Introduction
Bladder cancer (BC) is a common urinary system
cancer worldwide (1). Among cases
of BC, 90% are of urothelial carcinoma, 5% are of squamous cell
carcinoma, and the remaining 5% are of adenocarcinoma and
undifferentiated carcinoma (2).
Although the overall survival of patients with BC has significantly
improved with current therapies of transurethral resection,
intravesical chemotherapy, and Bacillus Calmette-Guérin (BCG)-based
immunotherapy, a large proportion (~60-70%) of patients with BC
have recurring non-invasive or in situ tumors (3), which progress into muscle invasive BC
(MIBC; in ~50% of all patients with BC) (4). Therefore, the development of novel
therapeutic agents is required to advance BC treatment.
Hanahan and Weinberg in 2011 proposed ten hallmarks
of cancer that are crucial for the transformation from normalcy to
neoplastic growth states and their ability to form malignant tumors
and distant metastatic focus (5).
This substantially affects the development of new therapeutic
strategies for the treatment of various cancers (6). One of the major features of cancer is
invasion and metastasis, which was demonstrated to be controlled by
the epithelial-mesenchymal transition (EMT) state (7). EMT is a dynamic process in which an
epithelial phenotype converts into a mesenchymal phenotype to
enable the invasion and metastasis of cancer cells (8). Autophagy also plays a critical role
in several features of cancer cells; most importantly, autophagy is
implicated at various stages of tumorigenesis (9). Autophagy is a double-edged sword; it
not only promotes tumor progression but also serves as a survival
mechanism in cancer cells to evade apoptotic cell death against
therapeutic agents (10). Notably,
BC cells were reported to exhibit a higher basal level of
autophagic flux with increased LC3-II expression compared with
cells of breast, prostate and kidney cancer (11). The inhibition of autophagy activity
using pharmacological inhibitors-such as chloroquine,
hydroxychloroquine, and bafilomycin A1-blocks BC cell invasiveness
(12), triggers cell apoptosis
in vitro, and decreases tumor growth in vivo
(13). Therefore, the targeting of
autophagic flux may be an efficient new strategy for overcoming
autophagy-related resistance to current therapies against BC.
MicroRNA (miRNA or miR), a small molecule of
noncoding RNAs (~21-25 nucleotides), acts in conjunction with
RNA-induced silencing complex to bind on the 3′-untranslated region
(UTR) of target messenger RNA. It prevents the formation of the
translation initiation complex, and this results in translation
inhibition (14). Previous studies
have reported that up to 60% of protein-coding genes can be
regulated by miRNAs at the translational level (15). In fact, miRNAs are involved in the
regulation of numerous cellular and developmental processes
(16). Furthermore, the
dysregulation of miRNAs is associated with cancer (17,18),
Alzheimer's disease (19),
rheumatoid arthritis (20), and
cardiovascular diseases (21).
miRNAs can be divided into oncogenic (i.e., tumor-promoter) miRNA
and tumor-suppressor miRNA (22).
Both oncogenic and tumor-suppressor miRNAs play roles in most
features of cancer, including proliferation, apoptosis,
angiogenesis, invasion and metastasis (23). MiR-34a was reported to be a
tumor-suppressor miRNA by the majority of studies in the literature
(24,25). For instance, miR-34a epigenetically
and negatively regulates the functional properties of cancer stem
cells by targeting stemness factors including NOTCH, MYC, BCL-2,
and CD44 (26). MiR-34a suppresses
tumor progression by inhibiting cell cycle (27), invasiveness (28), metastasis (29) and cancer-specific immune evasion
(30), but how it regulates
autophagic flux is not completely understood in BC.
In the present study, the role of miR-34a was
investigate in numerous cancer-related activities, including cell
cycle arrest, cell motility inhibition and autophagy activity
suppression, and the emergence of miR-34a treatment as a novel
therapeutic strategy for BC was discussed.
Materials and methods
Cell culture
The BC cell lines 5637 and T24 were obtained from
the Bioresource Collection and Research Center (Hsinchu, Taiwan)
and cultured in RPMI-1640 and McCoy's 5A medium, respectively
(Gibco; Thermo Fisher Scientific, Inc.). Both culture media were
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.).
Transfection of miRNA mimic
MISSION synthetic negative control mimic and
hsa-miR-34a mimic were purchased from Sigma-Aldrich; Merck KGaA.
ViaFect transfection reagent (Promega Corporation) was used to
transfect 25 nM control mimic or hsa-miR-34a into BC cells at 37°C
for 24 h. Cell samples were then analyzed for p62, LC3-II and STX17
expression.
Establishment of hsa-miR-34a stable
cells
The pLenti-III-mir-GFP vector inserted with
hsa-miR-34a sequence was provided by Applied Biological Materials.
The procedure for establishing the miR-34a-expressing stable cells
was identical to that of a previous study (18).
Western blot analysis
The immunoblotting method was identical to that in a
previous study by the authors (31). Primary antibodies anti-p62
(1:3,000; cat. no. GTX100685), anti-LC3-II (1:3,000; cat. no.
GTX127375) (both from GeneTex, Inc.), anti-STX17 (1:1,000; cat. no.
31261), anti-cyclin D1 (1:1,000; cat. no. 2978), anti-cyclin E2
(1:1,000; cat. no. 4132) (all from Cell Signaling technology,
Inc.), or anti-GAPDH (1:5,000; cat. no. ab8245; Abcam) were used to
detect indicated protein levels.
Gap closure assay
A three-well silicone insert (Ibidi GmbH) was used
to perform wound-like scratches in each well. Detached miR-34a
stable cells were removed with phosphate-buffered saline wash and
cultured in the serum-free medium for 24 h. Microscope images of
migratory cells were captured (Zeiss AG) and the width of the gap
in each scratch was measured using ImageJ software v1.44p (National
Institutes of Health).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNAs were isolated using easy-BLUE™ Total RNA
Extraction Kit. The mRNA and miRNA were reverse transcribed to cDNA
using the M-MLV Reverse Transcriptase kit (Invitrogen; Thermo
Fisher Scientific, Inc.) and Mir-X™ miRNA First-Strand Synthesis
kit (Clontech Laboratories, Inc.) according to the manufacturer's
instructions, respectively. RT-qPCR analyses were conducted using a
SYBR Green Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) and conducted on a StepOnePlus sequence detection
system (Thermo Fisher Scientific, Inc.). The thermal cycling
conditions were identical to those in previous studies (32,33).
RT-qPCR primers for indicated mRNAs detection were listed in
Table I. For miR-34a detection,
the forward primer of miR-34a was 5′-TGG CAG TGT CTT AGC TGG TTG
T-3′ (mRQ 3 as reverse primer supplied with the Mir-X™ miRNA
First-Strand Synthesis kit). The relative gene expression was
analyzed by using the comparative Ct (2−ΔΔCq) method
with genes normalized to GAPDH (mRNA), β-Actin (mRNA) or snRNA U6
(miRNA) (34).
 | Table ISequences of primers used for reverse
transcription-quantitative PCR. |
Table I
Sequences of primers used for reverse
transcription-quantitative PCR.
| Gene name | Species | Primer sequence
(5′→3′) |
|---|
| KIAA1632
(EPG5) | Human | F:
CCTTCTGTATCTTCACCGTCCG |
| Human | R:
GAAGTCAGCCACCTCGGTCAAA |
| SNAP29 | Human | F:
CAGCCACCCAAACCTTAGAAAGC |
| Human | R:
CGATCTTCTGGTGATAGGCTCG |
| Syntaxin 17 | Human | F:
TCGTGGGAAACCTTAGAAGCGG |
| Human | R:
GCAGCACTGTTGACATGGTCTG |
| RAB7L1 | Human | F:
TGCTCTGAAGGTTCTCCAGTGG |
| Human | R:
GGCAGAGGCATCCCGATAATAC |
| Cyclin D1 | Human | F:
TCTACACCGACAACTCCATCCG |
| Human | R:
TCTGGCATTTTGGAGAGGAAGTG |
| Cyclin E2 | Human | F:
CTTACGTCACTGATGGTGCTTGC |
| Human | R:
CTTGGAGAAAGAGATTTAGCCAGG |
| Snail | Human | F:
ACTGCAACAAGGAATACCTCAG |
| Human | R:
GCACTGGTACTTCTTGACATCTG |
| Twist | Human | F:
GCCAGGTACATCGACTTCCTCT |
| Human | R:
TCCATCCTCCAGACCGAGAAGG |
| ZEB1 | Human | F:
GGCATACACCTACTCAACTACGG |
| Human | R:
TGGGCGGTGTAGAATCAGAGTC |
| ZEB2 | Human | F:
AATGCACAGAGTGTGGCAAGGC |
| Human | R:
CTGCTGATGTGCGAACTGTAGG |
| E-cadherin | Human | F:
CGAGAGCTACACGTTCACGG |
| Human | R:
GGGTGTCGAGGGAAAAATAGG |
| N-cadherin | Human | F:
TGCGGTACAGTGTAACTGGG |
| Human | R:
GAAACCGGGCTATCTGCTCG |
| CDH11 | Human | F:
ACCCTCACCATCAAAGTCTG |
| Human | R:
TCAGGGTCACAAACAATACT |
| Vimentin | Human | F:
AGTCCACTGAGTACCGGAGAC |
| Human | R:
CATTTCACGCATCTGGCGTTC |
| β-actin | Human | F:
CACCATTGGCAATGAGCGGTTC |
| Human | R:
AGGTCTTTGCGGATGTCCACGT |
| GAPDH | Human | F:
GTCTCCTCTGACTTCAACAGCG |
| Human | R:
ACCACCCTGTTGCTGTAGCCAA |
Colony formation assay
The miR-34a stable cells were seeded on a six-well
plate at a density of 3×103 cells/well. A colony
consists of at least 50 stable cells. After 7 days of colony
formation, colonies were fixed with 3.7% formaldehyde for 20 min
and then stained with 0.05% crystal violet (w/v) for another 20 min
at room temperature. Colonies were indicated by the absorbance of
crystal violet extraction with 10% acetic acid.
Luciferase reporter assay
A DNA fragment containing 3′-UTRs of STX17 targeted
by miR-34a were constructed into a pmirGLO vector (Promega
Corporation). BC cells seeded on a 12-well plate were transfected
with empty vector or 3′-UTR reporter constructs (1
µg/µl) using a ViaFect transfection reagent (Promega
Corporation) for 24 h. Subsequently, cells were treated with
control or miR-34a mimic for another 24 h. Finally, luciferase
activity was measured using a Dual-Luciferase kit (Promega
Corporation). Firefly luciferase activity was normalized to
Renilla luciferase activity.
Flow cytometry
The miR-34a stable cells (3×105) were
seeded in six-well plates. Cell samples were then stained with
propidium iodide (Cell Signaling Technology, Inc.) at room
temperature for 30 min. Cell cycle distribution analysis was
assessed using an Accuri C5 flow cytometer (BD Biosciences) and
quantified using the CellQuest Pro software (BD Biosciences).
Immunofluorescence staining
The BC cells were placed on a chamber slide
(Sigma-Aldrich; Merck KGaA) for treatment. The immunofluorescence
staining methods were identical to those in a previous study by the
authors (33). Primary anti-p62
and anti-LC3-II antibodies (both 1/100 dilution) were used to
detect p62 and LC3-II proteins expression in BC cells. In addition,
an acridine orange (AO) staining assay (Sigma-Aldrich; Merck KGaA)
was used to visualize autophagic vacuoles after miR-34a mimic
treatment. Lysosome alteration in miR-34a mimic-treated BC cells
was detected using LysoTracker Red DND-99 (Thermo Fisher
Scientific, Inc.). All fluorescent imaging was performed using a
Nikon Ti2 microscopy system (Nikon Corporation).
Statistical analysis
All experiments were performed in triplicate and
analyzed by using GraphPad Prism 9 (GraphPad Software, Inc.).
Differences were analyzed using unpaired t-test. One-way analysis
of variance followed by Bonferroni's post hoc comparison tests were
used to compare the means of three or more groups. The results are
presented in terms of the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
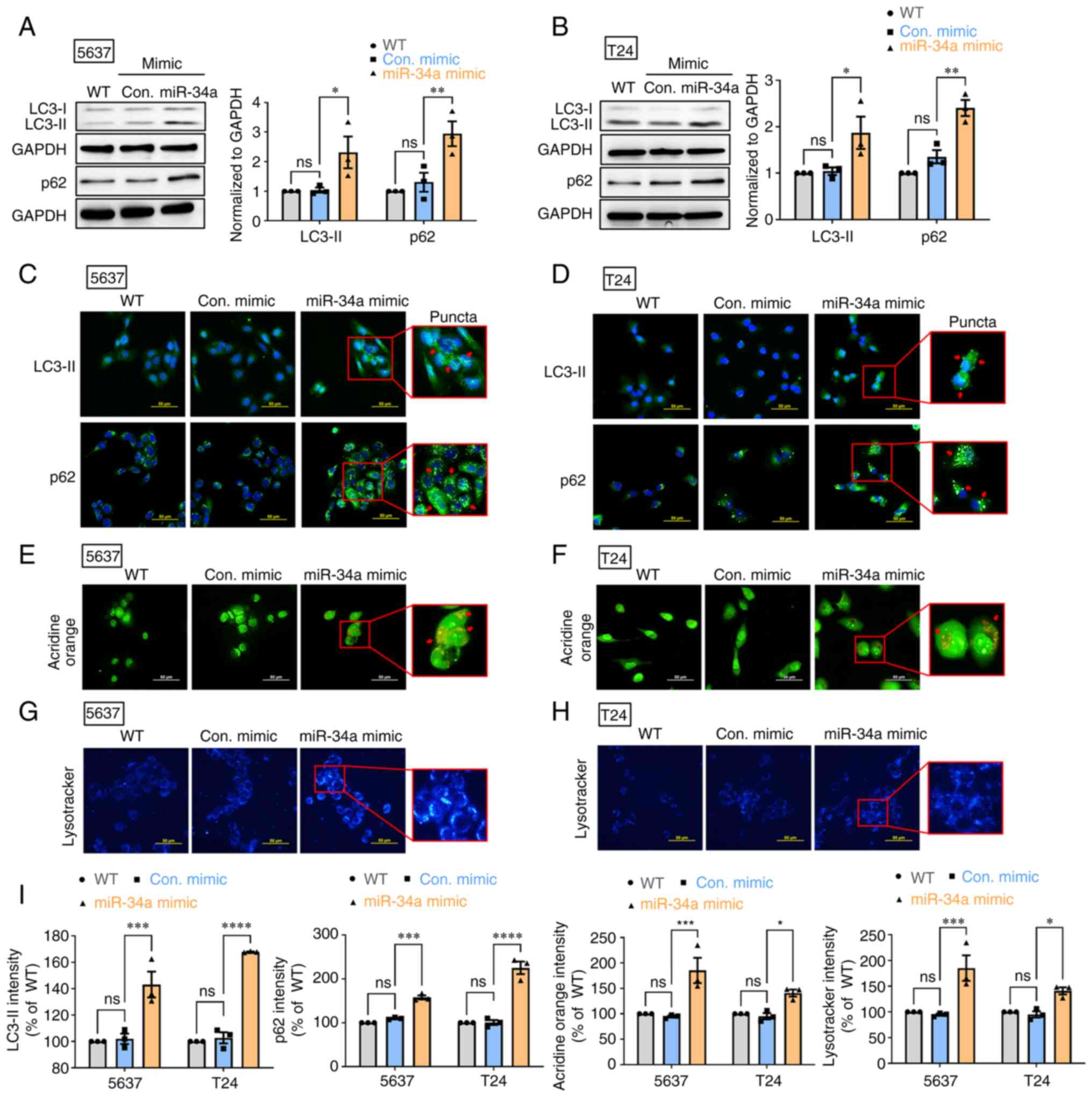
MiR-34a blocks autophagic flux in human
BC cells
Autophagy has been reported to exert a protective
mechanism in BC (35). First, it
was aimed to determine whether miR-34a suppresses protective
autophagy in human BC cells (5637 and T24). The treatment of 5637
and T24 cells with miR-34a mimic induced autophagy marker LC3-II
and p62 protein expression (Fig. 1A
and B). The successful transfection of miR-34a in mimic was
confirmed by RT-qPCR assay (Fig.
S1A). It was found that miR-34a promoted LC3-II and p62
accumulation in cytoplasm to form aberrant LC3-II and p62-positive
puncta (Fig. 1C and D).
Subsequently, AO staining was performed to visualize autophagic
vacuoles after miR-34a treatment. The findings indicated that
miR-34a increased the number of autophagic vacuoles in BC cells
(Fig. 1E and F). The results from
LysoTracker Blue staining confirmed the accumulation of lysosomes
after miR-34a treatment in BC cells (Fig. 1G and H). Fluorescence intensity of
LC3-II, p62, AO and LysoTracker Blue in BC cells is represented in
Fig. 1I. These findings jointly
suggested that miR-34a causes defects in autophagic flux, which
leads to LC3-II and p62 accumulation in the cytoplasm of BC
cells.
MiR-34a inhibits STX17 expression by
integrating with the 3′-UTR regions of STX17 mRNA
The autophagosomelysosome fusion step is
indispensable to autophagic flux and helps cells degrade components
in the cytoplasm (36). Analyzing
a miRNA database, four candidates target of miR-34a were
identified: Ectopic P-granules 5 autophagy tethering factor (EPG5
or known as KIAA1632), synaptosome associated protein 29 (SNAP29),
syntaxin 17 (STX17), and a member of RAS oncogene family-like 1
(RAB7L1; Fig. 2A), which are all
implicated in the process of autophagosome-lysosome fusion
(37). The transfection of BC
cells with miR-34a mimic was observed to decrease STX17 mRNA and
protein expression (Fig. 2B-E);
however, no such effect was identified in mRNA levels of EPG5,
SNAP29 and RAB7L1 expression (Fig. 2B
and C). Subsequently, a luciferase reporter assay was used to
verify that miR-34a directly binds on 3′-UTR regions of STX17 mRNA.
As indicated in Fig. 2F, miR-34a
strongly inhibited luciferase activity; this indicated that miR-34a
directly binds on 3′-UTR of STX17 to inhibit STX17 mRNA
translation.
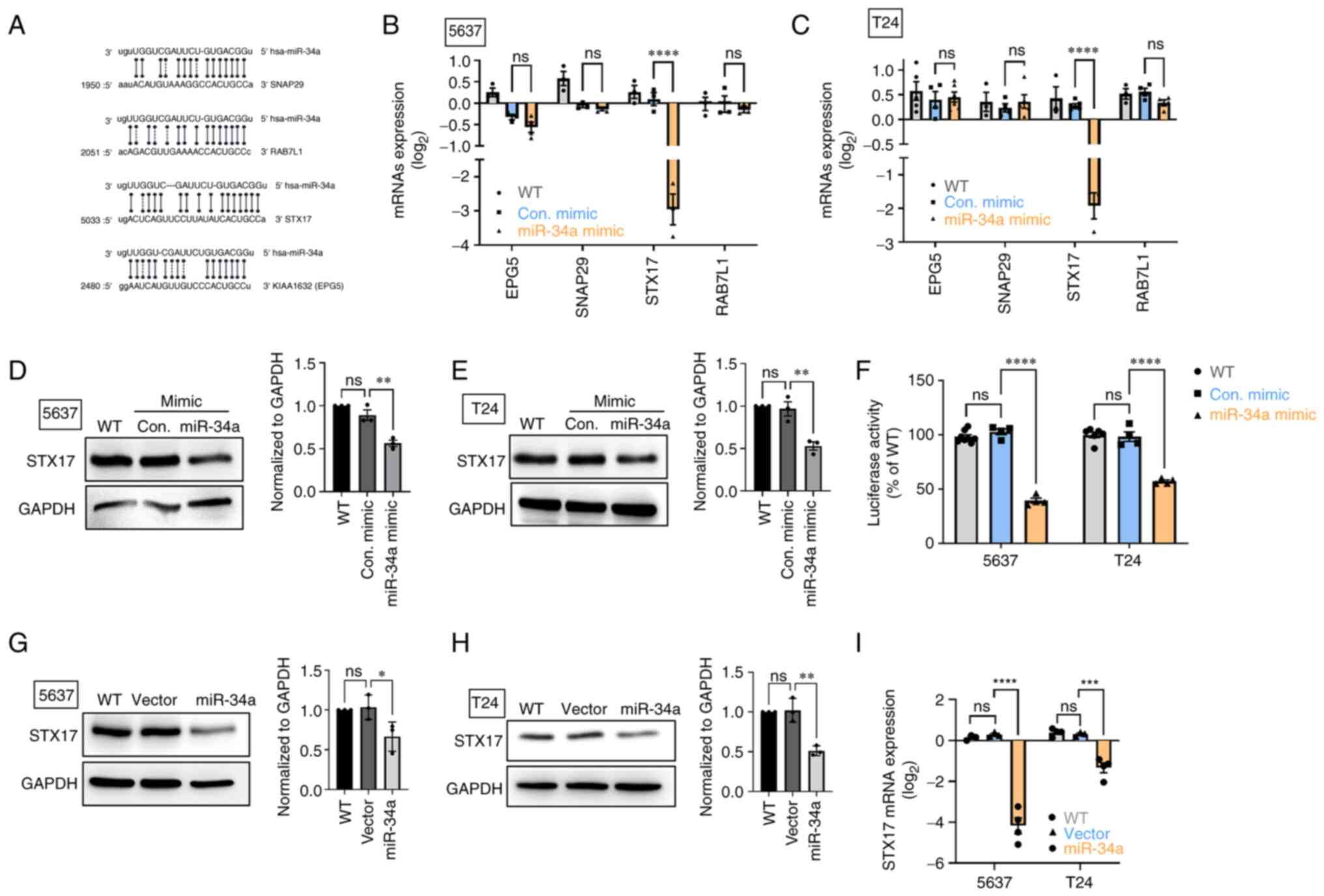 | Figure 2STX17 is involved in the mechanism of
miR-34a-mediated autophagy activity. (A) Positions of the putative
miR-34a binding sites in 3′UTR of SNAP29, RAB7L1, STX17 and EPG5.
(B and C) RT-qPCR assay revealed SNAP29, RAB7L1, STX17 and EPG5
mRNA expression after miR-34a mimic treatment. (D and E) The STX17
protein expression was detected using a western blot assay. (F)
Transfection of bladder cancer cells with STX17-3′-UTR plasmid (1
µg/µl) and miR-34a mimic (100 nM) for 24 h, then
relative luciferase/Renilla activities were measured. (G-I) The
basal levels of STX17 protein and mRNA expression in control stable
and miR-34a stable cells were detected using western blotting and
RT-qPCR assay, respectively. All data are expressed as the mean ±
SD in triplicate samples. *P<0.05,
**P<0.01, ***P<0.001 and
****P<0.0001 relative to the control group. STX17,
syntaxin 17; miR, microRNA; UTR, untranslated region; RT-qPCR,
reverse transcription-quantitative PCR; WT, wild-type; Con,
control; ns, not significant. |
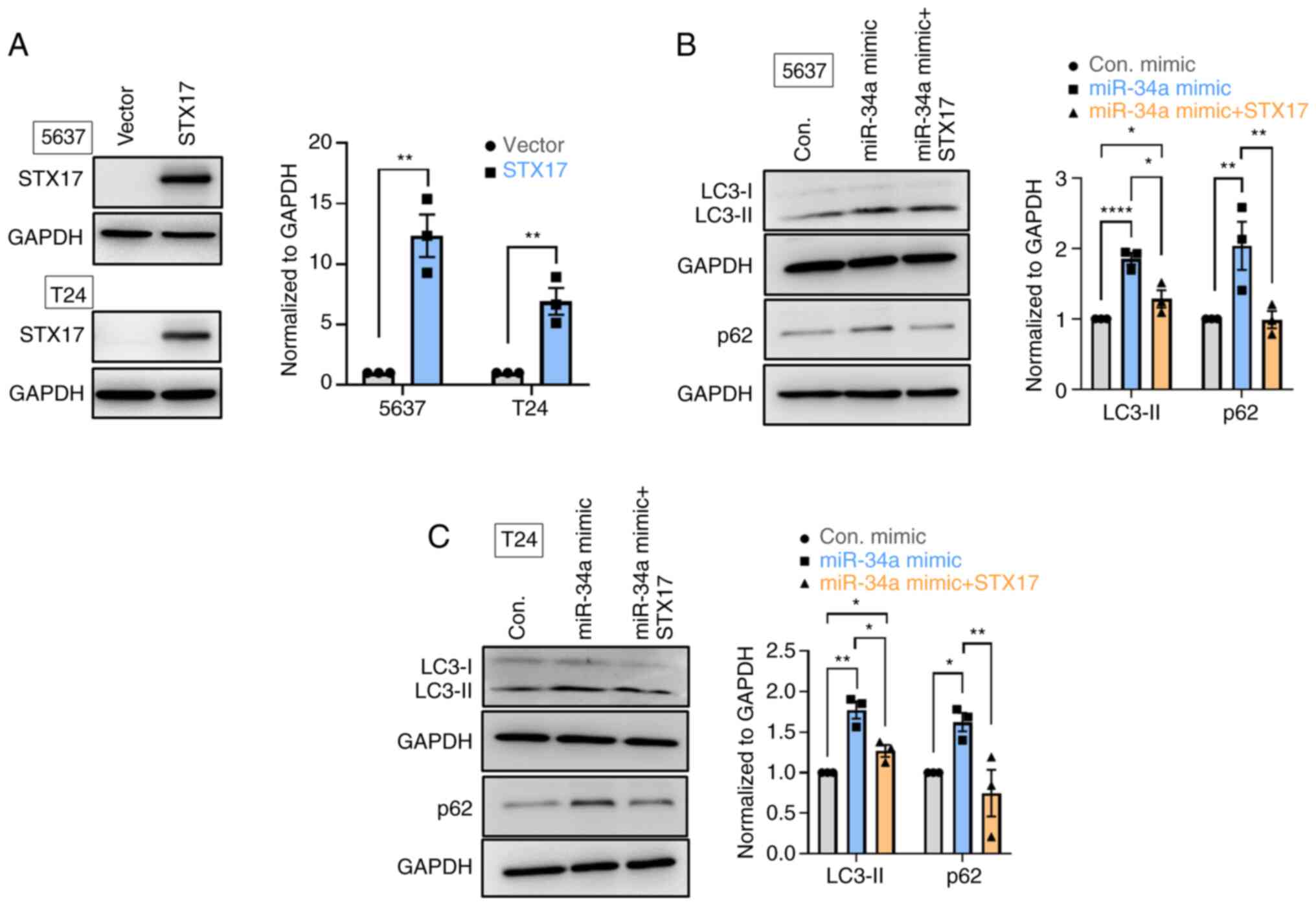
Stable cells that constitutively express miR-34a
were established to verify the effect of miR-34a overexpression in
regulating STX17 expression. The resulting data indicated that
miR-34a exhibited lower mRNA expression and a lower STX17 protein
level compared with an empty vector (Fig. 2G-I). Therefore, miR-34a serves as a
novel autophagic inhibitor through the inhibition of the
autophagosome-lysosome fusion step in BC.
MiR-34a inhibits STX17-mediated autophagy
activity and promotes chemosensitivity
STX17 overexpression was used to verify the function
of STX17 in the miR-34a-inhibited autophagic flux in BC cells. The
overexpression efficiency of STX17 was verified by comparisons with
a vector at protein levels (Fig.
3A). Upregulated expression of LC3-II and reduced p62
expression was observed, confirming autophagic activity during
STX17 overexpression (Fig. S1B).
It was found that miR-34a promoted LC3-II and p62 accumulation in
both 5637 and T24 cells; however, STX17 overexpression inhibited
such accumulation (Fig. 3B and C).
To explore the therapeutic potential of miR-34a, miR-34a and
first-line chemotherapeutic drugs were jointly administered for BC.
Cell viability results indicated that miR-34a promotes the
chemosensitivity of cisplatin, doxorubicin, epirubicin, and
mitomycin C in BC cells; while STX17 overexpression suppresses this
phenomenon (Fig. 4A-D). Colony
formation assay showed that the combinatory treatment of each
chemotherapeutic drug and miR-34a further reduces cell survival as
compared with chemotherapeutic drug alone (Fig. 4E-H). Therefore, the inhibition of
STX17-regulated autophagy by miR-34a sensitizes BC to
chemotherapeutic drug-induced cell death.
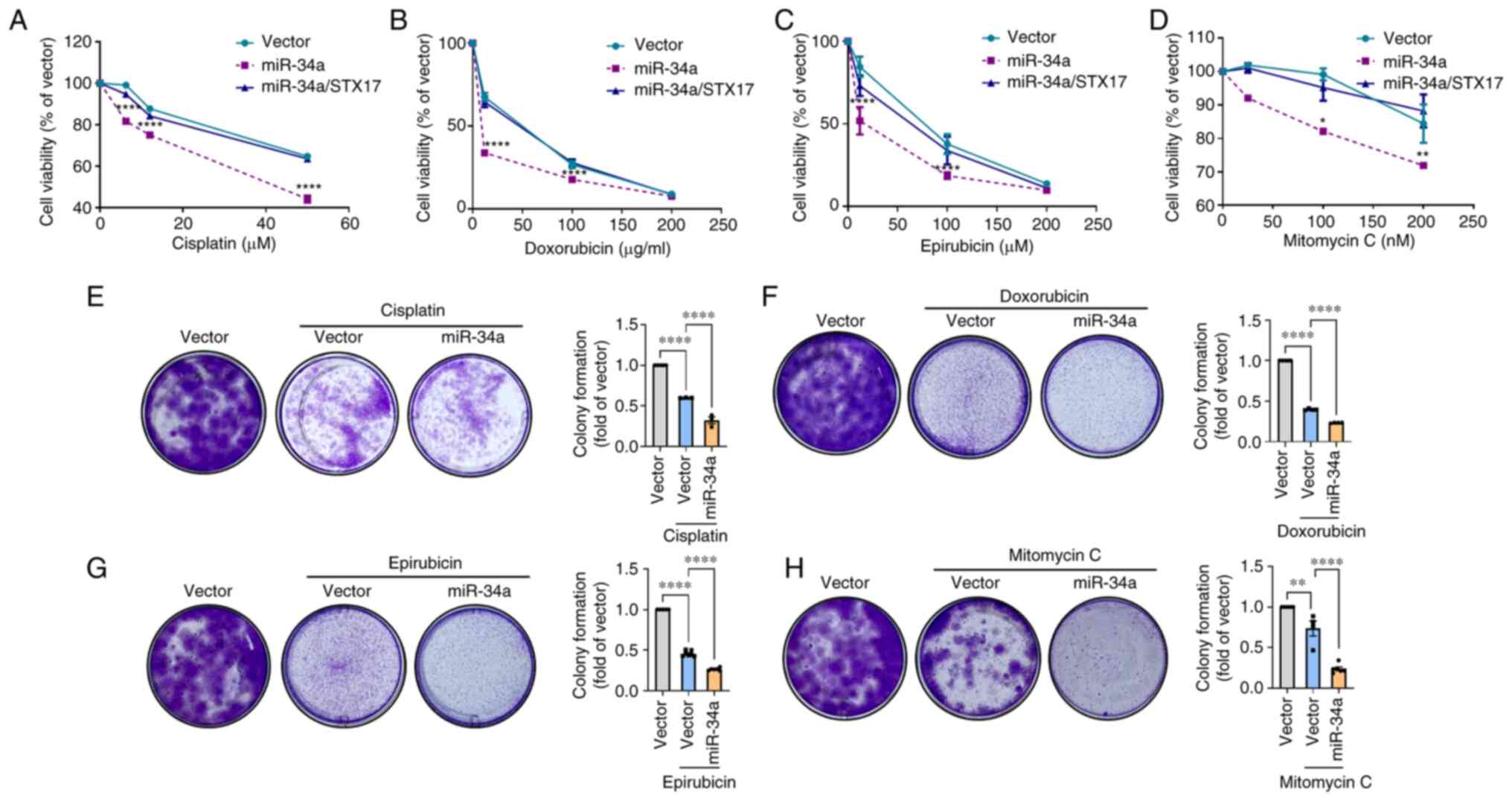 | Figure 4MiR-34a confers chemosensitivity in
BC cells. (A-D) The miR-34a stable cells were transfected with or
without STX17 plasmid (1 µg/µl) for 24 h followed by
exposing to various concentrations of chemotherapeutic drugs,
including cisplatin (0-50 µM), doxorubicin (0-200
µg/ml), epirubicin (0-200 µM) and mitomycin C (0-200
nM). After 24 h of treatment, resazurin-based cell viability was
measured. (E-H) The miR-34a stable cells were treated with
chemotherapeutic drugs, including cisplatin (1.5 µM),
doxorubicin (0.1 µg/ml), epirubicin (0.1 µM) and
mitomycin C (12.5 nM) for 7 days. Cell survival was detected by
colony formation assay. All data are expressed as the mean ± SDs in
triplicate samples. *P<0.05, **P<0.01,
and ****P<0.0001 relative to the control group. miR,
microRNA; BC, bladder cancer; STX17, syntaxin 17. |
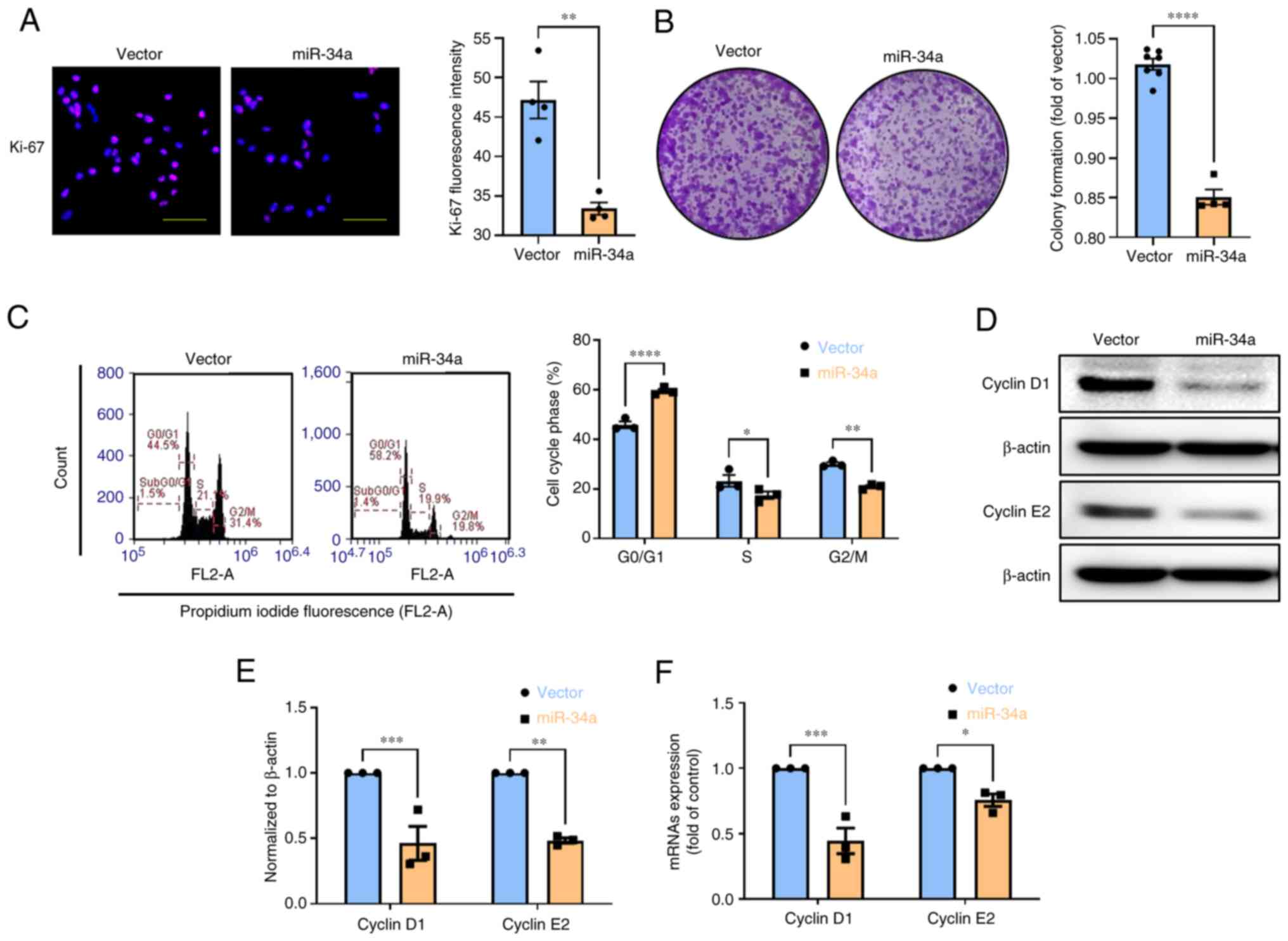
MiR-34a triggers G0/G1 cell cycle arrest
in BC cells
Chronic proliferation and cell migration and
invasion play a critical role in the features of cancer (5), therefore, the antitumor function of
miR-34a on these mechanisms deserves to be thoroughly investigated
in BC. Overexpression of BC cells with miR-34a significantly
decreased proliferation marker Ki-67 expression and colony
formation (Fig. 5A and B), while
STX17 overexpression suppresses this phenomenon (Fig. S1C). Compared with the vector,
miR-34a induced an increase in the G0/G1-phase population from
45.86-59.45%. A decrease in the S-phase and G2/M-phase population
from 23.26-17.48% and from 30.06-20.95%, respectively, was observed
(Fig. 5C). Mechanistically,
miR-34a suppresses the protein and mRNA expression of cyclin D1 and
cyclin E2; this affects G0/1 arrest in BC cells (Fig. 5D-F). Collectively, these data
revealed that miR-34a inhibits cell proliferation through G0/G1
cell cycle arrest in BC.
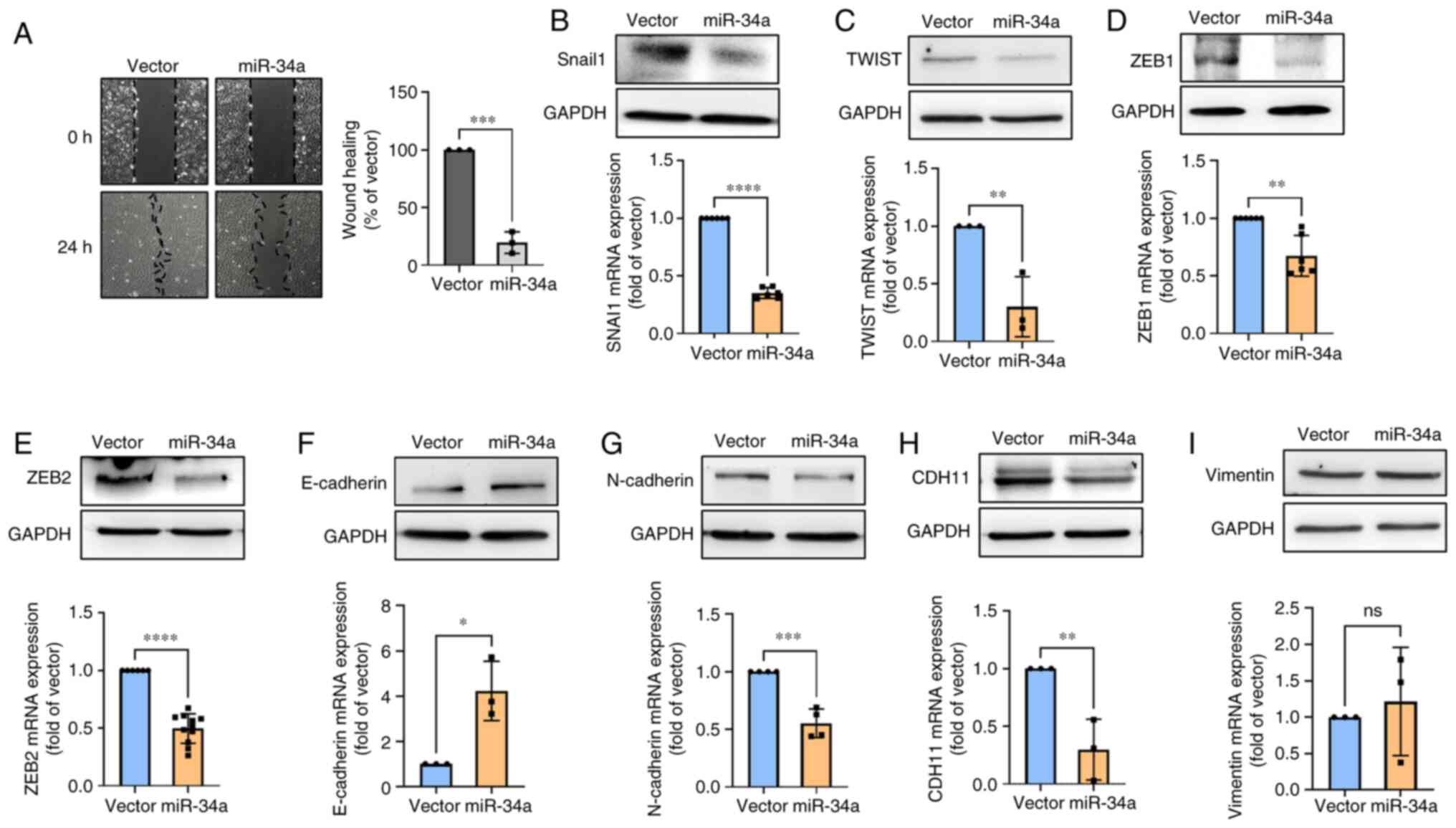
Mir-34a suppresses cell motility through
EMT inhibition
EMT is associated with migration and invasion in
various cancers (38,39). Given the importance of these
mechanisms, it was investigated whether miR-34a inhibits cell
motility through EMT suppression. As revealed in the gap closure
assay, miR-34a group presented the inhibitory action on cell
motility, and the vector group had no such effect (Fig. 6A). However, STX17 overexpression
impeded miR-34a-inhibited cell motility (Fig. S1D), indicating that autophagy is
involved in miR-34a-regulated cell motility of BC. It was found
that miR-34a strongly suppressed the levels of EMT-transcription
factors expression, including snail1, twist, and ZEB1/2 (Fig. 6B-E). Further results indicated that
the expression of the epithelial marker (E-cadherin) was
upregulated (Fig. 6F), whereas the
mesenchymal markers (N-cadherin and cadherin-11) was reduced in BC
(Fig. 6G and H). Vimentin
exhibited no such effect (Fig.
6I). Hence, the present results suggested that miR-34a plays a
critical role in inhibiting cell motility through EMT
suppression.
Discussion
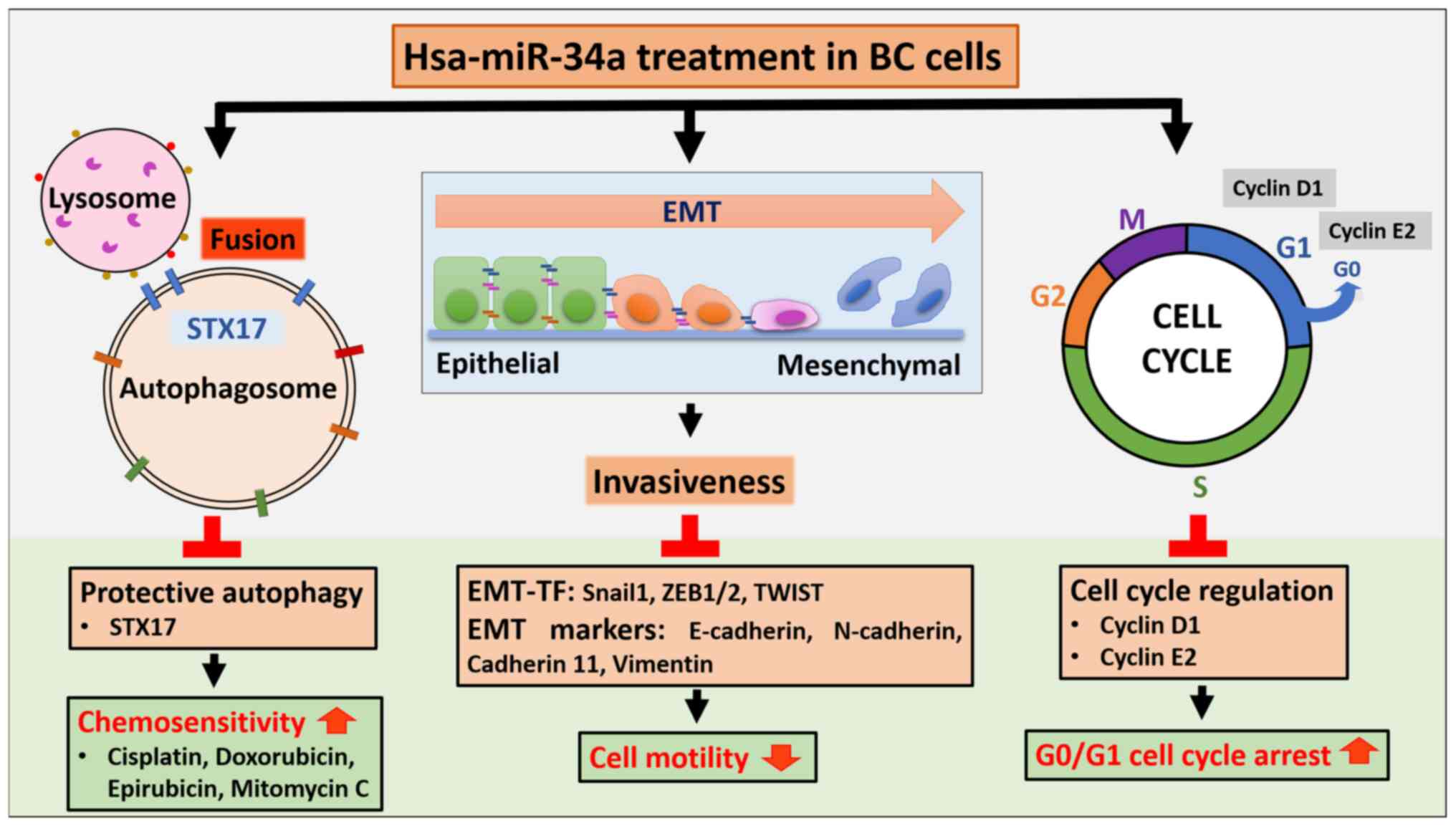
The findings of the present study indicated that
miR-34a exerts potent anticancer effects through three mechanisms:
i) MiR-34a triggers G0/G1 arrest and thus inhibits cell
proliferation; ii) miR-34a suppresses cell motility by EMT
downregulation; and iii) miR-34a directly inhibits STX17 expression
to block autophagic flux in BC, which in turn confers
chemosensitivity.
Multiple targets of miR-34a are verified in human
cancers (25). For example,
miR-34a leads to cell cycle arrest by targeting cyclins and cyclin
dependent-kinases (CDK1, CDK4, CDK6, cyclin D1 and cyclin E2)
(27,40). MiR-34a also negative regulates EMT
activity by targeting EMT-transcription factors ZEB1 (41), Snail (42) and Twist (43) in various cancers, showing the broad
potency of miR-34a as a tumor suppressor. Moreover, miR-34a has
been reported to be involved in tumor-mediated immunosuppression
(44,45). Mechanistically, the overexpression
of miR-34a in cells from acute myeloid leukemia blocks PD-L1
surface expression, which interrupts PD-L1-specific T cell
apoptosis. This indicates that miR-34a may serve a similar function
as anti-CTLA-4 and anti-PD-1/PD-L1 immune checkpoint inhibitors do
(45). Furthermore, miR-34a
inhibits cancer cell proliferation, migration and invasion by
upregulating the tumor-suppressor gene PTEN in BC cells (46) and downregulating proto-oncogene
c-Met in uveal melanoma cells (47). A previous study by the authors
highlights that miR-34a interacts with the 3′-UTR regions of MMP-2
mRNA to silence cell invasiveness in BC (24). Clinically, miR-34a is also
associated with BC based on our analysis of The Cancer Genome Atlas
database-regardless of overall survival, ethnicity, tumor
histology, molecular subtype, clinical stage and nodal metastasis
status-and can thus serve as a BC biomarker (35). These findings considerably advance
our understanding of miR-34a's function as a potential therapeutic
drug for or diagnostic marker of human BC or other types of
cancer.
Protective autophagy induced by antitumor agents was
found to function as a survival pathway to promote therapeutic
resistance in various cancers (35). Numerous preclinical studies have
demonstrated that lysosomal inhibitors of autophagy, such as
chloroquine and hydroxychloroquine (which have already been
approved for clinical use by the US Food and Drug Administration),
sensitize cancer cells to conventional therapy (48-50).
In addition, autophagy has been reported to facilitate focal
adhesion and cell motility through the collaboration of LC3 with
paxillin (51). Autophagy supports
to regulate cell cycle by the degradation of a number of cell cycle
proteins and other signaling adaptors, including cyclins,
cyclin-dependent kinase, hypoxia-inducible factor 1-alpha and
senescence associated protein (52). Therefore, interrupting autophagy
activity by a combination of autophagy inhibitors in cancer therapy
to improve clinical outcomes for patients has become a potential
strategy for future cancer treatments (53). The present findings demonstrated
that miR-34a can serve as a novel autophagic inhibitor by
suppressing STX17 expression in BC cells. The cellular function of
STX17 is known for promoting autophagosomal fusion with endosome or
lysosome (54); however, miR-34a
can directly bind on STX17 3′-UTR to inhibit its protein
translation, which in turn blocks autophagic flux in cancer cells.
It was also observed that miR-34a mimic enhanced the efficacy of
chemotherapeutic drugs, including cisplatin, doxorubicin,
epirubicin and mitomycin C, against BC cells. However, STX17
overexpression rescued cell survival and motility inhibited by
miR-34a (Fig. S1C and D), showing
that autophagy is involved in miR-34a-regulated antitumor functions
of BC. A similar study reported that miR-30a-3p enhances
cisplatin-based chemosensitivity through the inhibition of critical
autophagy-related proteins (ATG5, ATG12 and Beclin1) in BC and has
high efficacy against MIBC through the suppression of matrix
metalloproteinases-mediated cell aggressiveness (18). These preclinical results
demonstrated the potential of protective autophagy-targeting miRNA
therapy.
In 2013, the first and the only clinical trial
(NCT01829971) of liposomal mimic of miR-34a (MRX34) was conducted
to evaluate its safety when used against several solid tumors,
including primary liver cancer, small cell lung cancer, lymphoma,
melanoma, multiple myeloma, renal cell carcinoma and non-small cell
lung cancer. Unfortunately, the clinical trial was terminated due
to the development of five serious immune-related adverse events
upon MRX34 treatment in patients (55). In addition, numerous clinical
trials have been conducted to identify miRNAs as novel diagnostic
and prognostic biomarkers of i) non-Hodgkin's lymphoma and acute
leukemia (NCT05477667), ii) hormone-sensitive breast cancer
(NCT01612871) (56), and iii)
non-MIBC (NCT03591367). Similar clinical trials are ongoing. Such
extensive research efforts suggest the potential of miRNA therapy
for cancer.
In conclusion, the present findings revealed new
clinical potential for miR-34a as an antitumor agent in the
treatment of human BC through blocking the cell cycle, inhibiting
motility and impairing protective autophagy. MiR-34a can be
administered with anti-BC chemotherapeutic drugs to improve their
efficacy (Fig. 7).
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study available from the corresponding author on reasonable
request.
Authors' contributions
TIH and ACC conceptualized the study. TFT and KYC
curated data. TFT and CYH performed formal analysis. TIH, Ye-CC and
ACC conducted investigation. KYC, Yu-CC and ACC developed
methodology. Yu-CC, HEC and CYH were involved in project
administration. Ye-CC, PCC and TFT managed resources. PHC, Yu-CC
and PCC performed software analysis. HEC and TIH supervised the
study. PCC and ACC wrote the original draft. ACC and TIH wrote,
reviewed and edited the manuscript. ACC and TIH confirm the
authenticity of all the raw data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Ministry of Science and
Technology, Taiwan (grant nos. MOST-110 -2314-B-341-001-MY2,
MOST-110-2314-B-341-004 and MOST-111-2314-B-341-004) and Shin Kong
Wu Ho-Su Memorial Hospital (grant nos. 2021SKHBDR003 and
2022SKHBND002).
References
|
1
|
Sanli O, Dobruch J, Knowles MA, Burger M,
Alemozaffar M, Nielsen ME and Lotan Y: Bladder cancer. Nat Rev Dis
Primers. 3:170222017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen Z, Liu Q, Chen R, Liu Z, Li M, Ling
Q, Wu L, Yang J, Liu X, Wang T, et al: Clinical analysis of small
cell carcinoma of the bladder in Chinese: Nine case reports and
literature reviews. World J Surg Oncol. 15:332017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao L, Sun J, Wang K, Tai S, Hua R, Yu Y,
Fan Y and Huang J: Development of a new recurrence-free survival
prediction nomogram for patients with primary non-muscle-invasive
bladder cancer based on preoperative controlling nutritional status
score. Cancer Manag Res. 13:6473–6487. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Breau RH, Karnes RJ, Farmer SA, Thapa P,
Cagiannos I, Morash C and Frank I: Progression to detrusor muscle
invasion during urothelial carcinoma surveillance is associated
with poor prognosis. BJU Int. 113:900–906. 2014. View Article : Google Scholar
|
|
5
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Medina MA, Oza G, Sharma A, Arriaga LG,
Hernández Hernández JM, Rotello VM and Ramirez JT: Triple-negative
breast cancer: A review of conventional and advanced therapeutic
strategies. Int J Environ Res Public Health. 17:20782020.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roche J: The epithelial-to-mesenchymal
transition in cancer. Cancers (Basel). 10:522018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mittal V: Epithelial mesenchymal
transition in tumor metastasis. Annu Rev Pathol. 13:395–412. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang T, Song X, Yang Y, Wan X, Alvarez
AA, Sastry N, Feng H, Hu B and Cheng SY: Autophagy and hallmarks of
cancer. Crit Rev Oncog. 23:247–267. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
White E and DiPaola RS: The double-edged
sword of autophagy modulation in cancer. Clin Cancer Res.
15:5308–5316. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lin YC, Lin JF, Wen SI, Yang SC, Tsai TF,
Chen HE, Chou KY and Hwang TI: Inhibition of high basal level of
autophagy induces apoptosis in human bladder cancer cells. J Urol.
195:1126–1135. 2016. View Article : Google Scholar
|
|
12
|
Chou KY, Chen PC, Chang AC, Tsai TF, Chen
HE, Ho CY and Hwang TI: Attenuation of chloroquine and
hydroxychloroquine on the invasive potential of bladder cancer
through targeting matrix metalloproteinase 2 expression. Environ
Toxicol. 36:2138–2145. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Quan Y, Lei H, Wahafu W, Liu Y, Ping H and
Zhang X: Inhibition of autophagy enhances the anticancer effect of
enzalutamide on bladder cancer. Biomed Pharmacother.
120:1094902019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O'Brien J, Hayder H, Zayed Y and Peng C:
Overview of MicroRNA biogenesis, mechanisms of actions, and
circulation. Front Endocrinol (Lausanne). 9:4022018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Friedman RC, Farh KK, Burge CB and Bartel
DP: Most mammalian mRNAs are conserved targets of microRNAs. Genome
Res. 19:92–105. 2009. View Article : Google Scholar :
|
|
16
|
Fu G, Brkić J, Hayder H and Peng C:
MicroRNAs in human placental development and pregnancy
complications. Int J Mol Sci. 14:5519–5544. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar :
|
|
18
|
Hwang TI, Chen PC, Tsai TF, Lin JF, Chou
KY, Ho CY, Chen HE and Chang AC: Hsa-miR-30a-3p overcomes the
acquired protective autophagy of bladder cancer in chemotherapy and
suppresses tumor growth and muscle invasion. Cell Death Dis.
13:3902022. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Angelucci F, Cechova K, Valis M, Kuca K,
Zhang B and Hort J: MicroRNAs in Alzheimer's disease: Diagnostic
markers or therapeutic agents? Front Pharmacol. 10:6652019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang CC, Tseng TT, Liu SC, Lin YY, Law
YY, Hu SL, Wang SW, Tsai CH and Tang CH: S1P increases VEGF
production in osteoblasts and facilitates endothelial progenitor
cell angiogenesis by inhibiting miR-16-5p expression via the
c-Src/FAK signaling pathway in rheumatoid arthritis. Cells.
10:21682021. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Çakmak HA and Demir M: MicroRNA and
cardiovascular diseases. Balkan Med J. 37:60–71. 2020.PubMed/NCBI
|
|
22
|
Zhang B, Pan X, Cobb GP and Anderson TA:
microRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar
|
|
23
|
Dalmay T and Edwards DR: MicroRNAs and the
hallmarks of cancer. Oncogene. 25:6170–6175. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chou KY, Chang AC, Tsai TF, Lin YC, Chen
HE, Ho CY, Chen PC and Hwang TI: MicroRNA-34a-5p serves as a tumor
suppressor by regulating the cell motility of bladder cancer cells
through matrix metalloproteinase-2 silencing. Oncol Rep.
45:911–920. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang L, Liao Y and Tang L: MicroRNA-34
family: A potential tumor suppressor and therapeutic candidate in
cancer. J Exp Clin Cancer Res. 38:532019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li WJ, Wang Y, Liu R, Kasinski AL, Shen H,
Slack FJ and Tang DG: MicroRNA-34a: Potent tumor suppressor, cancer
stem cell inhibitor, and potential anticancer therapeutic. Front
Cell Dev Biol. 9:6405872021. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen F and Hu SJ: Effect of microRNA-34a
in cell cycle, differentiation, and apoptosis: A review. J Biochem
Mol Toxicol. 26:79–86. 2012. View Article : Google Scholar
|
|
28
|
Wang X, Zhao Y, Lu Q, Fei X, Lu C, Li C
and Chen H: MiR-34a-5p inhibits proliferation, migration, invasion
and epithelial-mesenchymal transition in esophageal squamous cell
carcinoma by targeting LEF1 and inactivation of the Hippo-YAP1/TAZ
signaling pathway. J Cancer. 11:3072–3081. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yang S, Li Y, Gao J, Zhang T, Li S, Luo A,
Chen H, Ding F, Wang X and Liu Z: MicroRNA-34 suppresses breast
cancer invasion and metastasis by directly targeting Fra-1.
Oncogene. 32:4294–4303. 2013. View Article : Google Scholar
|
|
30
|
Wu X, Cheng YL, Matthen M, Yoon A,
Schwartz GK, Bala S, Taylor AM and Momen-Heravi F: Down-regulation
of the tumor suppressor miR-34a contributes to head and neck cancer
by up-regulating the MET oncogene and modulating tumor immune
evasion. J Exp Clin Cancer Res. 40:702021. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ho CY, Chang AC, Hsu CH, Tsai TF, Lin YC,
Chou KY, Chen HE, Lin JF, Chen PC and Hwang TI: Miconazole induces
protective autophagy in bladder cancer cells. Environ Toxicol.
36:185–193. 2021. View Article : Google Scholar
|
|
32
|
Chuang JY, Chang AC, Chiang IP, Tsai MH
and Tang CH: Apoptosis signal-regulating kinase 1 is involved in
WISP-1-promoted cell motility in human oral squamous cell carcinoma
cells. PLoS One. 8:e780222013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tsai HC, Chang AC, Tsai CH, Huang YL, Gan
L, Chen CK, Liu SC, Huang TY, Fong YC and Tang CH: CCN2 promotes
drug resistance in osteosarcoma by enhancing ABCG2 expression. J
Cell Physiol. 234:9297–9307. 2019. View Article : Google Scholar
|
|
34
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
35
|
Lin JF, Lin YC, Tsai TF, Chen HE, Chou KY
and Hwang TI: Cisplatin induces protective autophagy through
activation of BECN1 in human bladder cancer cells. Drug Des Devel
Ther. 11:1517–1533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Parzych KR and Klionsky DJ: An overview of
autophagy: Morphology, mechanism, and regulation. Antioxid Redox
Signal. 20:460–473. 2014. View Article : Google Scholar :
|
|
37
|
Lőrincz P and Juhász G:
Autophagosome-lysosome fusion. J Mol Biol. 432:2462–2482. 2020.
View Article : Google Scholar
|
|
38
|
Chang AC, Lien MY, Tsai MH, Hua CH and
Tang CH: WISP-1 promotes epithelial-mesenchymal transition in oral
squamous cell carcinoma cells via the miR-153-3p/Snail axis.
Cancers (Basel). 11:19032019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang C, Dou R, Wei C, Liu K, Shi D, Zhang
C, Liu Q, Wang S and Xiong B: Tumor-derived exosomal
microRNA-106b-5p activates EMT-cancer cell and M2-subtype TAM
interaction to facilitate CRC metastasis. Mol Ther. 29:2088–2107.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sun F, Fu H, Liu Q, Tie Y, Zhu J, Xing R,
Sun Z and Zheng X: Downregulation of CCND1 and CDK6 by miR-34a
induces cell cycle arrest. FEBS Lett. 582:1564–1568. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xu Y, Guo B, Liu X and Tao K: miR-34a
inhibits melanoma growth by targeting ZEB1. Aging (Albany NY).
13:15538–15547. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Siemens H, Jackstadt R, Hünten S, Kaller
M, Menssen A, Götz U and Hermeking H: miR-34 and SNAIL form a
double-negative feedback loop to regulate epithelial-mesenchymal
transitions. Cell Cycle. 10:4256–4271. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Imani S, Wei C, Cheng J, Khan MA, Fu S,
Yang L, Tania M, Zhang X, Xiao X, Zhang X and Fu J: MicroRNA-34a
targets epithelial to mesenchymal transition-inducing transcription
factors (EMT-TFs) and inhibits breast cancer cell migration and
invasion. Oncotarget. 8:21362–21379. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Tsai TF, Chang AC, Chen PC, Ho CY, Chen
HE, Chou KY and Hwang TI: Autophagy blockade potentiates
cancer-associated immunosuppression through programmed death
ligand-1 upregulation in bladder cancer. J Cell Physiol.
237:3587–3597. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang X, Li J, Dong K, Lin F, Long M,
Ouyang Y, Wei J, Chen X, Weng Y, He T and Zhang H: Tumor suppressor
miR-34a targets PD-L1 and functions as a potential
immunotherapeutic target in acute myeloid leukemia. Cell Signal.
27:443–452. 2015. View Article : Google Scholar
|
|
46
|
Ding ZS, He YH, Deng YS, Peng PX, Wang JF,
Chen X, Zhao PY and Zhou XF: MicroRNA-34a inhibits bladder cancer
cell migration and invasion, and upregulates PTEN expression. Oncol
Lett. 18:5549–5554. 2019.PubMed/NCBI
|
|
47
|
Yan D, Zhou X, Chen X, Hu DN, Dong XD,
Wang J, Lu F, Tu L and Qu J: MicroRNA-34a inhibits uveal melanoma
cell proliferation and migration through downregulation of c-Met.
Invest Ophthalmol Vis Sci. 50:1559–1565. 2009. View Article : Google Scholar
|
|
48
|
Helmy SA, El-Mesery M, El-Karef A, Eissa
LA and El Gayar AM: Chloroquine upregulates TRAIL/TRAILR2
expression and potentiates doxorubicin anti-tumor activity in
thioacetamide-induced hepatocellular carcinoma model. Chem Biol
Interact. 279:84–94. 2018. View Article : Google Scholar
|
|
49
|
Liu X, Sun K, Wang H and Dai Y: Inhibition
of autophagy by chloroquine enhances the antitumor efficacy of
sorafenib in glioblastoma. Cell Mol Neurobiol. 36:1197–1208. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Verbaanderd C, Maes H, Schaaf MB, Sukhatme
VP, Pantziarka P, Sukhatme V, Agostinis P and Bouche G: Repurposing
drugs in oncology (ReDO)-chloroquine and hydroxychloroquine as
anti-cancer agents. Ecancermedicalscience. 11:7812017. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sharifi MN, Mowers EE, Drake LE, Collier
C, Chen H, Zamora M, Mui S and Macleod KF: Autophagy promotes focal
adhesion disassembly and cell motility of metastatic tumor cells
through the direct interaction of paxillin with LC3. Cell Rep.
15:1660–1672. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zheng K, He Z, Kitazato K and Wang Y:
Selective autophagy regulates cell cycle in cancer therapy.
Theranostics. 9:104–125. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liu T, Zhang J, Li K, Deng L and Wang H:
Combination of an autophagy inducer and an autophagy inhibitor: A
smarter strategy emerging in cancer therapy. Front Pharmacol.
11:4082020. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Itakura E, Kishi-Itakura C and Mizushima
N: The hairpin-type tail-anchored SNARE syntaxin 17 targets to
autophagosomes for fusion with endosomes/lysosomes. Cell.
151:1256–1269. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Hong DS, Kang YK, Borad M, Sachdev J,
Ejadi S, Lim HY, Brenner AJ, Park K, Lee JL, Kim TY, et al: Phase 1
study of MRX34, a liposomal miR-34a mimic, in patients with
advanced solid tumours. Br J Cancer. 122:1630–1637. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Jacot W, Dalenc F, Lopez-Crapez E,
Chaltiel L, Durigova A, Gros N, Lozano N, Lacaze JL, Pouderoux S,
Gladieff L, et al: PIK3CA mutations early persistence in cell-free
tumor DNA as a negative prognostic factor in metastatic breast
cancer patients treated with hormonal therapy. Breast Cancer Res
Treat. 177:659–667. 2019. View Article : Google Scholar : PubMed/NCBI
|