/ml) for two weeks. These HCT-8 cells
were then treated using stepwise dose-dependent 5-FU (product no.
F6627; Sigma-Aldrich-Merck KGaA) concentrations (0, 0.01, 0.1, 0.5,
2, and 10 µM) over 5 months. Subsequently, the acquired
drug-resistant HCT-8R cells were cultured and stabilized in a
10-µM 5-FU-containing medium.
Alkaline comet assay
Alkaline comet assay analysis was performed with a
commercial Comet Assay Kit (cat. no. KGA240-100; Nanjing KeyGEN
BioTECH Co., Ltd.) following the protocol as previously described
(14). Briefly, 1×105
cells/ml were mixed with molten LM agarose at 37°C at a ratio of
1:10 (vol/vol) and pipetted onto a COMET slide. The slides were
placed for 10 min in the dark at 4°C and were immersed in
pre-chilled lysis solution. The slides were then removed from the
lysis buffer, washed in Tris-HCl buffer and transferred to a
horizontal electrophoresis chamber. Voltage (20 V) was applied for
20 min. After being washed in distilled water, the slides were
immersed in 70% ethanol for 5 min and allowed to air dry. Slides
were stained with propidium iodide (PI), in the dark, at room
temperature for 10 min, and then analyzed by fluorescence
microscopy (Nikon 80I; Nikon Corporation). A total of 70-90 cells
were evaluated in each sample using the COMET Assay Software
Project (CASP software version casp1.2.3b1; CaspLab).
Wound healing assay
The wound healing assay was performed as previously
described with some modifications (15). Various cells (control, METTL3-KD or
METTL3-overexpressing HCT-8) were seeded in a 6-well plate at a
concentration of 5×105 cells/well until they reached 80%
of confluence. The cultured cells were scratched with a
200-µl sterile pipette tip in a line. The cells were then
washed three times with PBS and cultured with RMPI-1640 medium
without FBS for another 12 h or 24 h. The scratches were imaged
using a fluorescence microscope at a magnification of ×200 (Nikon
80I; Nikon Corporation) and analyzed by ImageJ software (1.52v;
National Institutes of Health).
Apoptosis assay through flow
cytometry
For the apoptosis assay, cells were first treated
with different concentrations of 5-FU close to the IC50
value. For the HCT-8 cells, the concentrations were 2 and 4
µg/ml, and for HCT-8R cells, 60 µg/ml was used. The
cells were then trypsinized, washed, and resuspended in 1 ml of
phosphate-buffered saline (PBS) containing 5% FBS. Subsequently,
the cells were washed again twice with ice-cold PBS, then fixed in
3 ml ice-cold ethanol. Following centrifugation at 3,000 × g for 3
min at room temperature, the cells were collected and stained with
both Annexin V-FITC and PI (Annexin V-FITC/PI Double Staining
Apoptosis Detection Kit; cat. no. KGA106; Nanjing KeyGEN BioTECH
Co., Ltd. at room temperature for 15 min according to the
manufacturer's instructions. Subsequently, apoptosis was analyzed
using BD FACSverse flow cytometer and FACSSuite Clinical software
(BD Biosciences).
Colony-forming assay
The colony-forming assay was conducted using HCT-8
stable cell lines in which METTL3 was knocked down or overexpressed
as previously described (14).
Briefly, ~500 cells were plated in a 6-well plate and incubated for
approximately 15 days at 37°C. For HCT-8R cell colony formation
assay, the cultured cells were treated with 0, 10 and 20
µg/ml 5-FU, which were lower than the IC50 value.
Colonies in each well were fixed with 4% paraformaldehyde, and then
washed using PBS and stained with 0.05% crystal violet at room
temperature for 30 min. Stained plates were washed again and dried
before scoring the colonies. Images of the colony-forming units
(>50 cells) were captured and recorded using a fluorescence
microscope (Nikon 80I) and Adobe Photoshop CC 2017 software.
Immunofluorescence staining
For immunofluorescence assays, HCT-8 stable cell
lines were cultured in 6-well plates containing an acid-treated
cover slide per well overnight to make these cells adhere to the
slide. Cells (~5×104 cells per slide) on cover slides
were then washed with PBS thrice. Subsequently, these cells were
fixed in PBS-containing 4% formaldehyde at room temperature for 30
min, then washed with PBS again. Following permeabilization with
0.1% Triton X-100 for 10 min, the cells were blocked with 3% BSA at
room temperature for 1 h, and then incubated with a primary
antibody against phosphorylated (p)-γ-H2AX (1:300; product no.
9718S; Cell Signaling Technology, Inc.), KI67 (1:300; cat. no.
27309-1-AP; ProteinTech Group, Inc.) and 53BP1 (1:300; product code
ab175933; Abcam) overnight at 4°C. Finally, washing with PBST was
performed twice, after which these cells were incubated with
fluorescent secondary antibodies at room temperature for 90 min.
The secondary antibodies were as follows: Alexa Fluor™ 594 Tyramide
SuperBoost™ Kit, goat anti-mouse IgG (1:200 dilution; cat. no.
B40942), and Alexa Fluor™ 488 Tyramide SuperBoost™ Kit, goat
anti-rabbit IgG (1:200 dilution; cat. no. B40943; both from
Invitrogen; Thermo Fisher Scientific, Inc.). The cells were then
stained with DAPI at 37°C for 15 min. The mounted slides were
visualized under a fluorescence microscope (Nikon 80I; Nikon
Corporation).
m6A dot blot assays
Briefly, total RNA was isolated using the TRIzol
(Sigma-Aldrich; Merck KGaA) kit, following the manufacturer's
instructions. RNA concentration was measured using a NanoDrop
ultrafine Ultraviolet spectrophotometer (ND-1000; Thermo Fisher
Scientific, Inc.). A total of 2 µg RNA samples were then
spotted onto a positively-charged nylon membrane (GE Healthcare;
Cytiva), and air-dried at room temperature for 5 min. The membranes
were subsequently UV cross-linked, washed with PBST containing 0.1%
Tween-20 (cat. no. T104863; Aladdin) for 5 min, blocked with 5%
nonfat milk in TBST containing 0.1 % Tween-20 at room temperature
for 90 min, and incubated with an anti-m6A antibody
(1:1,000 dilution; cat. no. A17924; ABclonal Biotech Co., Ltd.)
overnight at 4°C. Subsequently, the HRP-conjugated anti-rabbit IgG
secondary antibody (1:10,000 dilution; cat. no. AS014; ABclonal
Biotech Co., Ltd.) was added to the membrane with gentle shaking
for 1.5 h at room temperature, followed by development with
enhanced chemiluminescence (cat. no. FD8020; Hangzhou Fude
Biological Technology Co., Ltd.). A total of 1% methylene blue
staining agent (cat. no. M196501; Aladdin) was used at room
temperature for 1 h for mRNA loading.
Drug sensitivity assay
Approximately 3,000 METTL3 stabilized knockdown or
overexpression of HCT-8/HCT-8R cells were seeded per well for at
least four parallel experiments. Cells were treated with multiple
dilutions of 5-FU for 24 h. The cell viability of 5-FU-treated
cells was then assayed using the Cell Counting Kit-8 (CCK-8) assay
(cat. no. IV08-1000T; Invigentech, Inc.) according to the
manufacturer's instructions. Using CCK-8 treated for 90 min, the
cell survival curves of METTL3-KD HCT-8 or HCT-8R cells were
determined with various concentrations of 5-FU treatment (METTL3-KD
HCT-8 cells: 0, 0.5, 1, 1.5, 3 µg/ml; METTL3-KD HCT-8R
cells: 0, 10, 20, 40, 60 µg/ml) due to their different
sensitivity. In addition, HCT-8 and HCT-8R cells were treated with
0, 3, 6, 9, 12 µg/ml 5-Fu. In subsequent experiments,
RAD51AP1-KD HCT-8 cells were treated with different concentrations
of 5-Fu (0, 0.5, 1, 3, 12 µg/ml). Data were expressed as the
percentage of growth relative to the untreated controls.
Reverse transcription-quantitative PCR
(RT-qPCR) assay
RNA extraction and real-time PCR Total RNA from
cells were isolated by TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed into cDNA with HiScript
II Q RT SuperMix kit (cat. no. R222-01; Vazyme Biotech Co., Ltd.).
RT-qPCR was performed using SYBR Green (cat. no. Q341-02; Vazyme
Biotech Co., Ltd.) and operated on an ABI StepOne PCR system. Each
reaction was repeated three times. Gene relative expression was
determined relative to β-actin in each reaction. The qPCR cycling
conditions were as follows: Denaturation at 95°C for 5 min,
followed by 40 cycles of denaturation at 95°C for 15 sec, primer
annealing and extension at 60°C for 1 min. The Cq value was
determined from the same threshold fluorescence value for the
analyzed genes. The relative quantities of mRNA were calculated
using the comparative quantification cycle method
(2−ΔΔCq) (16). The
primers for RT-qPCR are listed in Table SI.
Western blotting
Total cellular protein was extracted using RIPA
lysis buffer (P0013B; Beyotime Institute of Biotechnology) which
contained 50 mM Tris (pH 7.4), 150 mM NaCl, 1% Triton X-100, 1%
sodium deoxycholate and 0.1% SDS. Total protein concentration was
determined by bicinchoninic acid analysis (Hangzhou Fude Biological
Technology Co., Ltd), and then proteins in lysed samples (30
µg per well) were separated on a 10% acrylamide gel. Protein
was then transferred to polyvinylidene difluoride membranes.
Following 5% skimmed milk blocking for 90 min at room temperature,
the membranes were incubated overnight at 4°C with a primary
antibody. Subsequently, the membranes were incubated with a
secondary antibody for 90 min at room temperature following
extensive TBST (containing 0.1% Tween-20) washing. The blots were
filmed by enhanced chemiluminescence (cat. no. FD8020; Hangzhou
Fude Biological Technology Co., Ltd.) and developed via Tanon 4500
luminescent imaging workstation (Tanon Science and Technology Co.,
Ltd.). The following antibodies were used for western blotting:
Anti-METTL3 (1:1,000 dilution; cat. no. 15073-1-AP; ProteinTech
Group, Inc.), anti-RAD51AP1 (1:1,000 dilution; cat. no. 11255-1-AP;
ProteinTech Group, Inc.), anti-Flag (1:1,000 dilution; cat. no.
AP0007MH; Bioworld Technology, Inc.), anti-p-γH2AX (1:1,000
dilution; product no. 9718S; Cell Signaling Technology, Inc.),
anti-H2AX (1:1,000 dilution; cat. no. A11540; ABclonal Biotech Co.,
Ltd.), anti-caspase-3 (1:1,000 dilution; cat. no. 19677-1-AP;
ProteinTech Group, Inc.), anti-Bax (1:1,000 dilution; cat. no.
A7626; ABclonal Biotech Co., Ltd.) and anti-TBB5 (1:1,000 dilution;
cat. no. AM1031A; Abgent, Inc.). The HRP-conjugated secondary
antibody was obtained from ABclonal Biotech Co., Ltd. (1:10,000
dilution; cat. no. AS014; anti-rabbit IgG; and cat. no. AS003;
anti-mouse IgG). The densitometry of western blot protein bands was
analyzed by ImageJ software (1.52v; National Institutes of
Health).
Bioinformatics analysis
The m6A modification in gene exonic 5′
untranslated region (5′UTR), or 3′UTR mRNA regions was screened by
the m6A-Atlas, which is a comprehensive knowledgebase
for unraveling the m6A epitranscriptome (http://180.208.58.19/m6A-Atlas/). The
translational studies on the expression of METTL3 and RAD51AP1 were
conducted by LinkedOmics, which is a publicly available portal that
includes multi-omics data from all 32 TCGA Cancer types and 10
Clinical Proteomics Tumor Analysis Consortium (CPTAC) cancer
cohorts (http://www.linkedomics.org/)
(17).
Statistical analyses
Each experiment was performed at least three
independent times. Statistical analysis was conducted using
GraphPad Prism 8.0 (GraphPad Software, Inc.). Data were analyzed
for the assumption of normal distribution using the
Kolmogorov-Smirnov test and homogeneity test of variance using
Bartlett's test. The statistical evaluation was performed by
unpaired two-tailed Student's t-test between two groups (sh-METTL3
vs. sh-CN, METTL3 vs. Con, HCT-8 vs. HCT-8R and sh-RAD51AP1 vs.
sh-CN), and one-way analysis of variance (ANOVA) for relative
m6A densities and relative protein levels among multiple
groups. Tukey's test was employed to assess the post hoc
comparisons for variables that were different among multiple
groups. For correlation analysis between the expression of METTL3
and RAD51AP1 observed in CRC from the LinkedOmics database
(http://www.linkedomics.org), Pearson's
correlation test was used. P<0.05 was considered to indicate
statistically significant differences.
Results
METTL3 promotes the proliferation of CRC
cells
Elevated expression of METTL3, which has been
detected in human CRC tissues, was associated with poor prognosis
in patients with CRC (11,18). Therefore, to investigate the
regulatory effect of METTL3 on CRC, a METTL3-KD stable cell line
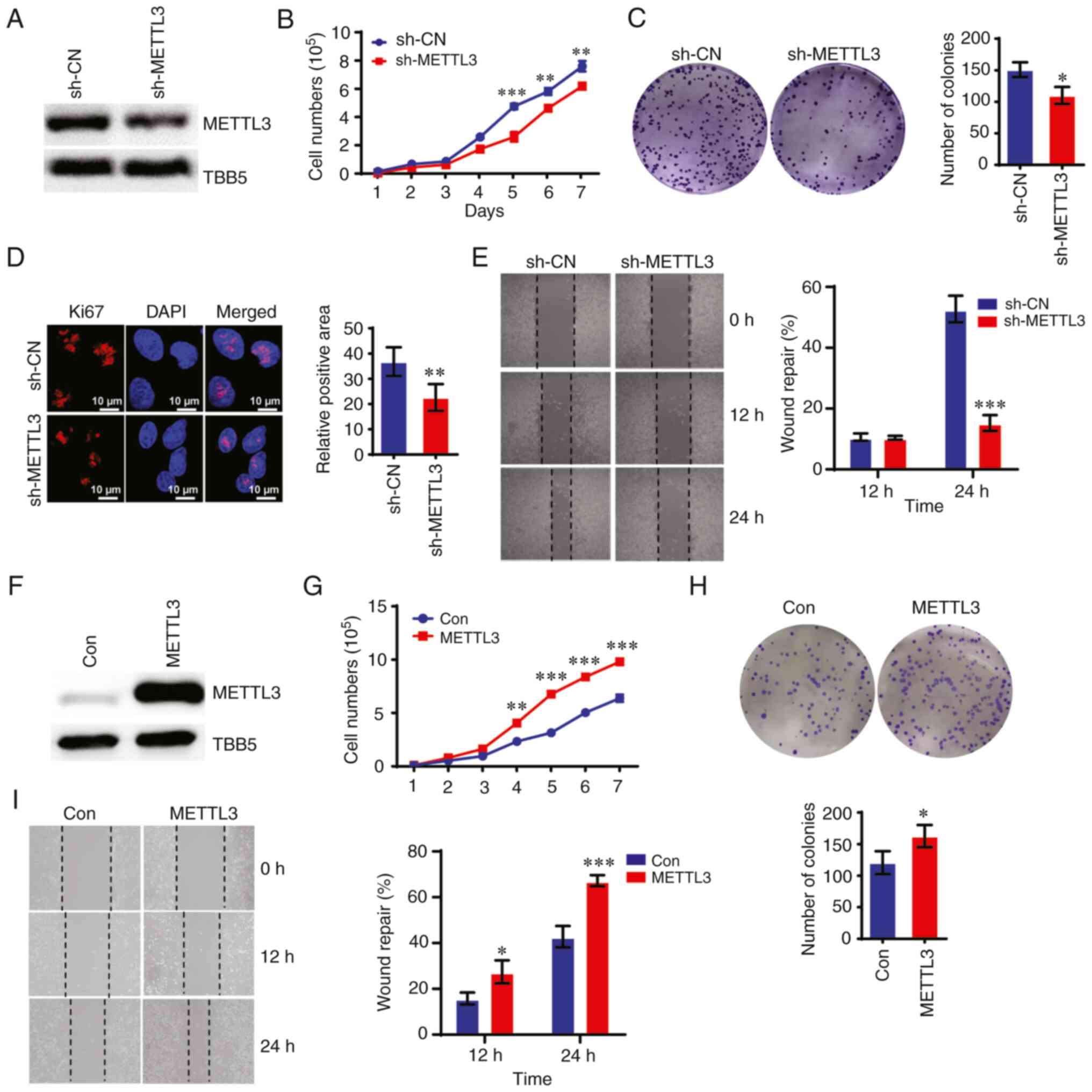
was constructed with METTL3 shRNA lentivirus in CRC HCT-8 cells.
Western blotting and RT-qPCR demonstrated that METTL3 expression
was significantly downregulated in sh-METTL3 cells compared with
that in control HCT-8 cells (Figs.
1A and S1A). Downregulating
METTL3 markedly suppressed the proliferation of HCT-8 cells, as
measured using CCK-8 and colony formation assays (Fig. 1B and C). Ki67 expression was
downregulated in sh-METTL3 cells compared with that in control
cells (Fig. 1D). The wound healing
assay showed that METTL3 depletion significantly impaired the
migration of HCT-8 cells compared with that of control cells
(Fig. 1E). By contrast,
overexpressing METTL3 promoted the proliferation, colony formation
ability and migration ability of CRC cells (Figs. 1F-I and S1B). As expected, knockdown of METTL3
reduced global m6A levels in HCT-8 cells, whereas
overexpression of METTL3 increased m6A levels in these
cells compared with control cells (Fig. S1C and D).
Knockdown of METTL3 sensitizes CRC cells
to 5-FU
5-FU is currently the most commonly used
chemotherapeutic agent to improve survival in patients with CRC
(19). Therefore, the present
study investigated whether knocking down METTL3 would affect 5-FU
treatment in CRC. The cell survival assay demonstrated that
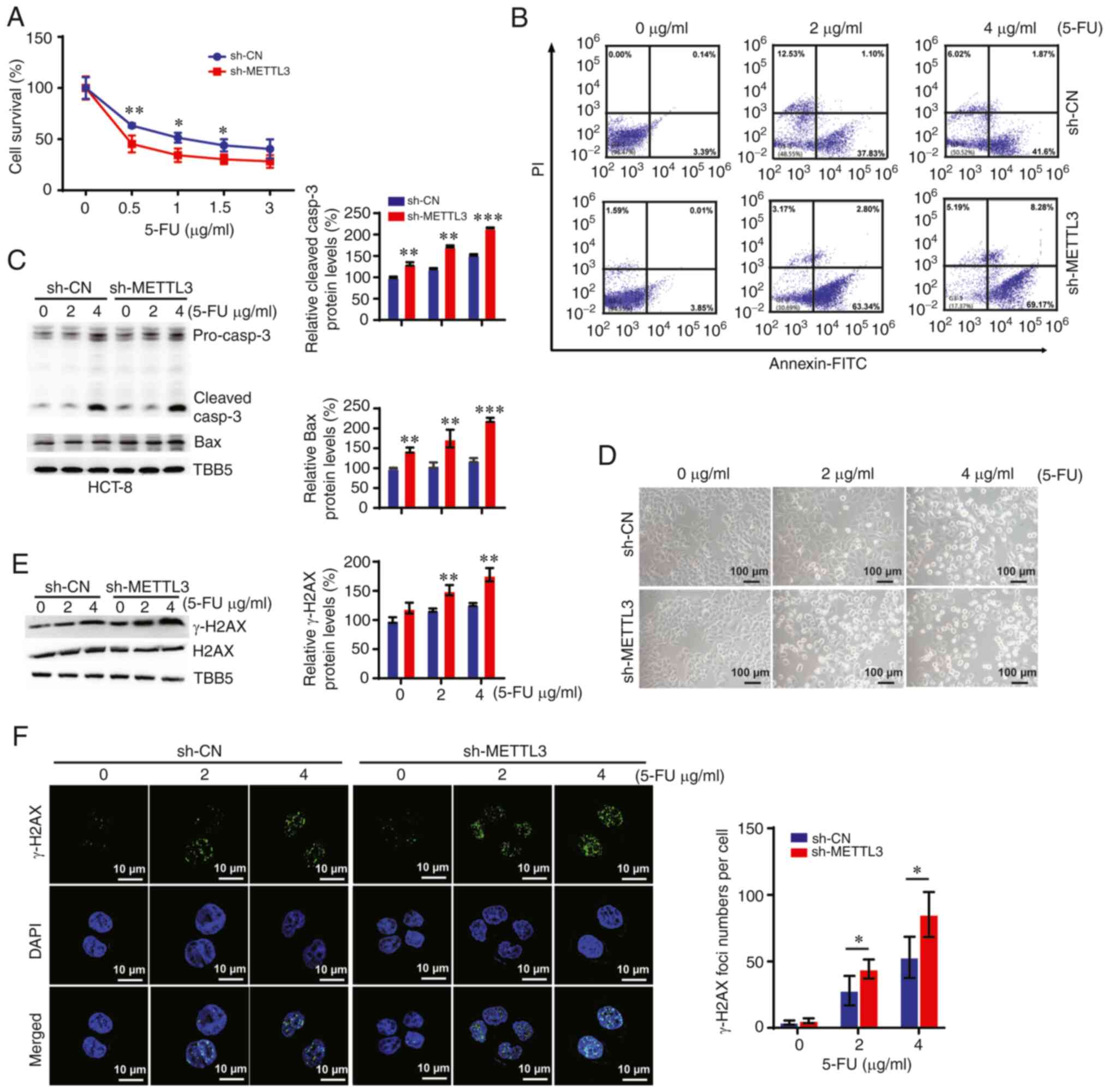
knocking down METTL3 sensitized CRC cells to 5-FU (Fig. 2A). Furthermore, downregulation of
METTL3 promoted apoptosis in a concentration-dependent manner
[quadrant (Q)2 indicates late apoptosis and Q4 indicates early
apoptosis] in HCT-8 cells (Fig.
2B). In addition, the expression of pro-apoptotic proteins
(caspase-3 and Bax) was upregulated in 5-FU-treated cells, which
was enhanced by METTL3 depletion (Fig.
2C). These results were verified using morphological analysis,
which showed that knockdown of METTL3 promoted 5-FU
treatment-induced cell death in CRC cells with higher apoptosis as
revealed by several smaller rounded cell fragments and cell loss
(Fig. 2D).
A previous study showed that 5-FU could cause DNA
damage, specifically, double-strand (and single-strand) breaks
during the S phase of the cell cycle due to the misincorporation of
5-fluorodeoxyuridine triphosphate (5-FdUTP) into DNA (20). Therefore, the present study
investigated whether METTL3 modified the levels of DNA damage in
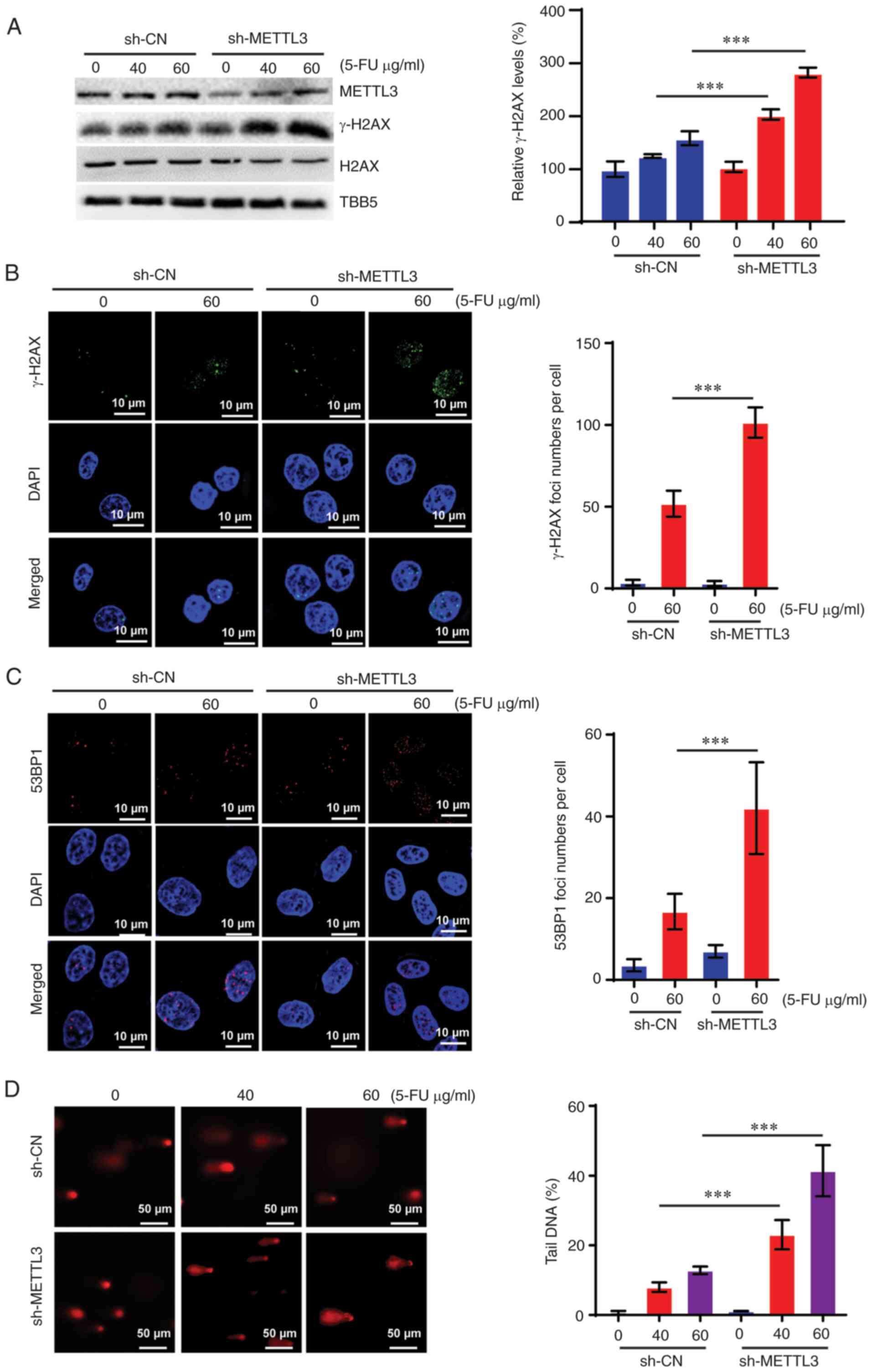
HCT-8 cells following 5-FU treatment. As shown in Fig. 2E, downregulation of METTL3 enhanced
the expression level of phosphorylated histone γ-H2AX (an
established marker of DNA damage), which was induced by 5-FU.
Furthermore, the present study detected γ-H2AX- and p53-binding
Protein 1 (53BP1)-positive foci in control and sh-METTL3
5-FU-treated HCT-8 cells via immunofluorescence assays. The present
data revealed that 5-FU treatment dose-dependently increased the
foci numbers of γ-H2AX and 53BP1, which were augmented upon
knockdown of METTL3 (Figs. 2F and
S2). By contrast, overexpression
of METTL3 enhanced DNA repair and decreased DNA damage in HCT-8
cells, which further impeded apoptosis and triggered the resistance
of HCT-8 cells to 5-FU (Fig.
S2B-E).
METTL3 and m6A levels are
upregulated in 5-FU resistant CRC cells
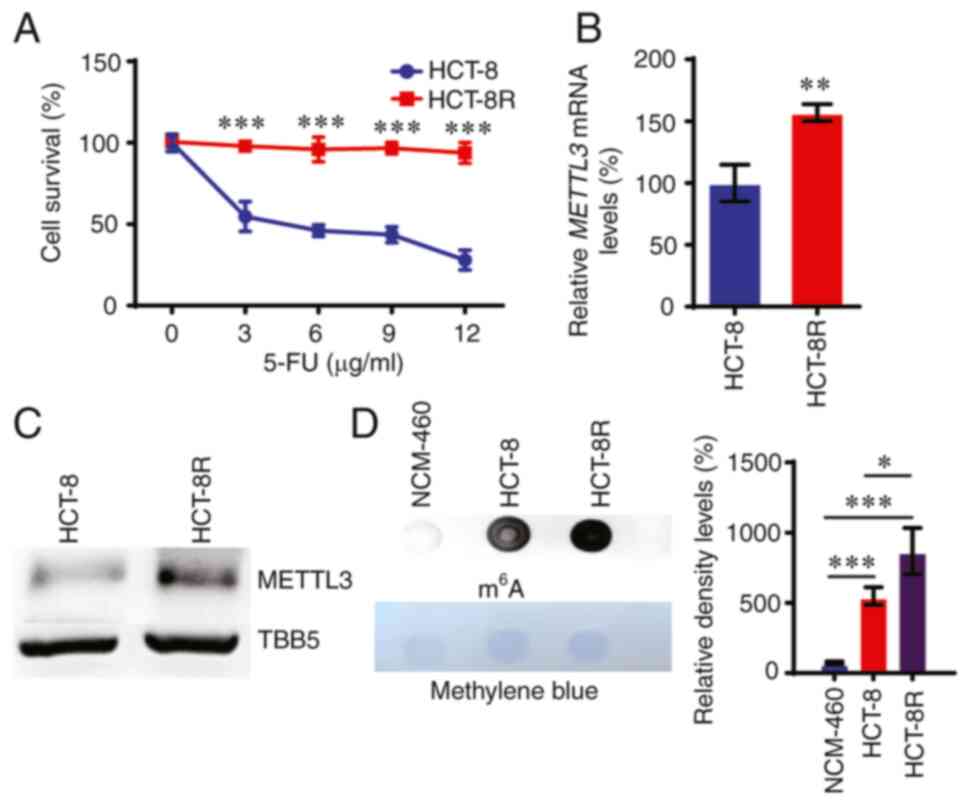
To investigate whether METTL3 contributes to
5-FU-induced resistance in CRC, 5-FU-resistant cells were obtained
using parental HCT-8 CRC cells treated with increasing
concentrations of 5-FU, as previously described (7). A cell survival assay was then used to
confirm the acquired resistance to 5-FU by HCT-8R cells (Fig. 3A). Next, the METTL3 and RNA
m6A expression levels were assessed in these HCT-8R
cells. As shown in Fig. 3B and C,
both the mRNA and protein levels of METTL3 were upregulated in
HCT-8R cells compared with those in control HCT-8 cells. Dot blot
assay showed that the total m6A RNA levels were elevated
in HCT-8R cells compared with those in HCT-8 control cells
(Fig. 3D).
Depletion of METTL3 overcomes
5-FU-induced resistance in CRC cells
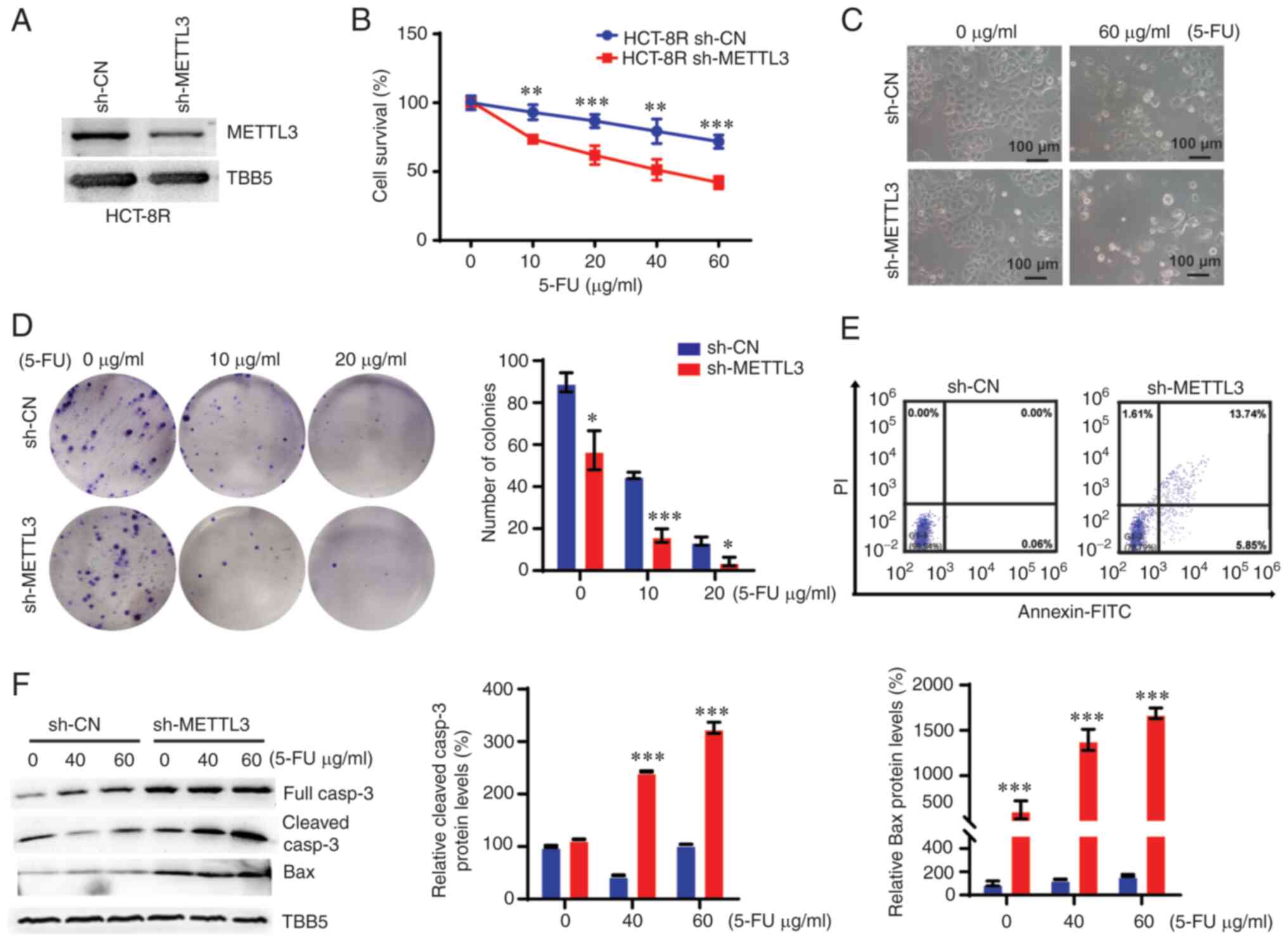
Since METTL3 was abundantly overexpressed in HCT-8R
cells, it was hypothesized that manipulating METTL3 could alter the
5-FU-induced resistance in CRC. To confirm this hypothesis, shRNA
was used to knock down the expression of METTL3 in HCT-8R cells
(Figs. 4A and S3). A cell survival assay and
morphological analysis demonstrated that downregulation of METTL3
overcame 5-FU-induced resistance in HCT-8R cells (Fig. 4B and C). A colony formation assay
also confirmed this role of METTL3 on 5-FU-induced resistance in
CRC cells (Fig. 4D). Furthermore,
the effect of METTL3 on 5-FU-induced apoptosis in HCT-8R cells was
detected. As expected, treatment with 10 µg/ml 5-FU for 12 h
did not promote apoptosis in 5-FU-resistant HCT-8R cells, whereas
the same treatment induced ~20% apoptosis in METTL3-KD HCT-8R cells
(Fig. 4E). Knocking down METTL3
increased the caspase-3 and Bax expression levels in HCT-8R cells
(Fig. 4F).
Knockdown of METTL3 enhances 5-FU-induced
DNA damage in HCT-8R cells
Elevated DNA repair abilities lead to drug
resistance, which severely limits the efficacy of chemotherapeutic
drugs in cancer cells (14,21).
The role of DNA repair in 5-FU response and resistance has been
observed in numerous studies (5,6). The
present study further investigated whether knocking down METTL3
would modulate DNA damage accumulation in HCT-8R cells. The present
data showed that the expression levels of γ-H2AX were significantly
increased in METTL3-KD HCT-8R cells compared with those in HCT-8R
control cells treated with 40 or 60 µg/ml 5-FU (Fig. 5A). Consistently, an increasing
number of positive γ-H2AX and 53BP1 nuclei foci were detected in
METTL3-KD HCT-8R cells following 5-FU treatment (Fig. 5B and C). Comet assays also revealed
that METTL3-KD HCT-8R cells had higher levels of spontaneous DNA
strand break than HCT-8R control cells upon 5-FU treatment
(Fig. 5D). Therefore, these
results indicated that downregulating METTL3 increased DNA damage
accumulation in 5-FU-resistant CRC cells treated with 5-FU.
METTL3 regulates RAD51AP1 expression in
CRC cells
A previous study by our research group identified
that the protein levels of essential enzymes participating in the
base excision repair (BER) pathway did not show significant changes
in HCT-8R cells versus HCT-8 cells (7). Several studies have verified the
effect of METTL3 on HR repair (10,22,23).
Thus, the present study applied a comprehensive strategy to
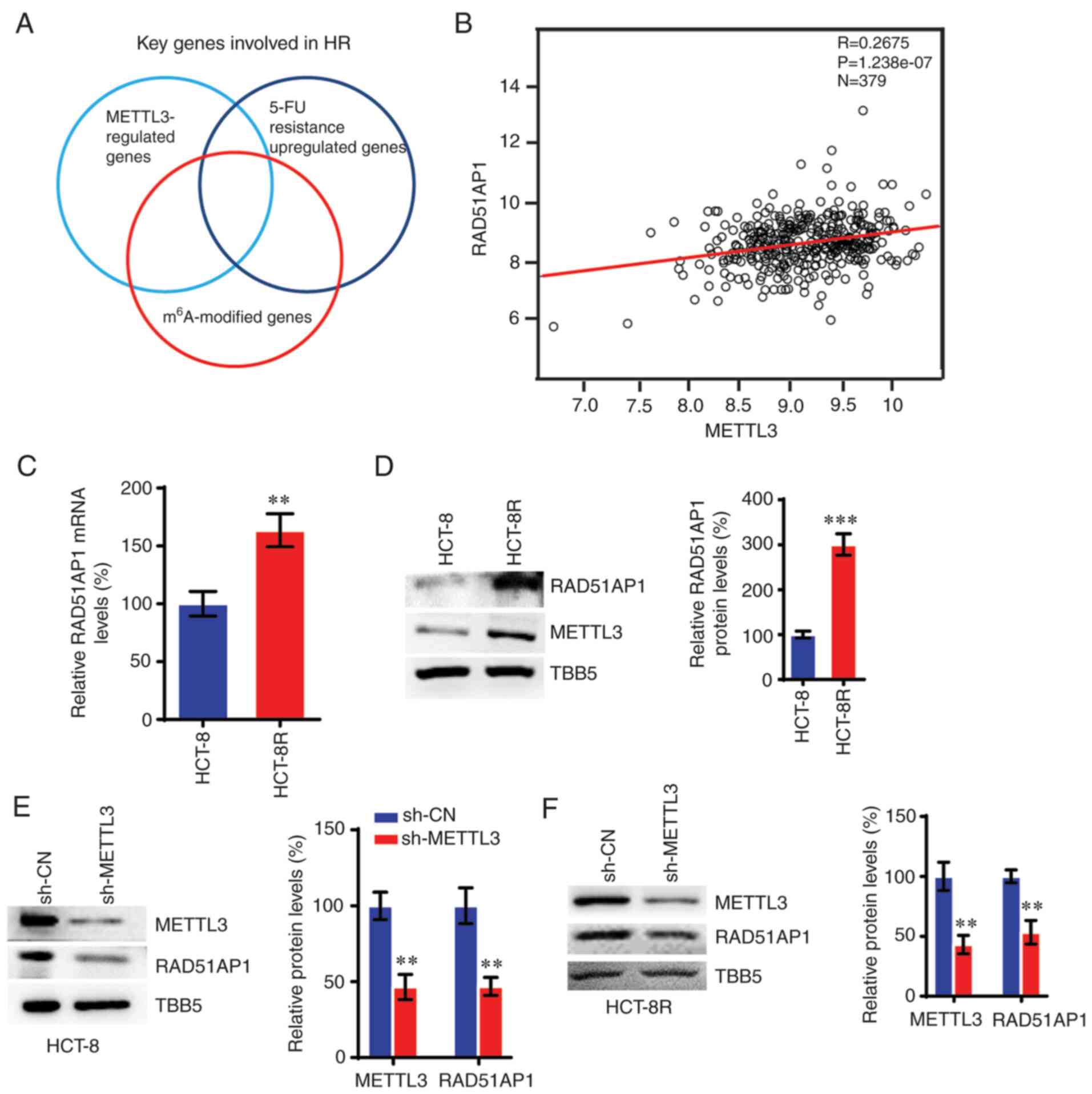
identify potential genes involved in the METTL3-mediated response
to 5-FU in HCT-8R cells (Fig. 6A).
RT-qPCR was used to assess key genes involved in HR in order to
detect which genes were modulated in METTL3-KD or 5-FU-resistant
cells versus control cells (Fig.
S4). Subsequently, HR genes that were up- or down-regulated in
METTL3-KD and 5-FU-resistant cells were further screened for
modification of m6A in their exonic, 5′UTR or 3′UTR mRNA
regions using the m6A-Atlas, which is a comprehensive
knowledgebase for unraveling the m6A epitranscriptome
(24). This screening strategy led
us to focus on RAD51AP1, which plays a key role in HR repair by
interacting with and enhancing the recombinase activity of RAD51
(25,26). A previous study showed that
RAD51AP1 promoted the invasion of RNA transcripts into donor DNA at
the double standard break sites in transcribed regions, and
stimulated HR repair through the formation of DNA-RNA (DR)-loops
(27). A positive correlation
between the expression of METTL3 and RAD51AP1 was observed in CRC
cells (Fig. 6B), which was
consistent with the results of METTL3. The expression levels of
RAD51AP1 increased in 5-FU-resistant HCT-8R cells compared with
those in control HCT-8 cells (Fig. 6C
and D). Knockdown of METTL3 also resulted in decreased RAD51AP1
expression in both HCT-8 and HCT-8R cells (Fig. 6E and F).
Downregulation of RAD51AP1 promotes DNA
damage and sensitizes CRC cells to 5-FU
Considering that the expression pattern of RAD51AP1
is regulated by METTL3, and that METTL3 is involved in
5-FU-dependent responses, the present study investigated whether
RAD51AP1 regulated 5-FU-dependent chemotherapy responses. A cell
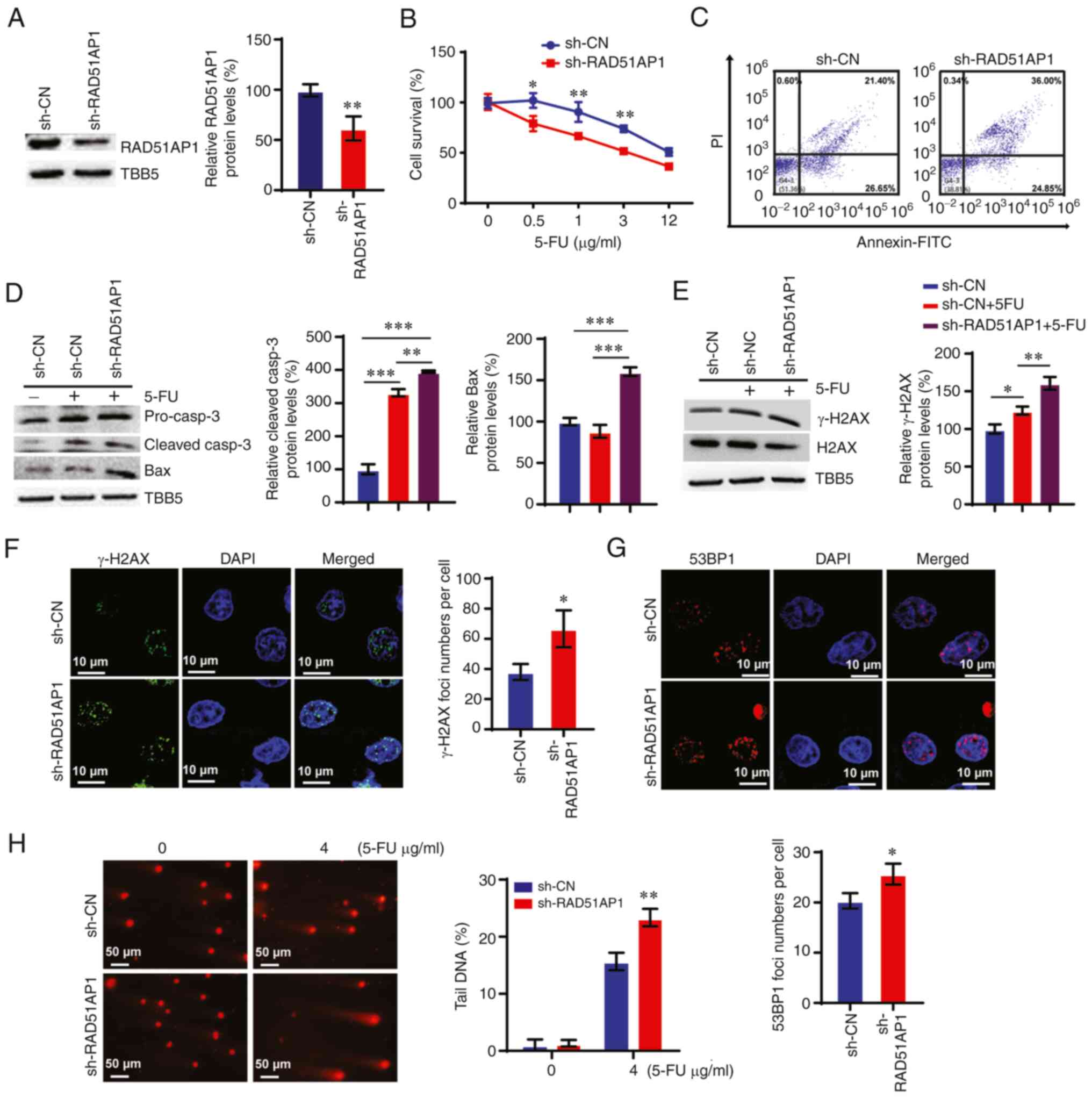
survival assay demonstrated that knockdown of RAD51AP1
significantly sensitized HCT-8 cells to 5-FU (Fig. 7A and B). Furthermore, knocking down
RAD51AP1 promoted apoptosis in HCT-8 cells with 5-FU treatment
(Fig. 7C). Consistently, the
protein levels of caspase-3 and Bax were increased by RAD51AP1
downregulation (Fig. 7D). The
present study further detected DNA damage in RAD51AP1-KD cells. The
data revealed that knocking down RAD51AP1 enhanced 5-FU-induced
γ-H2AX expression levels in HCT-8 cells (Fig. 7E). The number of positive nuclei
foci of both γ-H2AX and 53BP1 increased in RAD51AP1-KD HCT-8 cells
subjected to 5-FU treatment (Fig. 7F
and G). Furthermore, comet assays revealed higher levels of
spontaneous DNA strand breaks in RAD51AP1-KD cells compared with
those in control cells following 5-FU treatment (Fig. 7H).
Knockdown of RAD51AP1 overcomes
5-FU-resistance in CRC cells
As aforementioned, RAD51AP1 was elevated in HCT-8R
cells. Therefore, the present study further analyzed whether
RAD51AP1 contributed to 5-FU resistance in CRC cells. The data
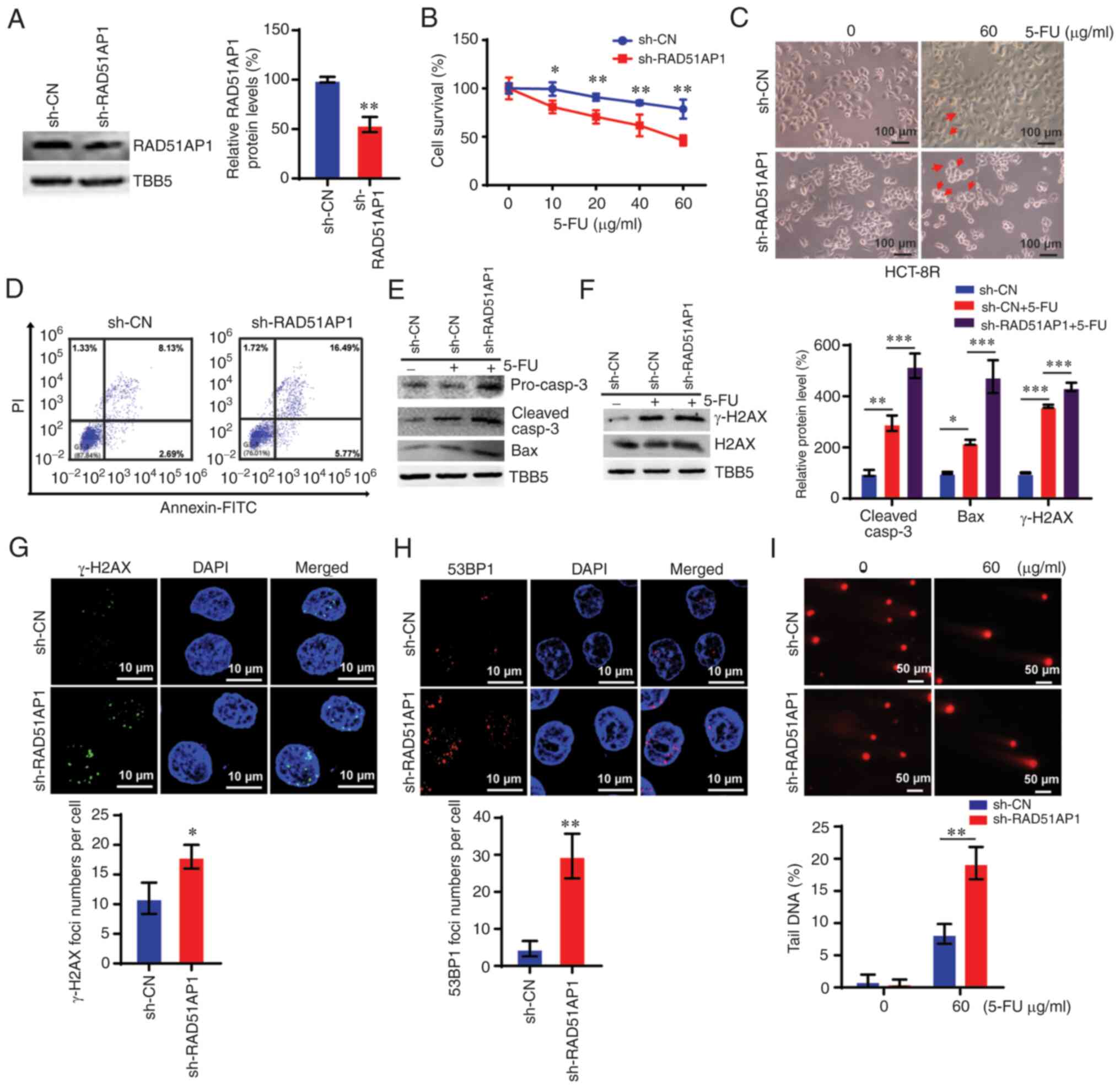
showed that knocking down RAD51AP1 suppressed 5-FU resistance in
HCT-8R cells (Fig. 8A and B). Cell
morphology analysis of RAD51AP1-KD HCT-8R cells verified the above
data, which showed more apoptotic bodies and cell loss in the
RAD51AP1-KD cells with 60 µg/ml 5-FU treatment (Fig. 8C). Flow cytometric analysis also
demonstrated increased apoptosis in RAD51AP1-KD HCT-8R cells
(Fig. 8D). By downregulating
RAD51AP1 in HCT-8R cells, the expression of caspase-3 and Bax was
upregulated (Fig. 8E). Knocking
down RAD51AP1 markedly enhanced the DNA damage levels in HCT-8R
cells subjected to 5-FU treatments (Fig. 8F-I). These data indicated that
knocking down RAD51AP1 overcame 5-FU resistance in CRC cells by
enhancing DNA damage accumulation and promoting cell apoptosis.
Discussion
5-FU is one of the most commonly used CRC
chemotherapy drugs in the clinics, which can effectively kill
gastrointestinal tumor cells or inhibit their proliferation.
However, its long-term application can induce drug resistance of
tumor cells, leading to failures in clinical treatments (23). Thus patients with CRC urgently need
new therapeutic targets to improve the clinical efficacy of 5-FU.
Presently, the mechanism of chemotherapy resistance in CRC remains
unclear; however, the current study observed that the METTL3 and
m6A levels were highly expressed in 5-FU-resistant CRC
cells. Knockdown of METTL3 sensitized CRC cells to 5-FU and
promoted 5-FU treatment-induced DNA damage in CRC cells, leading to
cell death. Downregulation of METTL3 overcame 5-FU-induced
resistance in CRC cells. Additionally, it was also found that
METTL3 was positively correlated with the expression of the
RAD51AP1 protein in CRC cells. Similarly, downregulation of
RAD51AP1 significantly sensitized CRC cells to 5-FU, and overcame
5-FU-resistantance in CRC cells by enhancing DNA damage
accumulation and promoting cell apoptosis. The above experimental
results confirmed the effect of METTL3 on 5-FU resistance in CRC
cells.
Increasing evidence has shown that m6A
and its key methyl transferase METTL3 influence the occurrence and
development of tumors. Li et al (28) observed that METTL3 served as an
oncogene in CRC. As reported, METTL3 maintained the expression of
SRY-box 2 (SOX2) in CRC cells through a m6A-insulin like
growth factor 2 mRNA binding protein 2-dependent mechanism, which
was consistent with the present results. CSC is a tumor cell line
with strong carcinogenic and metastatic potential, which is
considered a reason for chemotherapy resistance. CSC-labeled SOX2
has been confirmed to have carcinogenic effects. Therefore,
inhibiting METTL3 can reduce the expression of its surface antigen,
thereby enhancing the chemotherapy response of CRC in vivo
and in vitro. Enhanced chemotherapy-based responses reduce
the frequency of stem cells, which effectively stops the recurrence
and metastasis of malignant tumors. By contrast, Deng et al
(29) reported the tumor
suppressing effect of METTL3 during CRC cell proliferation,
migration, and invasion through the p38/ERK pathway. The authors
proposed that the main reason why METTL3 played a dual role in
cancer regulation was the difference in targeting pathways and
cancer heterogeneity. The results of the present study showed that
although METTL3 promoted the proliferation of CRC cells, the
proliferative and metastatic abilities of HCT-8 cells were
significantly reduced upon downregulation of METTL3. The present
study further demonstrated that inhibition of METTL3 sensitized
HCT-8 cells to 5-FU and overcome 5-FU resistance in the CRC cells
through regulation of RAD51AP1 expression. Although knockdown of
METTL3 could not reverse the drug resistance completely, the
present data at least demonstrated that suppression of METTL3 could
partially overcome the 5-FU-resistance in HCT-8R cells. These data
indicated that METTL3 may combine with other genes together to
confer drug resistance in CRC. However, understanding the role and
mechanism of METTL3 in colon cancer requires further research,
including the collection of additional clinical tissue specimens
for analysis.
In summary, the present results showed that METTL3
was an oncogene in colon cancer, which regulated the proliferation
and metastasis of colon cancer. However, since METTL3 and its
regulated HR repair protein, RAD51AP1, are highly expressed in
5-FU-resistant CRC cells, silencing them enhanced 5-FU-induced DNA
damage in HCT-8R cells, improved chemotherapy response and overcame
5-FU resistance. Therefore, METTL3 could be used as an important
indicator to predict the level of differentiation and metastasis of
CRC cells, thereby providing new insights and targets for future
treatments and drug developmental processes. However, the
regulatory role of METTL3 in the occurrence and development of CRC
as well as its molecular mechanism should be further
investigated.
Supplementary Data
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
ZH, MF, YZ and ML designed and supervised the study,
and wrote the manuscript. ML, MX, EL and ZZ performed the
experiments and analyzed the data. ML, YT, ZG, EL and MF provided
technical and material support. ML, MX and ZZ confirm the
authenticity of all the raw data. ML, YT, YZ, ZG, MF and ZH
conceived, wrote, and edited the manuscript as well as contributed
to funding acquisition. All authors have read and agreed to the
published version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National Natural Science
Foundation of China (grant no. 32171407), the Priority Academic
Program Development of Jiangsu Higher Education Institutions and
Nanjing Medical Science and Technique Development Foundation (grant
no. QRX17187) and the Postgraduate Research & Practice
Innovation Program of Jiangsu Province.
Abbreviations:
|
METTL3
|
methyltransferase-like 3
|
|
FBS
|
fetal bovine serum
|
|
HR
|
homologous recombination
|
|
PBS
|
phosphate-buffered saline
|
|
PI
|
propidium iodide
|
References
|
1
|
Xi Y and Xu P: Global colorectal cancer
burden in 2020 and projections to 2040. Transl Oncol.
14:1011742021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global cancer statistics 2020:
GLOBOCAN estimates of incidence and mortality worldwide for 36
cancers in 185 countries. CA Cancer J Clin. 71:209–249. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Biller LH and Schrag D: Diagnosis and
treatment of metastatic colorectal cancer: A review. Jama.
325:669–685. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xie YH, Chen YX and Fang JY: Comprehensive
review of targeted therapy for colorectal cancer. Signal Transduct
Target Ther. 5:222020. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sethy C and Kundu CN: 5-Fluorouracil
(5-FU) resistance and the new strategy to enhance the sensitivity
against cancer: Implication of DNA repair inhibition. Biomed
Pharmacother. 137:1112852021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wyatt MD and Wilson DM III: Participation
of DNA repair in the response to 5-fluorouracil. Cell Mol Life Sci.
66:788–799. 2009. View Article : Google Scholar :
|
|
7
|
He L, Zhu H, Zhou S, Wu T, Wu H, Yang H,
Mao H, SekharKathera C, Janardhan A, Edick AM, et al: Wnt pathway
is involved in 5-FU drug resistance of colorectal cancer cells. Exp
Mol Med. 50:1–12. 2018. View Article : Google Scholar
|
|
8
|
Liu S, Zhuo L, Wang J, Zhang Q, Li Q, Li
G, Yan L, Jin T, Pan T, Sui X, et al: METTL3 plays multiple
functions in biological processes. Am J Cancer Res. 10:1631–1646.
2020.PubMed/NCBI
|
|
9
|
Zeng C, Huang W, Li Y and Weng H: Roles of
METTL3 in cancer: Mechanisms and therapeutic targeting. J Hematol
Oncol. 13:1172020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li E, Xia M, Du Y, Long K, Ji F, Pan F, He
L, Hu Z and Guo Z: METTL3 promotes homologous recombination repair
and modulates chemotherapeutic response in breast cancer by
regulating the EGF/RAD51 axis. Elife. 11:e752312022. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen H, Gao S, Liu W, Wong CC, Wu J, Wu J,
Liu D, Gou H, Kang W, Zhai J, et al: RNA N(6)-Methyladenosine
Methyltransferase METTL3 facilitates colorectal cancer by
activating the m(6)A-GLUT1-mTORC1 axis and is a therapeutic target.
Gastroenterology. 160:1284–1300.e16. 2021. View Article : Google Scholar
|
|
12
|
Xiang Y, Laurent B, Hsu CH, Nachtergaele
S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al: RNA m(6)A
methylation regulates the ultraviolet-induced DNA damage response.
Nature. 543:573–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wiese C, Dray E, Groesser T, San Filippo
J, Shi I, Collins DW, Tsai MS, Williams GJ, Rydberg B, Sung P and
Schild D: Promotion of homologous recombination and genomic
stability by RAD51AP1 via RAD51 recombinase enhancement. Mol Cell.
28:482–490. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu X, Liu R, Wang M, Kumar AK, Pan F, He
L, Hu Z and Guo Z: MicroRNA-140 impedes DNA repair by targeting
FEN1 and enhances chemotherapeutic response in breast cancer.
Oncogene. 39:234–247. 2020. View Article : Google Scholar
|
|
15
|
Grada A, Otero-Vinas M, Prieto-Castrillo
F, Obagi Z and Falanga V: Research techniques made simple: Analysis
of collective cell migration using the wound healing assay. J
Invest Dermatol. 137:e11–e16. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(T)(-Delta Delta C) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Vasaikar SV, Straub P, Wang J and Zhang B:
LinkedOmics: Analyzing multi-omics data within and across 32 cancer
types. Nucleic Acids Res. 46:D956–D963. 2018. View Article : Google Scholar :
|
|
18
|
Peng W, Li J, Chen R, Gu Q, Yang P, Qian
W, Ji D, Wang Q, Zhang Z, Tang J and Sun Y: Upregulated METTL3
promotes metastasis of colorectal cancer via miR-1246/SPRED2/MAPK
signaling pathway. J Exp Clin Cancer Res. 38:3932019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
McQuade RM, Stojanovska V, Bornstein JC
and Nurgali K: Colorectal cancer chemotherapy: The evolution of
treatment and new approaches. Curr Med Chem. 24:1537–1557. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
De Angelis PM, Svendsrud DH, Kravik KL and
Stokke T: Cellular response to 5-fluorouracil (5-FU) in
5-FU-resistant colon cancer cell lines during treatment and
recovery. Mol Cancer. 5:202006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li L, Kumar AK, Hu Z and Guo Z: Small
molecule inhibitors targeting key proteins in the DNA damage
response for cancer therapy. Curr Med Chem. 28:963–985. 2021.
View Article : Google Scholar
|
|
22
|
Zhang C, Chen L, Peng D, Jiang A, He Y,
Zeng Y, Xie C, Zhou H, Luo X, Liu H, et al: METTL3 and
N6-Methyladenosine promote homologous recombination-mediated repair
of DSBs by modulating DNA-RNA hybrid accumulation. Mol Cell.
79:425–442.e7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu D, Horton JR, Yang J, Hajian T, Vedadi
M, Sagum CA, Bedford MT, Blumenthal RM, Zhang X and Cheng X: Human
MettL3-MettL14 RNA adenine methyltransferase complex is active on
double-stranded DNA containing lesions. Nucleic Acids Res.
49:11629–11642. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tang Y, Chen K, Song B, Ma J, Wu X, Xu Q,
Wei Z, Su J, Liu G, Rong R, et al: m6A-Atlas: A comprehensive
knowledgebase for unraveling the N6-methyladenosine (m6A)
epitranscriptome. Nucleic Acids Res. 49:D134–D143. 2021. View Article : Google Scholar :
|
|
25
|
Dunlop MH, Dray E, Zhao WX, San Filippo J,
Tsai MS, Leung SG, Schild D, Wiese C and Sung P: Mechanistic
insights into RAD51-associated Protein 1 (RAD51AP1) action in
homologous DNA repair. J Biol Chem. 287:12343–12347. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pires E, Sung P and Wiese C: Role of
RAD51AP1 in homologous recombination DNA repair and carcinogenesis.
DNA Repair (Amst). 59:76–81. 2017. View Article : Google Scholar
|
|
27
|
Ouyang J, Yadav T, Zhang JM, Yang H,
Rheinbay E, Guo H, Haber DA, Lan L and Zou L: RNA transcripts
stimulate homologous recombination by forming DR-loops. Nature.
594:283–288. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li T, Hu PS, Zuo Z, Lin JF, Li X, Wu QN,
Chen ZH, Zeng ZL, Wang F, Zheng J, et al: METTL3 facilitates tumor
progression via an m(6)A-IGF2BP2-dependent mechanism in colorectal
carcinoma. Mol Cancer. 18:1122019. View Article : Google Scholar
|
|
29
|
Deng R, Cheng YK, Ye SB, Zhang J, Huang R,
Li P, Liu H, Deng Q, Wu X, Lan P and Deng Y: m(6)A
methyltransferase METTL3 suppresses colorectal cancer proliferation
and migration through p38/ERK pathways. Onco Targets Ther.
12:4391–4402. 2019. View Article : Google Scholar :
|