Introduction
Proviral integration of Moloney virus 2 (PIM2)
belongs to a family of serine/threonine kinases consisting of three
members: PIM1, PIM2 and PIM3 (1).
PIMs are highly conserved proteins (2), and play roles in various cellular
processes, such as cell growth, proliferation, apoptosis and
regulation of signal transduction (3). PIM2 is overexpressed in hematopoietic
cancers (3), and is considered to
be an oncogene involved in multiple signaling pathways (4). PIM2 can phosphorylate and activate
substrates that control cancer progression and tumorigenesis
(4). PIM2 inhibits apoptosis by
phosphorylating downstream targets, including eukaryotic
translation initiation factor 4E-binding protein 1 (4EBP1),
tuberous sclerosis complex 2 (TSC2), and BCL2-associated agonist of
cell death (BAD) (5-8). In hematological malignancies, PIM2
promotes oncogenic progression as a pro-survival factor (9).
PIM2 has been demonstrated to be a possible
therapeutic target in hematological malignancies (7,10-15),
as the inhibition of PIM2 induces apoptosis and inhibits cancer
cell proliferation in vitro and in vivo (7). Ongoing clinical trials for PIM2
inhibitors (https://clinicaltrials.gov/; Identifier: NCT01456689
and NCT01588548) (16) have
yielded discouraging results with not sufficient efficacy or
dose-limiting toxicity in hematological malignancies, and a phase 1
trial (NCT03715504) commended in April, 2019 in solid tumors.
Previous studies reported that PIM2 also plays important roles in
tumor progression, epithelial to mesenchymal transition,
chemotherapy resistance (17), and
aerobic glycolysis (18) in solid
tumors. However, the importance and the strategy for targeting PIM2
in solid cancers has not been fully elucidated. Recently, it has
been shown that a novel pan-PIM inhibitor, JP11646, demonstrates
anticancer activity in multiple myeloma (7). The present study examined the utility
of targeting PIM2 in multiple solid cancers, and investigated the
antitumor efficacy and mechanisms of action of JP11646.
Materials and methods
Bioinformatics analysis
PIM2 expression was compared between tumors and
normal tissues in The Cancer Genome Atlas (TCGA) dataset. Clinical
and gene expression data from RNA sequence were downloaded through
the cBioportal (19,20) website. The expression levels of
PIMs were compared among the cell lines using the CCLE dataset
(21,22).
Cells, cell culture and reagents
Human cancer cell lines, including pharyngeal
carcinoma FaDu (HTB-43), ovarian cancer SK-OV-3 (HTB-77), breast
cancer MDA-MB-231 (CRM-HTB-26) and BT549 (HTB-122), prostate
adenocarcinoma PC-3 (CRL-1435), liver cancer HepG2 (HB-8065),
pancreatic ductal adenocarcinoma (PDAC) MIAPaCa-2 (CRM-CRL-1420)
and PANC-1 (CRL-1469), colorectal cancer HT-29 (HTB-38) and DLD-1
(CCL-221), and non-small cell lung cancer (NSCLC) H1650 (CRL-5883),
H661 (HTB-183), H460 (HTB-177) and A549 (CCL-185) cell lines were
obtained from ATCC. The FaDu, SK-OV-3, MDA-MB-231, PC-3, HepG2,
MIAPaCa-2, PANC-1 and HT29 cells were cultured in Dulbecco's
modified Eagle's medium (DMEM) (Gibco; Thermo Fisher Scientific,
Inc.) with 10% fetal bovine serum (FBS) (Gibco; Thermo Fisher
Scientific, Inc.); the BT549, DLD-1, H1650, H661, H460 and A549
cells were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific,
Inc.) with 10% FBS in a humidified incubator at 37°C in 5%
CO2. All cell lines were used within 20 passages after
revival. All cell lines were shown to be mycoplasma-free using the
PlasmoTest kit (InVivoGen, Inc.). The MDA-MB-231 cells stably
overexpressing PIM2 and controls were generated by the transfection
of either 2 µg/ml PIM2-p3xFlag-CMV-14, which was kindly
provided by Dr Jeremy Don (Bar-Ilan University, Ramat Gan, Israel)
or empty vector (MilliporeSigma) using Lipofectamine LTX (Thermo
Fisher Scientific, Inc.) into MDA-MB-231 cells. PIM2 stably
overexpressing and control clones were selected with G418 for >2
weeks and used in further experiments. Human PIM2 specific siRNA
(sense, 5′-ACC UUC UUC CCG ACC CUC Att-3′ and antisense, 5′-UGA GGG
UCG GGA AGA AGG Utt-3′) or non-targeting siRNA (#4390843, Thermo
Fisher Scientific, Inc.) was transfected into the BT549 cells at a
final concentration of 30 nM using DharmaFECT 1 Transfection Agent
(GE Dharmacon), according to the manufacturer's instructions. The
cells transfected with the siRNA were collected at 72 h following
transfection and changes in protein expression were examined using
western blot analysis.
The novel pan-PIM inhibitor, JP11646, was obtained
from Jasco Pharmaceuticals, LLC and its structure is illustrated in
Fig. S1 (23,24).
Another pan-PIM inhibitor (AZD1208) (25) and the proteasome inhibitor,
bortezomib, were obtained from Selleck Chemicals.
Drug sensitivity assay in in vitro
A total of 5,000 cells were seeded per well of a
96-well plate and incubated overnight at 37°C. Several
concentrations of JP11646, ranging from 0.005 to 10 µM, were
added to each well, and the cells were incubated for 72 h at 37°C.
Cell proliferation was measured using the CellTiter 96®
AQueous One Solution Cell Proliferation Assay kit (Promega
Corporation), according to the manufacturer's protocol. The
half-maximal growth inhibitory (GI50) values were
calculated using non-linear regression.
Western blot analysis
The breast cancer cell lines, MDA-MB-231 and BT549,
were cultured with either the vehicle (H2O) or JP11646
(100 or 200 nM) for 24 h. The BT549 cells were treated with 1
µM AZD1208 for 24 h. The MDA-MB-231 cells were pre-treated
with 1 nM Bortezomib for 12 h, and then treated with 200 nM JP11646
for 24 h. Cells were lysed using RIPA lysis buffer (Cell Signaling
Technology, Inc.), and lysates were quantified using the Micro BCA
Protein Assay kit (Thermo Fisher Scientific, Inc.) and equal amount
of proteins were separated by electrophoresis using 4-12% gradient
gel and transferred to a nitrocellulose membrane. The membranes
were blocked with 5% milk for 1 h at room temperature, and then
incubated with primary antibodies (PIM1; cat. no. 3247, 1:1,000,
34, 44 kDa, PIM2; cat. no. 4730, 1:1,000, 34,38,40 kDa, PIM3; cat.
no. 4165, 1:1,000, 35 kDa, cleaved PARP; cat. no. 5625, 1:1,000, 89
kDa, 4EBP; cat. no. 9452, 1:1,000, 20 kDa, p-4EBPSer65;
cat. no. 9451, 1:1,000, 20 kDa, TSC2; cat. no. 4308, 1:1,000, 200
kDa, or GAPDH; cat. no. 5174, 1:1,000, 37 kDa; all from Cell
Signaling Technology, Inc. and p-TSC2Ser1798; cat. no.
sc-293149, 1:1,000, 200 kDa from Santa Cruz Biotechnology, Inc.) at
4°C overnight. Bands were developed with HRP-labeled
anti-rabbit secondary antibodies (1:2,500; cat. no. W4011, Promega
Corporation) for 3 h at room temperature, followed by the Clarity
Western ECL detection system (Bio-Rad Laboratories, Inc.).
Chemiluminescence signals were acquired using a ChemiDoc MP imager
(Bio-Rad Laboratories, Inc.).
Apoptosis assay
The breast cancer cell lines, MDA-MB-231 and BT549,
were treated with either the vehicle (DMSO, MilliporeSigma),
JP11646 (100 or 200 nM) or AZD1208 (1 µM) for 48 h. The
cells were washed with phosphate-buffered saline (PBS) and
suspended in Annexin V binding buffer (BioLegend, Inc.), followed
by stained with Annexin V (BioLegend, Inc.) and propidium iodide
(MilliporeSigma). The apoptotic rate was analyzed using FACS
LSRFortessa (BD Biosciences).
In vivo xenograft model
Approval from the Roswell Park Cancer Institution
Animal Care and Use Committee was obtained for the experiments in
the present study. The animals were accommodated at a constant
temperature of 22°C and 50-60% humidity with a 12-h light/dark
cycle, and standard conditions with free access to food and water.
A total of 114 CB17 SCID mice (female, 6-8 weeks-old, weighing
18-22 g) were purchased in-house from the Roswell Park
Comprehensive Cancer Center. For the experiment with MDA-MB-231
overexpressing PIM2, 1×106 cell suspensions in a mixture
of 2 µl PBS and 18 µl Matrigel (Corning, Inc.) were
injected into the chest mammary fat pads and tumor growth were
observed for up to 28 days (2 groups; control and PIM2
overexpression, n=7 mice per group). For JP11646 treatment, cell
suspensions (1×106 of MDA-MB-231, 3×106 of
MIAPaCa-2, 2×106 of PANC-1, 5×106 of HepG2,
5×106 of A549, 5×106 of HT29 and
5×106 of H1650) in a mixture of 50 µl PBS and 50
µl Matrigel were injected subcutaneously into the mouse
flanks, or in the case of MDA-MB-231 cells, into the abdomen
mammary fat pads. When the average tumor sizes reached 100
mm3, the mice were randomized and treated with the
vehicle, standard care agent, or JP11646 (For MDA-MB-231: 2 groups;
control and JP11646, for HepG2, MIAPaCa-2, PANC1, A549, H1650 and
HT29: 3 groups; control, JP11646 and standard care agent, n=5 per
group). JP11646 was prepared fresh (2.5 mg/ml) in a proprietary
carrier solution of 30% modified β-cyclodextrin (Ligand
Pharmaceuticals Inc.). The vehicle (proprietary carrier solution of
30% modified β-cyclodextrin) or 15 mg/kg JP11646 were administered
by intraperitoneal injection continuously for 2 days a week. The
details of standard care agents and the vehicles are summarized in
Table SI. The control group
received two different vehicles. Tumor size and mice conditions
were monitored 2-3 times a week using calipers, and the tumor
volume was estimated using the following equation: Volume=(length)
x (width)2/2. The humanitarian endpoints were set by the
institutional IACUC, and the animals were monitored closely by
independent veterinarian technicians who evaluated whether any
endpoints had been reached. The maximum tumor dimension was set as
one of the institutional endpoints which prevents the tumors from
reaching >10% of the animal body weight, as weighing tumors is
not practical as the experiment is proceeding. When the maximum
tumor diameter reached 2 cm (maximum observed dimension and volume:
MDA-MB-231, 20 mm and 3,062.5 mm3; HepG2, 20.1 mm and
2,231.2 mm3; MIAPaCa-2, 20.4 mm and 3,459.6
mm3; PANC-1, 20.5 mm and 2,007.7 mm3; A549,
21.6 mm and 3,179 mm3; H1650, 20.8 mm and 2,306.4
mm3; HT29, 20.4 mm and 2,844.7 mm3), or other
humanitarian endpoints were observed, such as weight loss (≥20%) or
tumors with ulcers, the experiment was terminated and the animals
were euthanized with CO2 inhalation (30-70%) as per
institutional guidelines for the humanitarian care of animals
(MDA-MB-231, 24 days; HepG2, 23 days; MIAPaCa-2, 29 days; PANC-1,
40 days; A549, 29 days; H1650, 18 days; HT29, 12 days).
Statistical analysis
Data are presented as the mean ± standard
error of the mean. Comparisons between two groups were performed
using an unpaired Student's t-test, and those among more than two
groups were performed using one-way ANOVA followed by Tukey's post
hoc test. All statistical analyses were performed using GraphPad
Prism7 (GraphPad Software, Inc.).
Results
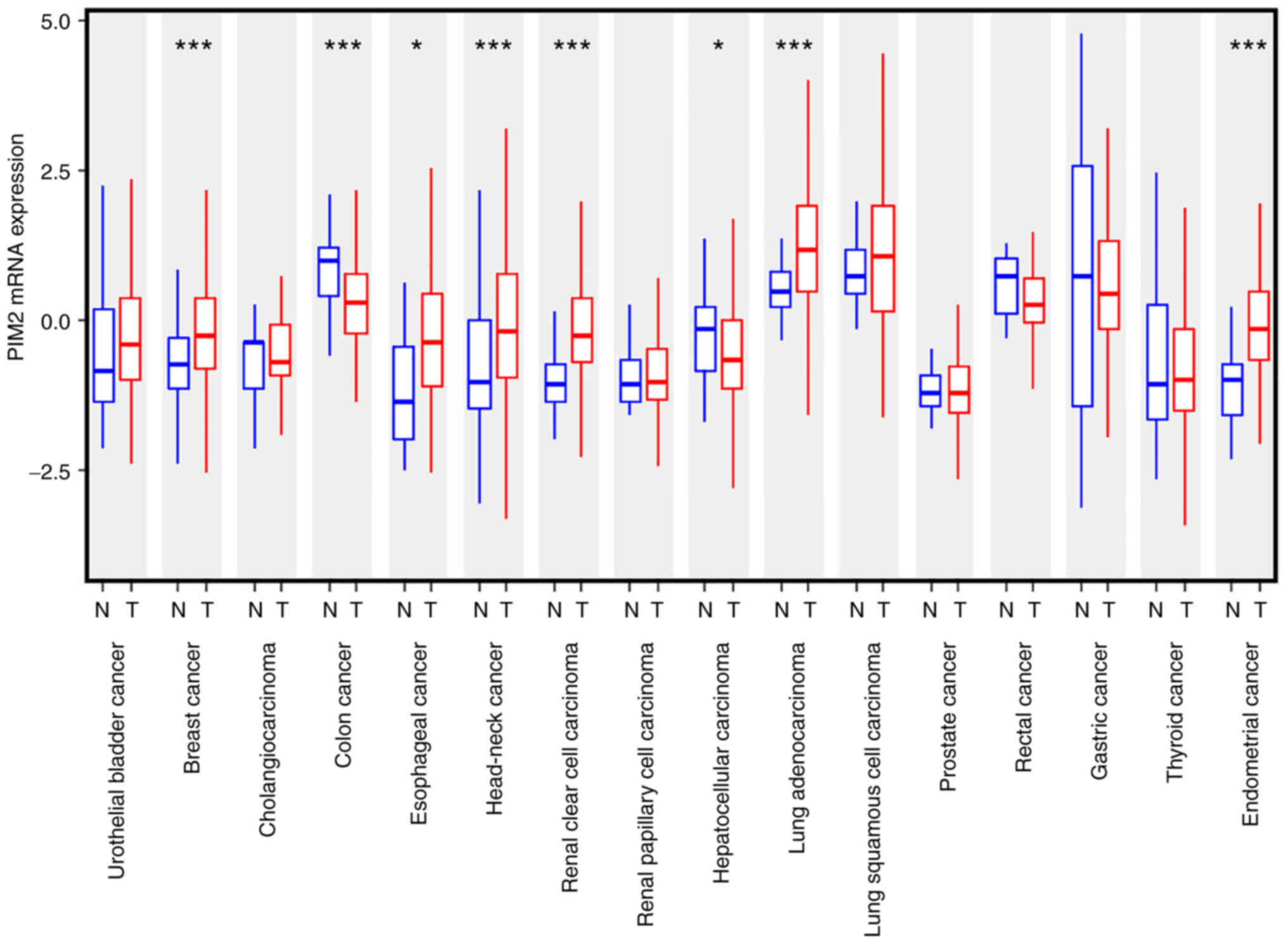
PIM2 is overexpressed in various solid
cancers
It was hypothesized that PIM2 is a factor in solid
tumor progression. Thus, TCGA datasets were interrogated to compare
the PIM2 mRNA levels between cancerous and normal tissues. PIM2 was
overexpressed as compared to normal tissue in several types of
solid cancers, including breast, esophageal, head and neck, renal
clear cell carcinoma, lung adenocarcinoma and endometrial cancer
(Fig. 1). On the other hand, its
expression was lower in hepatocellular carcinoma and colorectal
cancer, and the authors were not able to investigate its expression
in PDAC as there was no normal tissue mRNA expression data of
pancreatic cancer in TCGA cohort (Fig.
1). Thus, the present study investigated whether PIM2 plays a
role in tumor growth and whether it is a potential therapeutic
target in solid cancers.
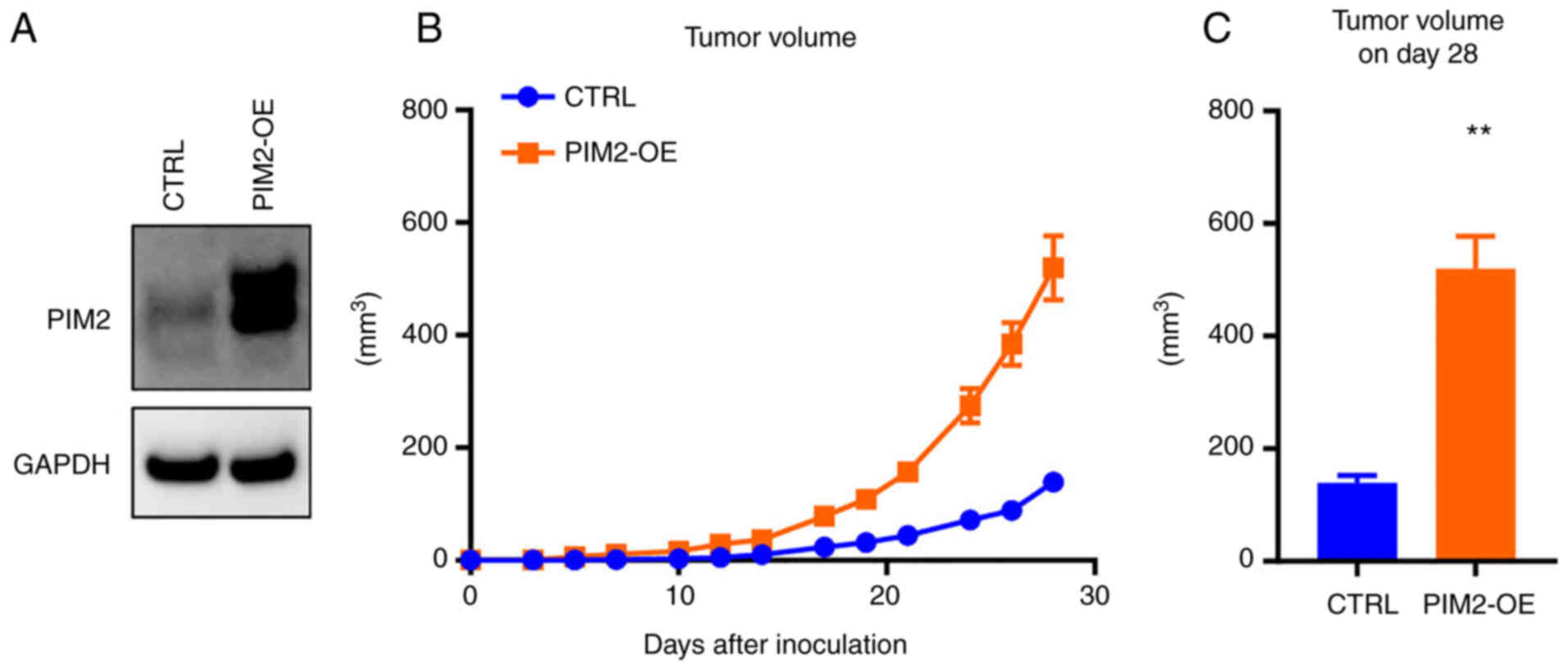
PIM2 overexpression promotes tumor growth
in vivo
To investigate whether PIM2 plays a role in
promoting tumor growth in breast cancer, either empty or PIM2
inserted p3xFlag-CMV-14 were transfected and PIM2 overexpression
was confirmed in the MDA-MB-231 breast cancer cells (Fig. 2A). Control or PIM2-overexpressing
cells were injected into mice mammary fat pads. Tumor growth
generated by PIM2-overexpressing cells occurred more rapidly
compared with the controls (Fig. 2B
and C), suggesting that PIM2 promotes the progression of
MDA-MB-231 tumors.
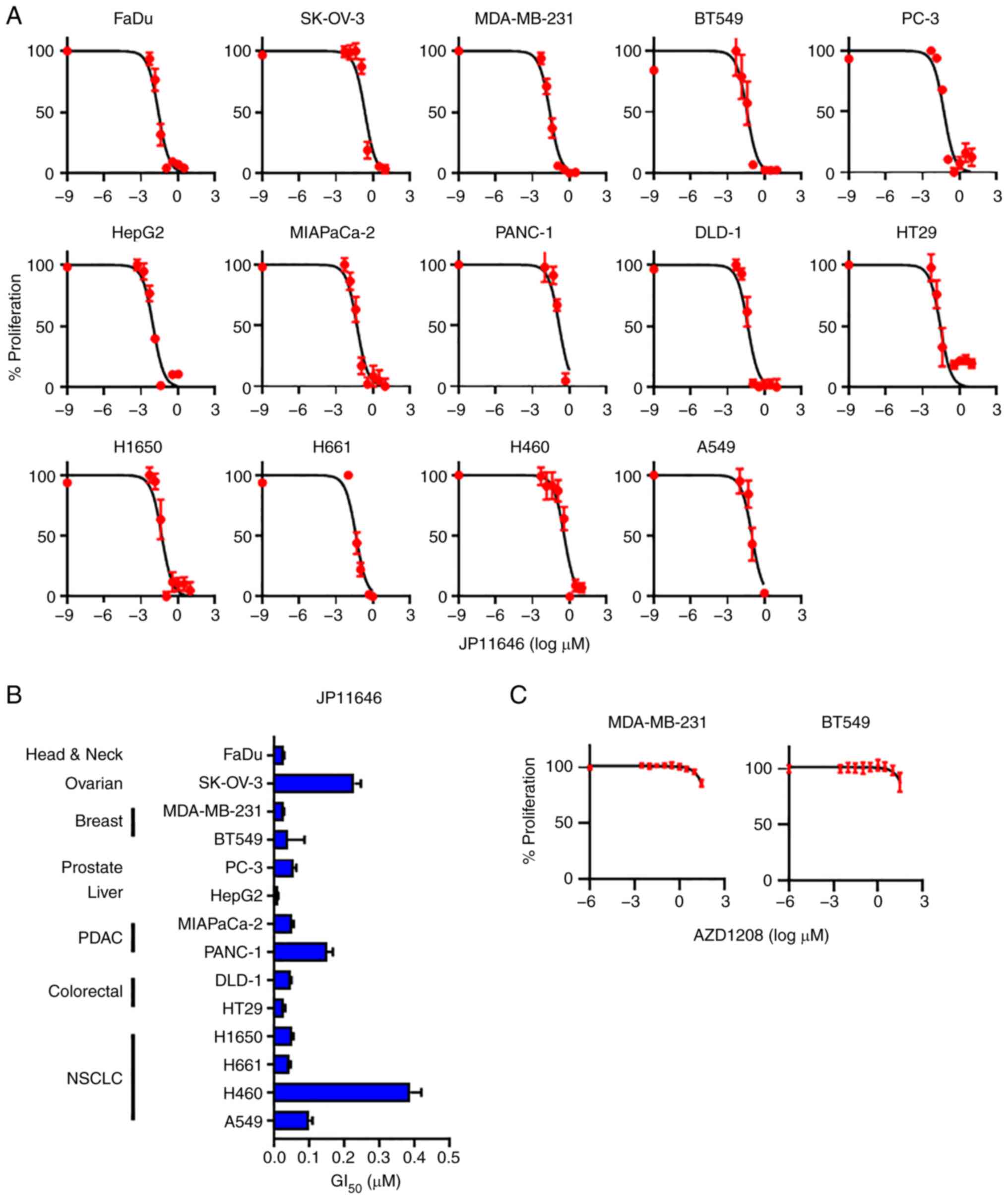
Pan-PIM inhibitor, JP11646, inhibits cell
growth in multiple solid cancers
Subsequently, the present study examined whether the
novel pan-PIM inhibitor, JP11646, inhibits cell proliferation in
solid cancers. Although the GI50 values varied among the
cancer cell lines, JP11646 suppressed cancer cell proliferation in
a concentration-dependent manner in all cell lines tested,
including in head and neck cancer FaDu, ovarian cancer SK-OV-3,
breast cancer MDA-MB-231 and BT549, prostate cancer PC-3, liver
cancer HepG2, PDAC MIAPaCa-2 and PANC1, colorectal cancer DLD-1 and
HT29, and NSCLC H1650, H661, H460 and A549 cell lines (Fig. 3A and B), although the
GI50 values were not associated with PIM2 expression
(Fig. S2). Another pan-PIM
inhibitor (AZD1208) was also tested, whose GI50 value
for the acute myeloid leukemia cell line was 0.02 µM
(26). It exhibited minimal
efficacy in decreasing cell proliferation at only the highest dose
of 30 µM in both the MDA-MB-231 and BT549 cells (Fig. 3C).
JP11646 treatment downregulates PIM2
protein expression
The present study then investigated the mechanisms
underlying the effects of JP11646 on cancer cell proliferation.
Previous research has demonstrated that JP11646 treatment induces
the apoptosis of multiple myeloma cells, being associated with
selective PIM2 downregulation (7),
while the anti-apoptotic role of PIM2 is considered to be due to
its phosphorylation of downstream targets, including 4EBP1, TSC2
and BAD (9,27). The present study was able to
recapitulate the observation that treatment with JP11646 resulted
in the selective downregulation of PIM2, but not of PIM1 or PIM3
protein expression (Fig. 4A). The
induction of cleaved PARP by JP11646 treatment was also confirmed
in both MDA-MB-231 and BT549 cells (Fig. 4A), suggesting the induction of
apoptosis. To examine the effects of PIM2 downregulation on
apoptosis induction and the phosphorylation of downstream targets,
BT549 cells were treated with either siRNA or the second pan-PIM
inhibitor, AZD1208. PIM2 knockdown using siRNA also increased
cleaved PARP expression together with the decreased phosphorylation
of 4EBP1 and TSC2 (Fig. 4B). In
addition, the results revealed the decreased phosphorylation of
known PIM2 targets, 4EBP1 and TSC2, at 1 µM of AZD1208,
indicating the inhibition of PIM2 kinase activity (Fig. 4B). However, AZD1208 did not
decrease the PIM2 protein level and did not upregulate cleaved
PARP, likely reflecting a lack of apoptosis induction (Fig. 4B).
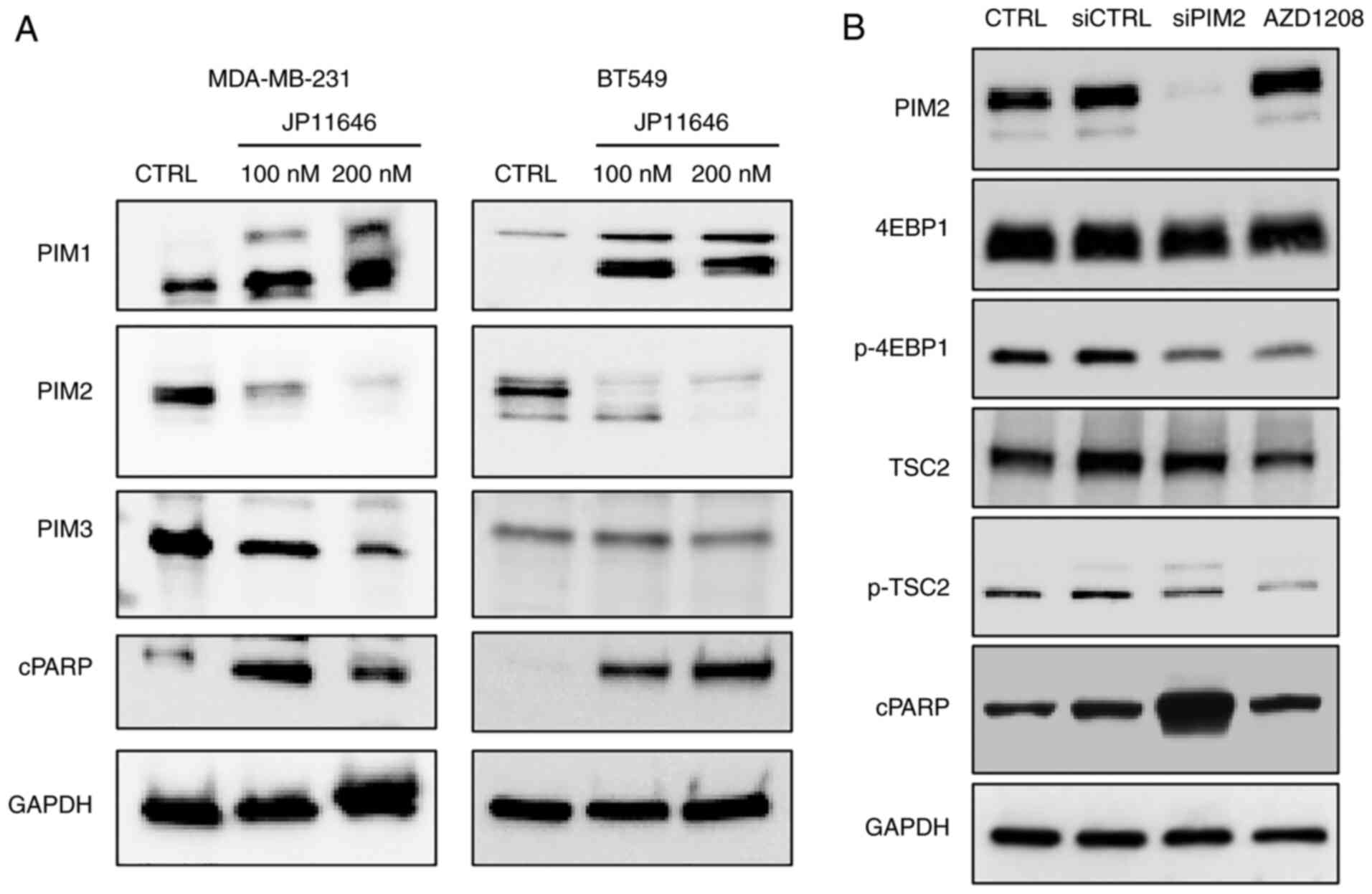 | Figure 4JP11646 treatment downregulates the
PIM2 protein level. (A) Western blot analyses of PIMs (PIM1; 34, 44
kDa, PIM2; 34,38,40 kDa, PIM3; 35 kDa) and cleaved PARP (89 kDa)
from JP11646-treated MDA-MB-231 and BT549 cells. (B) Western blot
analyses of PIM2, 4EBP1 (20 kDa), p-4EBP (20 kDa), TSC2 (200 kDa),
p-TSC2 (200 kDa) and cleaved PARP in BT549 cells transfected with
siRNA or treated with AZD1208 (1 µM). PIM, proviral
integration of Moloney virus; cPARP, cleaved PARP. |
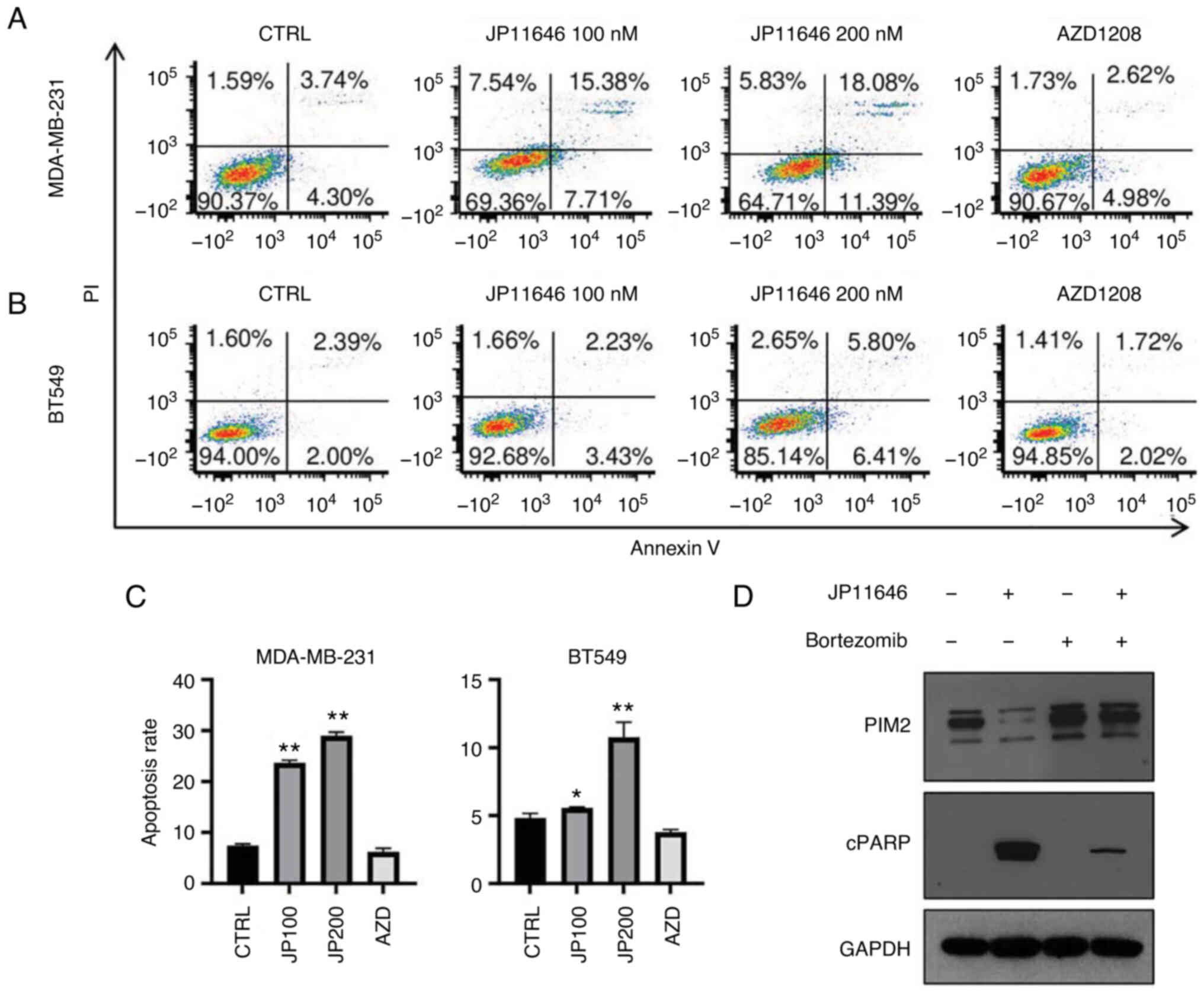
Proteasome-dependent PIM2 protein
degradation induces the apoptosis of breast cancer cells
The present study further examined the effects of
targeting PIM2 on apoptosis induction; the MDA-MB-231 and BT549
cells were treated with either JP11646 or the second pan-PIM
inhibitor, AZD1208. Consistent with the induction of cleaved PARP,
JP11646 treatment significantly increased the apoptotic rate, and
AZD1208 treatment did not result in a change over the baseline
levels in both MDA-MB-231 and BT549 (Fig. 5A-C). The present study then further
examined the mechanisms of PIM2 downregulation by JP11646. The
addition of the proteasome inhibitor, bortezomib, with JP11646
prevented PIM2 downregulation and decreased cleaved PARP levels,
suggesting that proteasome activity leading to PIM2 degradation is
required for JP11646 to induce cell death through the apoptosis of
MDA-MB-231 cells (Fig. 5D),
although it did not rescue cell viability 72 h after treatment
(Fig. S3).
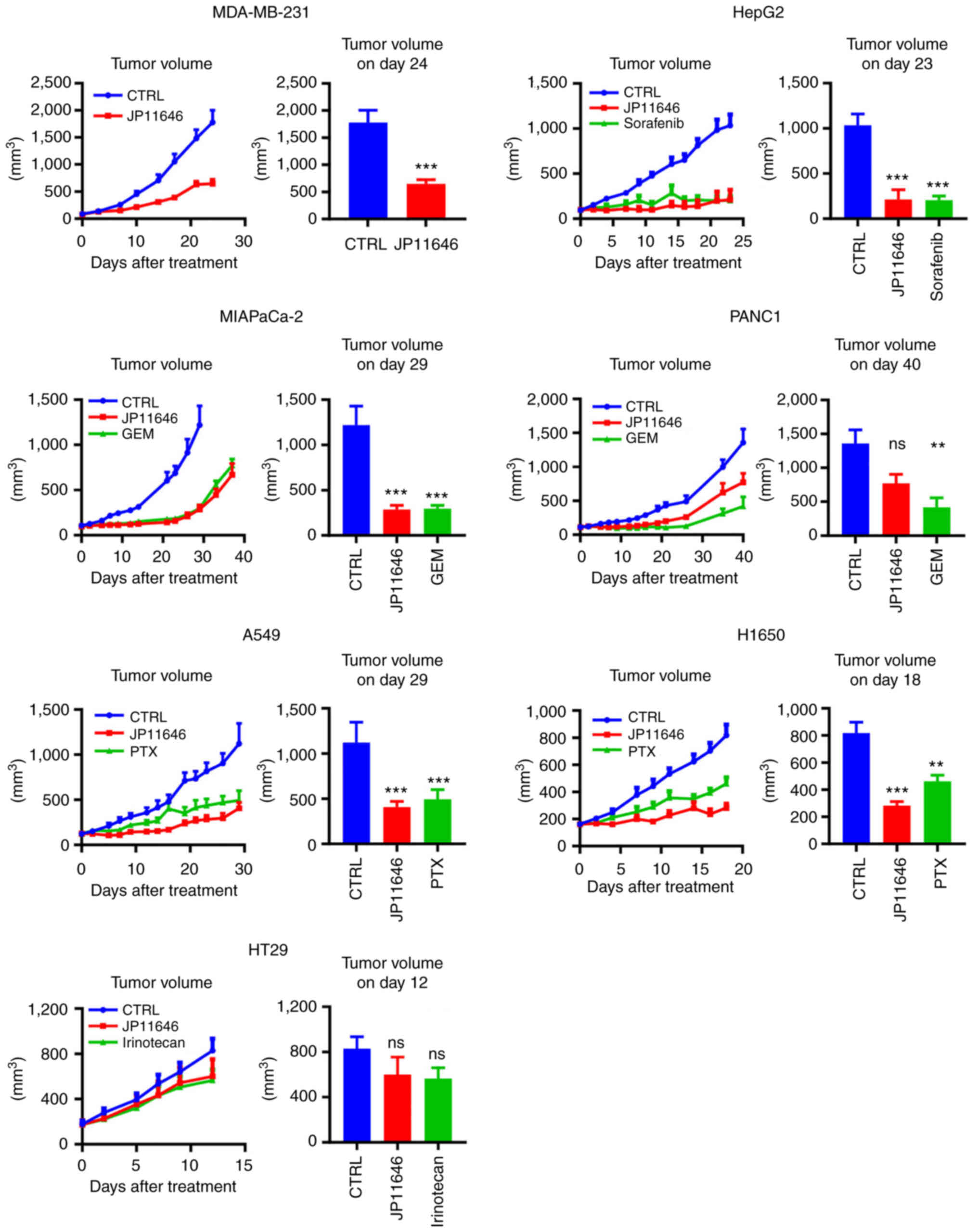
PIM2 inhibition by JP11646 suppresses
tumor growth in vivo
Finally, the present study examined the efficacy of
JP11646 in multiple in vivo cancer models. JP11646
significantly suppressed tumor growth in five out of seven tested
xenograft tumor models. These included breast cancer MDA-MB-231,
liver cancer HepG2, PDAC MIAPaCa-2, and NSCLCs A549 and H1650 in
vivo models (Fig. 6). However,
JP11646 did not suppress tumor growth in PDAC PANC1 and colon
cancer HT29 xenograft models (Fig.
6). Standard therapeutic agents were used for some models for
the relative measure of JP11646 efficacy, sorafenib for liver
cancer, gemcitabine for PDAC, paclitaxel for NSCLC and irinotecan
for colorectal cancer. JP11646 resulted in comparable or improved
antitumor efficacy when compared to most standard therapies. At the
administered drug doses and schedules, no mouse exhibited a weight
loss >20% or any other detectable severe side-effect.
Discussion
In the present study, it was demonstrated that PIM2
promoted in vivo tumor growth and may thus represent a
potential therapeutic target. Targeting PIM2 using siRNA or the
pan-PIM inhibitor, JP11646, resulted in the downregulation of PIM2
and the upregulation of cleaved PARP expression in breast cancer
cells. JP11646-induced PIM2 degradation and the induction of
apoptosis were dependent on proteasome activity and were associated
with the inhibition of cancer cell proliferation in vitro
and tumor growth in vivo.
Consistent with previous findings (7), the present study demonstrated that
PIM2 downregulation induced apoptosis. The findings suggested that
treatment with JP11646 resulted in a more profound inhibition of
PIM2 signaling. It was found that the effect of JP11646 on PIM2 was
proteosome-dependent in MDA-MB-231 cells. A previous study
demonstrated the possibility of kinase-independent PIM2 activity
(7), which may explain the
enhanced anticancer efficacy of JP11646 as compared to other kinase
inhibitors. Although bortezomib did not rescue cell viability at 72
h after treatment, the mechanisms through which JP11464 leads to
cell death are not yet fully understood. This mechanism of JP11646
which leads to apoptosis by proteasome-dependent PIM2 protein
degradation warrants further investigation in multiple cancer types
in the future. It may lead to improvements in clinical efficacy,
which have not yet been achieved by the current clinical testing of
PIM inhibitors (16).
The present study also demonstrated the efficacy of
JP11646 in preclinical mouse models of multiple solid cancers. As
previously reported, it is necessary to demonstrate an efficacy in
in vivo preclinical models before translating this into
clinical practice (28-30). The present study broadly examined
the efficacy of JP11646 in variety of cancers independent of PIM2
expression based on the hypothesis that targeting PIM2 for
degradation highlights a potential novel mechanism. In total, five
out of seven JP11646-treated cancers exhibited a significant tumor
growth suppression compared to the controls with acceptable
side-effect profiles. These results were compared to
standard-of-care treatments with comparable results. This suggests
that targeting PIM2 by JP11646 may be a potential novel therapeutic
strategy for the treatment of solid cancers.
Even though the current experimental results
demonstrated the anticancer efficacy of JP11646 in multiple
cancers, JP11646 efficacy was not associated with PIM2 mRNA
expression. This could be due to differences in mRNA translation,
the dependence of cells on PIM2 signaling or another yet
undiscovered function of PIM2 protein. It may also be due to
differences in PIM2 targeting for degradation or the existence of
compensatory signaling pathways. The off-target effects of JP11646
have been extensively investigated (7). Although a previous study reported the
specificity of PIM2 inhibition by JP11646 (7), there is a possibility that the
response to JP11646 is more complex, as PIM2 signaling has been
shown to be involved in numerous important pathways in cancer cell
biology. Furthermore, JP11646 did not exhibit efficacy in some
mouse models. The resistance mechanisms to JP11646 are under
investigation. One possibility of resistance is the crosstalk
between cancer cells and the tumor microenvironment. Although the
present study used immune deficient mice, there are multiple
components other than immune cells. The tumor microenvironment
plays a role to support tumor progression (31); therefore, it may help cancer cells
to acquire resistance. It may lead to the inability of JP11646 to
achieve a necessary concentration intracellularly due to restricted
diffusion or efflux pumps. Further studies are thus warranted to
identify predictive markers of JP11646 sensitivity.
The present study demonstrated anticancer effects by
targeting PIM2. However, the detailed mechanisms of PIM2 function
and necessary inhibition have not yet been fully elucidated. The
molecular mechanisms of PIM2 function and the effect of the JP11646
inhibitor in multiple cancer types, as well as the efficacy of
other PIM2 inhibitors needs to be investigated in the future.
Further studies are warranted to fill this gap in knowledge.
In conclusion, the present study found that PIM2
promoted cancer progression in solid tumors. PIM2 inhibition by
JP11646 induced apoptosis via PIM2 protein degradation and
suppressed cancer cell proliferation in vitro and in
vivo. PIM2, if properly targeted, may serve as a novel
therapeutic target for the treatment of solid cancers.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. TCGA data were downloaded through cBioportal website
(https://www.cbioportal.org).
Authors' contributions
MGM, AAA and MO designed the study. EK, MGM, JM, MY
and YD performed the experiments. EK, MGM, JM, AAA, JC, CMB, KPL,
IHG, KT and MO. interpreted the results. JC and CMB provided the
resources. EK, KPL, IHG, KT and MO prepared the article. MO
provided supervision. EK and MO confirm the authenticity of all the
raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Since all the patients analyzed in the present study
were from de-identified publicly available cohorts, institutional
review board approval was waived. Informed consent was obtained by
the researchers of the original publication (32). The present study was performed in
accordance with the Declaration of Helsinki. All cellular
experiments were approved by the Roswell Park Comprehensive Cancer
Center Biosafety Committee. All animal experiments were conducted
under approved Institutional Animal Care and Use Committee (IACUC)
protocols and the Roswell Park Comprehensive Cancer Center
Laboratory Animal Shared Resource is accredited by the American
Association for the Accreditation of Laboratory Animal Care
(AAALAC).
Patient consent for publication
Not applicable.
Competing interests
M. Opyrchal has a Research support from Bayer and
Pfizer and is an advisory board at AstraZeneca and Novartis. The
other authors declare that they have no competing interests.
Acknowledgments
Flow cytometric analyses were conducted at Flow
& Image Cytometry Shared Resource and the animal experiments
were conducted at Laboratory Animal Shared Resource in Roswell Park
Comprehensive Cancer Center (Buffalo, NY, USA).
Funding
The present study was supported by the National Cancer Institute
(NCI) grant P30CA016056 involving the use of Roswell Park Cancer
Comprehensive Cancer Center Shared Resources.
Abbreviations:
|
PIM2
|
proviral integration of Moloney virus
2
|
|
4EBP1
|
4E-binding protein 1
|
|
TSC2
|
tuberous sclerosis complex 2
|
|
BAD
|
BCL2-associated agonist of cell
death
|
|
TCGA
|
The Cancer Genome Atlas
|
|
PDAC
|
pancreatic ductal adenocarcinoma
|
|
NSCLC
|
non-small cell lung cancer
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FBS
|
fetal bovine serum
|
|
PBS
|
phosphate-buffered saline
|
References
|
1
|
Cuypers HT, Selten G, Quint W, Zijlstra M,
Maandag ER, Boelens W, van Wezenbeek P, Melief C and Berns A:
Murine leukemia virus-induced T-cell lymphomagenesis: Integration
of proviruses in a distinct chromosomal region. Cell. 37:141–150.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Eichmann A, Yuan L, Bréant C, Alitalo K
and Koskinen PJ: Developmental expression of pim kinases suggests
functions also outside of the hematopoietic system. Oncogene.
19:1215–1224. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nawijn MC, Alendar A and Berns A: For
better or for worse: The role of pim oncogenes in tumorigenesis.
Nat Rev Cancer. 11:23–34. 2011. View
Article : Google Scholar
|
|
4
|
Jinesh GG, Mokkapati S, Zhu K and Morales
EE: Pim kinase isoforms: Devils defending cancer cells from
therapeutic and immune attacks. Apoptosis. 21:1203–1213. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yan B, Zemskova M, Holder S, Chin V, Kraft
A, Koskinen PJ and Lilly M: The PIM-2 kinase phosphorylates BAD on
serine 112 and reverses BAD-induced cell death. J Biol Chem.
278:45358–45367. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ren K, Gou X, Xiao M, He W and Kang J:
Pim-2 cooperates with downstream factor XIAP to inhibit apoptosis
and intensify malignant grade in prostate cancer. Pathol Oncol Res.
25:341–348. 2019. View Article : Google Scholar
|
|
7
|
Nair JR, Caserta J, Belko K, Howell T,
Fetterly G, Baldino C and Lee KP: Novel inhibition of PIM2 kinase
has significant anti-tumor efficacy in multiple myeloma. Leukemia.
31:1715–1726. 2017. View Article : Google Scholar :
|
|
8
|
Hideshima T, Nakamura N, Chauhan D and
Anderson KC: Biologic sequelae of interleukin-6 induced PI3-K/Akt
signaling in multiple myeloma. Oncogene. 20:5991–6000. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fox CJ, Hammerman PS, Cinalli RM, Master
SR, Chodosh LA and Thompson CB: The serine/threonine kinase Pim-2
is a transcriptionally regulated apoptotic inhibitor. Genes Dev.
17:1841–1854. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cervantes-Gomez F, Stellrecht CM, Ayres
ML, Keating MJ, Wierda WG and Gandhi V: PIM kinase inhibitor,
AZD1208, inhibits protein translation and induces autophagy in
primary chronic lymphocytic leukemia cells. Oncotarget.
10:2793–2809. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen LS, Yang JY, Liang H, Cortes JE and
Gandhi V: Protein profiling identifies mTOR pathway modulation and
cytostatic effects of pim kinase inhibitor, AZD1208, in acute
myeloid leukemia. Leuk Lymphoma. 57:2863–2873. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen LS, Redkar S, Taverna P, Cortes JE
and Gandhi V: Mechanisms of cytotoxicity to pim kinase inhibitor,
SGI-1776, in acute myeloid leukemia. Blood. 118:693–702. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yang Q, Chen LS, Neelapu SS, Miranda RN,
Medeiros LJ and Gandhi V: Transcription and translation are primary
targets of pim kinase inhibitor SGI-1776 in mantle cell lymphoma.
Blood. 120:3491–3500. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang Q, Chen LS, Neelapu SS and Gandhi V:
Combination of pim kinase inhibitor SGI-1776 and bendamustine in
B-cell lymphoma. Clin Lymphoma Myeloma Leuk. 13(Suppl 2):
S355–S362. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cervantes-Gomez F, Chen LS, Orlowski RZ
and Gandhi V: Biological effects of the pim kinase inhibitor,
SGI-1776, in multiple myeloma. Clin Lymphoma Myeloma Leuk. 13(Suppl
2): S317–S329. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cortes J, Tamura K, DeAngelo DJ, de Bono
J, Lorente D, Minden M, Uy GL, Kantarjian H, Chen LS, Gandhi V, et
al: Phase I studies of AZD1208, a proviral integration moloney
virus kinase inhibitor in solid and haematological cancers. Br J
Cancer. 118:1425–1433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Uddin N, Kim RK, Yoo KC, Kim YH, Cui YH,
Kim IG, Suh Y and Lee SJ: Persistent activation of STAT3 by
PIM2-driven positive feedback loop for epithelial-mesenchymal
transition in breast cancer. Cancer Sci. 106:718–725. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Han X, Ren C, Yang T, Qiao P, Wang L,
Jiang A, Meng Y, Liu Z, Du Y and Yu Z: Negative regulation of
AMPKalpha1 by PIM2 promotes aerobic glycolysis and tumorigenesis in
endometrial cancer. Oncogene. 38:6537–6549. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBioPortal. Sci Signal. 6:112013. View Article : Google Scholar
|
|
21
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The cancer cell line encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ghandi M, Huang FW, Jané-Valbuena J,
Kryukov GV, Lo CC, McDonald ER III, Barretina J, Gelfand ET,
Bielski CM, Li H, et al: Next-generation characterization of the
cancer cell line encyclopedia. Nature. 569:503–508. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Baldino CM, Caserta J, Lee CS, Nicewonger
R, Flanders Y and Dumas S: Aminopyrimidine kinase inhibitors. Jasco
Pharmaceuticals LLC; United States: 2013
|
|
24
|
Flanders Y, Dumas S, Caserta J, Nicewonger
R, Baldino M, Lee CS and Badlino CM: A versatile synthesis of novel
pan-PIM kinase inhibitors with initial SAR study. Tetrahedron Lett.
56:3186–3190. 2015. View Article : Google Scholar
|
|
25
|
Zhang X, Song M, Kundu JK, Lee MH and Liu
ZZ: PIM kinase as an executional target in cancer. J Cancer Prev.
23:109–116. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dakin LA, Block MH, Chen H, Code E,
Dowling JE, Feng X, Ferguson AD, Green I, Hird AW, Howard T, et al:
Discovery of novel benzylidene-1,3-thiazolidine-2,4-diones as
potent and selective inhibitors of the PIM-1, PIM-2, and PIM-3
protein kinases. Bioorg Med Chem Lett. 22:4599–4604. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tamburini J, Green AS, Bardet V, Chapuis
N, Park S, Willems L, Uzunov M, Ifrah N, Dreyfus F, Lacombe C, et
al: Protein synthesis is resistant to rapamycin and constitutes a
promising therapeutic target in acute myeloid leukemia. Blood.
114:1618–1627. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Katsuta E, DeMasi SC, Terracina KP,
Spiegel S, Phan GQ, Bear HD and Takabe K: Modified breast cancer
model for preclinical immunotherapy studies. J Surg Res.
204:467–474. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Katsuta E, Oshi M, Rashid OM and Takabe K:
Generating a murine orthotopic metastatic breast cancer model and
performing murine radical mastectomy. J Vis Exp. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Katsuta E, Rashid OM and Takabe K: Murine
breast cancer mastectomy model that predicts patient outcomes for
drug development. J Surg Res. 219:310–318. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Katsuta E, Rashid OM and Takabe K:
Clinical relevance of tumor microenvironment: immune cells,
vessels, and mouse models. Human Cell. 33:930–937. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Curtis C, Shah SP, Chin SF, Turashvili G,
Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et
al: The genomic and transcriptomic architecture of 2,000 breast
tumours reveals novel subgroups. Nature. 486:346–352. 2012.
View Article : Google Scholar : PubMed/NCBI
|