1. Introduction
Nuclear receptors (NRs) are ligand-activated
transcription factors involved in main cellular processes including
homeostasis, metabolism, growth, differentiation and development
(1). These receptors interact
with specific DNA sequences in the promoter and enhancer regions of
their target genes modulating their transcription by co-binding to
a series of co-factors/coregulators belonging to transcription
initiation machinery (2). The NR
superfamily can be divided into three classes based on the type of
ligand: Endocrine, metabolic and orphan NRs (3). The endocrine NR subfamily includes
estrogen receptors (ERs) αlpha (ERα or ESR1) and ERβ (ERβ or ESR2),
androgen receptors (AR), progesterone receptors, glucocorticoid
receptors, mineralocorticoid receptors, vitamin D receptors (VDRs),
retinoic acid receptors (RARs), as well as thyroid hormone
receptors (THRs) (1).
Numerous factors contribute to the regulation of
NRs, especially the recruitment of coregulatory proteins,
post-translational modifications and interactions with other
transcription factors.
Dysregulated NR signaling can promote a large series
of pathological processes. In particular, mutation or aberrant
expression of NR and/or of their coregulators may influence both
the development and progression of several human diseases including
cancer (4). Moreover, epigenetic
regulation, especially by activation of non-coding RNA molecules
(ncRNAs), represents an element of complexity in the modulation of
NR-dependent gene expression.
Long non-coding (lnc) RNAs are non-coding molecules
longer than 200 bp, able to interact with mRNA, DNA, protein and
microRNA (miRNA or miR) (5). They
have different regulatory functions in humans: i) Regulation of
histone modifications at the chromatin level by interaction with
histone-modified complexes or enzymes; ii) transcriptional
regulation and post-transcriptional regulation; iii) miRNA sponge;
iv) RNA stability; v) Protein relocalization and vi)
post-translation modifications (6).
During cancer evolution, lncRNAs may regulate cell
proliferation, apoptosis, migration, invasion, stem-like phenotype
and remodeling of the tumor microenvironment (TME) (7). Previous studies have shown that
lncRNAs can function as positive or negative regulators of
NR-dependent gene expression, and in turn, NRs can regulate
different lncRNAs. This crosstalk appears to be particularly
important in the modulation of oncogenic processes (8,9).
The steroid receptor RNA activator (SRA) was
identified as the first lncRNA able to bind and increase the
activity of steroid receptors (10) (Fig.
1). SRA is able to interact with numerous proteins acting as a
'scaffold' for the assembly of other coregulatory proteins that
direct NR-dependent transcription (11-14).
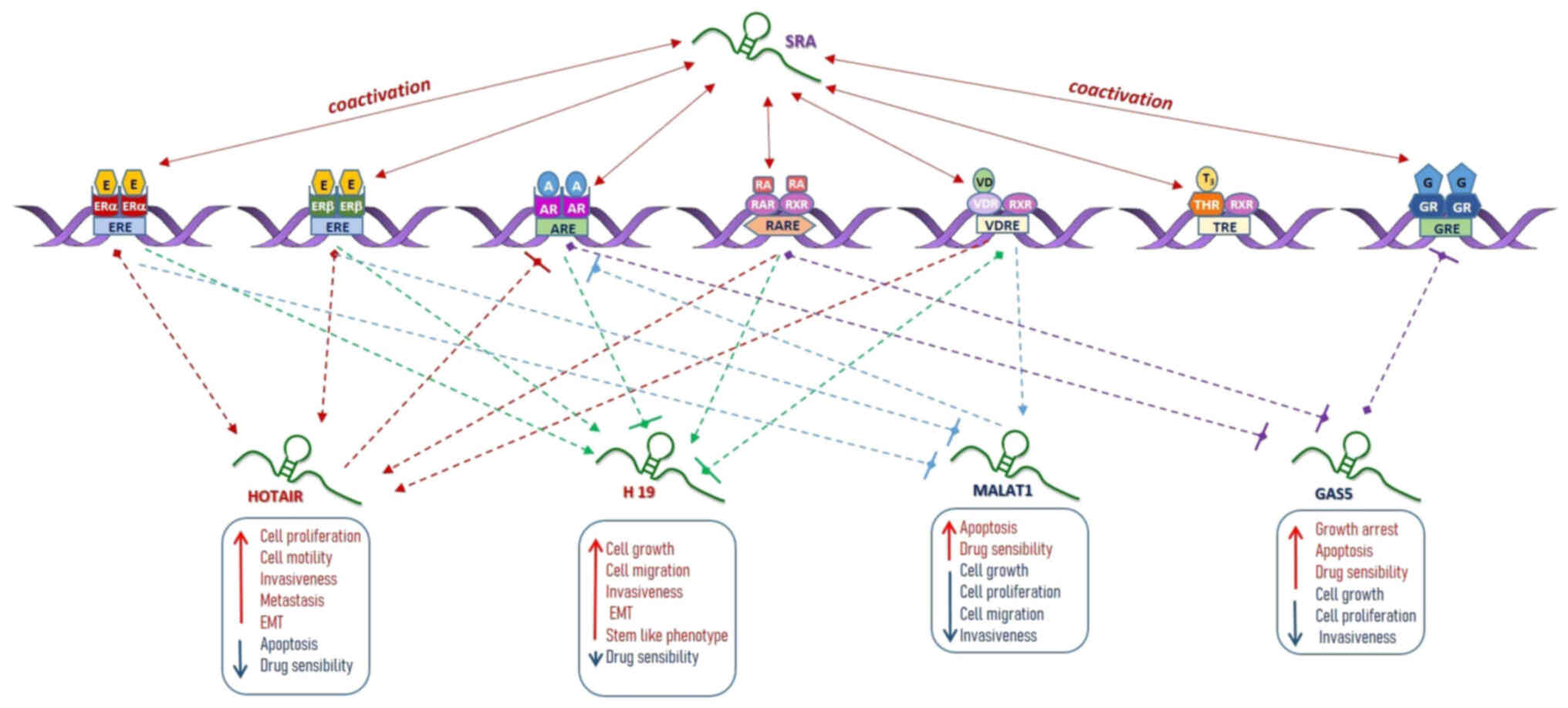 | Figure 1Schematic representation of the
reciprocal functional interactions between nuclear receptors and
the best-characterized lncRNAs in tumor cells. The lncRNAs steroid
receptor RNA activator represents a coactivator for all NRs, which
is able to act as a scaffold for the assembly of other coregulators
to modulate NR-dependent transcription, in human cancer. The
lncRNAs HOTAIR and H1 mostly behave as oncogenes in cancer cells.
HOTAIR is able to interact with most of the NRs. It contains in its
promoter multiple estrogen response elements and can be
transcriptionally induced by estradiol. HOTAIR crosstalk with ERα
and ERβ promotes cell proliferation, migration, invasion,
metastasis and EMT and reduces apoptosis and drug sensibility in
breast and prostate cancer cells. HOTAIR crosstalk with AR leads to
a formation of a suppressive complex that decreased AR
transcription and promotes cell invasion in prostate cancer cells.
HOTAIR crosstalk with RAR can induce myeloid cell differentiation
in acute myeloid leukemia cell lines. HOTAIR crosstalk with VDR
promotes cell growth and proliferation in keratinocyte carcinoma
cells. Similarly, the lncRNA H19 is able to interact with most NRs.
Its crosstalk with ERα can modulate cell growth, proliferation,
invasion, EMT features, as well as drug resistance through Notch
and c-Met activation in breast and prostate cancer cells. Lastly,
H19 can be regulated not only by estrogens but also by hypoxia. H19
crosstalk with ERβ can promote stem-like features, especially in
papillary thyroid cancer cells, while its crosstalk with RAR can
modulate telomerase activity in promyelocytic leukemia cells.
However, H19 expression is negatively regulated by androgen/AR
signaling in prostate cancer cells, and by VDR signaling in colon
cancer cells. Unlike HOTAIR and H19, lncRNAs MALAT1 and GAS5 can be
considered tumor suppressor genes. MALAT1 crosstalk with ERα and
ERβ leads to a reduction of its expression which translates into
effects on cell growth, proliferation and invasion feature,
especially in breast cancer cells. On the contrary, androgen/AR
signaling is able to induce MALAT1 expression promoting apoptosis
in breast cancer cells by miRNA-320b. Moreover, VDR silencing is
able to increase MALAT1 expression, thus inducing PI3K/Akt
signaling and promoting growth and proliferation in epidermal
keratinocytes. GAS5 crosstalk with AR and RAR can modulate cell
proliferation in prostate and glioblastoma cancer cells. GAS5 is
also capable to interact with GR inhibiting its transcription and
consequently promoting growth arrest and apoptosis in breast cancer
cells. lncRNA, long non-coding RNA; NR, nuclear receptor; HOTAIR,
HOX antisense intergenic RNA; ER, estrogen receptor; EMT,
epithelial-mesenchymal transition; AR, androgen receptor; RAR,
retinoic acid receptor; VDR, vitamin D receptor; MALAT1,
metastasis-associated lung adenocarcinoma transcript-1. |
In addition to the role of signaling by NR, numerous
studies revealed that SRA is involved in other physiological
functions: i) Modulation of adipogenesis via direct binding to and
promoting the transcriptional activity of peroxisome proliferator
activated receptor γ, ii) regulation of steroidogenesis and adrenal
biology by binding and activation of SF-1 as well as
dosage-sensitive sex reversal-adrenal hypoplasia protein, iii)
promotion of muscle differentiation by increasing the activity of
myogenic differentiation 1 protein (12), iv) induction of proliferation and
migration of vascular smooth muscle cells by stimulating the
phosphorylation of mitogen-activated protein kinase, extracellular
signal-regulated kinases and cAMP response element-binding protein
CREB (15), v) promotion of
melanin pigmentation process by modulation of p38 and
phosphoribosyl-anthranilate isomerase gene expression (16); and vi) modulation of insulin
signaling and regulation of β-oxidation (17).
Instead, aberrant SRA expression and mutant variants
have been identified in several pathological conditions, not only
in hormone-driven tumors (Fig. 1)
(12). SRA promotes cervical
cancer cell migration and invasion via upregulation of matrix
metalloproteinase-9, matrix metalloproteinase-2, vascular
endothelial growth factor A (VEGFA) expression and NOTCH receptor 1
signaling pathways (18). Park
et al (19) revealed that
SRA is involved in the proliferation, migration and invasion of
endometrial cancer cells by increasing the expression of EIF4E-BP1
and Wnt/β-catenin signaling activity. SRA mediates p38 activation,
cell invasion along with proliferation, regulates
epithelial-mesenchymal transition (EMT) and distant metastasis in
melanoma (20) and induces tumor
progression as well as drug resistance in colorectal cancer by
oxidative phosphorylation pathway genes (21). In hormone-driven cancers, SRA is
directly involved in the mechanisms underlying breast tumorigenesis
and tumor progression (22), and
their isoforms are capable of enhancing AR activities in prostate
cancer cells (23).
More recently, Kim et al (24) demonstrated that SRA is able to
regulate cell migration, proliferation, and invasion of ovarian
cancer cells by modulation of EMT and expression of NOTCH-related
genes.
Given the importance of NRs in hormone-driven
cancers, a deeper understanding of epigenetic regulatory mechanisms
in human tumors may offer new scenarios for new diagnostic and
predictive biomarkers. In the present review, it was proposed to
schematize the crosstalk existing between the main endocrine NR and
the lncRNAs in tumor initiation and progression.
2. Crosstalk between ERα and lncRNAs in
cancer
Two different ER subtypes were identified: i) The
NRs ERα and ERβ, and ii) the membrane receptor G protein-coupled ER
1 (GPER1). Their expression is tissue-specific. ERα, has 3 isoforms
and truncated isoforms can suppress ERα activity by
heterodimerizing with full-length isoforms (25,26). It is mainly expressed in female
sex organs, such as the breast, ovaries and uterus as well as
involved in the regulation of cell proliferation in addition to
apoptosis in cancer cells (27).
Mechanistically, ERα translocates from the cytoplasm to the
nucleus, upon binding to 17β-oestradiol (E2), and binds estrogen
response elements (EREs) in the ERα-dependent gene promoter
(28).
Breast cancer (BC)
Most of the data highlights in particular the
crosstalk between ERα and lncRNAs in BC. In BC, ERα can be
activated by estrogen in luminal subtypes, but its aberrant
activity in a hormone-independent manner has also been described in
basal-type subtypes (29).
Numerous estrogen-regulated lncRNAs have been identified in BC. HOX
antisense intergenic RNA (HOTAIR) is transcriptionally induced by
E2 and it is involved in the modulation of chromatin-modifying
enzymes in addition to the regulation of gene silencing (30). ERs along with a series of
coregulators (histone methylases MLL1, MLL3 and CREB-binding
protein/p300) bind to the promoter of HOTAIR, containing multiple
functional EREs, in an E2-dependent manner (30). This interaction potentially
contributes to BC evolution and progression (31,32). Gupta et al (33) described that aberrant expression
of HOTAIR can affect cell motility and matrix invasion in BC cells,
and its silencing in doxorubicin-resistant cells decreases cell
proliferation and increases apoptosis (Fig. 1) (33). However, estrogen promoted HOTAIR
through its membrane receptor GPR30 by the suppression of miR-148a
(34). This highlights that the
mutual modulation mechanism between ERα and HOTAIR is very complex
and implies the simultaneous activation of other molecular
networks.
Similar to HOTAIR, metastasis-associated lung
adenocarcinoma transcript-1 (MALAT1) is regulated by estrogen and
it associates with EREs along with the polycomb repressive complex
1 (PRC1) (35). MALAT1 binding to
chromatin and other components can be reduced in presence of
estrogen, indicating it has an inhibitory role in regulating
estrogen target genes. Zhao et al (36) demonstrated that high-concentration
E2 treatment largely decreased MALAT-1 RNA level. Silencing of
MALAT1 showed similar effects on the proliferation, migration and
invasion of BC cells. These data highlighted that E2 treatment
affects BC cells proliferation, migration and invasion in an
ERα-independent, via a dose-dependent way by decreasing the MALAT1
RNA level (Fig. 1) (36). Previously, a meta-analysis study
revealed that high MALAT1 expression is associated with poor
relapse-free survival (37) and
its aberrant expression can be related to Oncotype DX 21-gene test,
to predict the recurrence score, in patients with ER+
tumors and early BC (38). These
data strongly indicated the role of MALAT-1 as an important
prognostic marker in patients with BC.
Myocardial infarction-associated transcript (MIAT)
is overexpressed in ER+ BC tissues especially in
aggressive ductal BC (39). In
these cells, MIAT expression is induced by E2 through ERα
activation. Silencing MIAT inhibited cell cycle progression and
induced apoptosis, whereas its upregulation increased cell
proliferation in MCF-7 cells (39). Moreover, Luan et al
(40) underlined that MIAT
promotes BC progression and functions as competing endogenous RNA
to regulate dual specificity phosphatase 7 (DUSP7) gene expression
by sponging miR155-5p in BC. Diethylstilbestrol led to a dose- and
time-dependent upregulation of MIAT in MCF-7 cells that were
dependent on ERα. MIAT silencing and pharmacological inhibition
decreased cell proliferation by perturbing G1 to S phase cell cycle
transition (41). The functional
studies could suggest the targeting of MIAT for the optimization of
therapeutic strategies in Erα+ patients with BC.
The lncRNA LINC00472 promoter has an ERα binding
site and can be upregulated by ERα. When overexpressed, it can
interact with the NF-kB transcription factor modulating the
downstream pathway (42).
LINC00472 overexpression can perturb tumor growth and
reverse the aggressive phenotype of breast tumor cells in
vitro. A meta-analysis, performed on clinical studies,
described overexpression of LINC00472 in ER+
tumors, associated with improved outcomes of patients, and its
low/absent expression in ER− tumors (42).
Even LINC01016 can be to be directly regulated by
ERα, having an ERα binding site on its promoter, and thereby its
expression is rapidly induced by estrogen treatment. Silencing of
LINC00160 results in reduced proliferation (43). ER+ breast tumors
exhibit high expression levels of LINC01016 and patients with
higher LINC01016 expression are related to poor overall survival
(43). Understanding the
molecular mechanisms and molecular pathways involved in the
crosstalk between these lncRNAs and Erα could therefore suggest new
potential prognostic-predictive markers in patients characterized
by ERα+. However, all these data should be confirmed on
a large series of patients.
The lncRNA Down syndrome cell adhesion molecule
antisense RNA 1-AS1 (DSCAM-AS1) is an estrogen-responsive lncRNA
differentially expressed between benign and malignant BC cells
(44) and it can distinguish
features between luminal subtypes of BC (45). Its overexpression in patients with
BC was associated with poor overall survival (46,47). DSCAM-AS1 can induce G1/S
transition of cell cycle and represents a poor prognostic factor in
luminal patients with BC treated with hormone therapy (48). In vitro studies highlighted
that DSCAM-AS1 interacts with nuclear ribonucleoprotein (hnRNPL) in
BC cells, promoting tumor progression and inducing tamoxifen
resistance (49). More recently,
Yadav et al (50)
demonstrated that progesterone also can induce expression of
DSCAM-AS1 and its silencing mimics the effect of progesterone in
impeding cell migration and invasion in PR+ BC cells.
Additionally, DSCAM-AS1 sponges miR-130a that regulates the
expression of ESR1 by binding to its 3′-untranslated region (UTR),
thereby mediating the effect of progesterone in BC cells (50). These data underlined once again
that the reciprocal modulation mechanisms are very complex and that
they can also involve other mediators such as progesterone.
Numerous other lncRNAs can be deregulated under the
effect of estrogen and therefore by the modulation of Erα: i)
Eosinophil granule ontogeny transcript (EGOT) dysregulation is
associated with tumor size, lymph node metastasis progression and
worse survival in patients with BC. Estrogen can reduce the
expression of EGOT in a dose-dependent manner, thereby modulating
autophagy in BC cells (51). ii)
Two lncRNAs, lncRNA152 and lncRNA67, exhibited significant
overexpression in ER+ breast tumors in comparison to
non-cancerous breast tissue; moreover, after estrogen treatment, it
can regulate cell proliferation and mitogenic division in
ER+ cell lines (52).
iii) A recent study, analyzing lncRNAs that are either dysregulated
in response to estrogen treatment in BC cells, identified ~90
upregulated lncRNAs and 160 downregulated lncRNAs by E2 stimulus.
In ER+ patients with BC, lncRNA LINC00160 demonstrated a
strong correlation with the survival rate and its neighboring
E2-responsive coding gene RUNX1 (53).
Numerous other lncRNAs estrogen signaling-dependent
are associated with endocrine therapy resistance in BC. BC cells
can develop different mechanisms to promote intrinsic and acquired
resistance to endocrine therapy; between these, the
ligand-independent transactivation of the ERα is one of the
main mechanisms responsible for acquired resistance, in which
numerous lncRNAs play an essential role. In particular, it is known
that aberrant ERα signaling is strongly involved in
tamoxifen resistance and several lncRNAs could modulate and promote
this process (54). H19 lncRNA
level was higher in ER-positive BC tissues than in ER-negative
tumors suggesting that it is an estrogen-inducible gene and plays a
key role in estrogen-induced cell proliferation (55). Treatment of BC cells with
tamoxifen or fulvestrant increases H19 expression while its
silencing leads to drug resistance. H19 expression can be modulated
by Notch and c-MET inhibition can reverse resistance to tamoxifen
and fulvestrant in an H19-dependent manner in these cells (56). Moreover, altered ERα expression
can also modify H19 levels thereby modulating the apoptosis
response, through pro-apoptotic gene BCL2 interacting killer
(BIK), to chemotherapy in BC cells (57).
lncRNA in non-homologous end joining (NHEJ) pathway
1 (LINP1) expression is upregulated in tamoxifen-resistant BC cells
and its knockdown resulted in increased tamoxifen-induced
apoptosis. LINP1 transcription is suppressed in the presence of
estrogen, and tamoxifen treatment restores its expression (56).
BC anti-estrogen resistance 4 (BCAR4) is inversely
associated with the development of resistance to anti-estrogens in
BC cells and associated with worse survival and also a higher risk
of metastasis in other cancers (58). Recently, a polymorphic variant of
the BCAR4 gene associated with BC susceptibility has been
identified (59).
The lncRNA DLGAP1 antisense RNA 2 (DLGAP1-AS2) can
promote ERα signaling, binding to the AFF3 protein and thereby the
inhibition of the degradation of it. It is significantly
upregulated in BC and tamoxifen can induce DLGAP1-AS2 expression
(60). In the same manner, the
lncRNA ERLC1 is involved in ERα signaling in BC tissues.
Functionally, ERα induces ERLC1 transcription, which in turn
stabilizes the ESR1 transcript by interaction with miR-129
and FXR1. Its expression became aberrant in tamoxifen-resistant BC
cells, and its silencing restored sensitivity to tamoxifen and
increased the efficacy of palbociclib or fulvestrant therapy
(61).
The lncRNA estrogen inducible lncRNA (ERINA) is
aberrantly expressed especially in ER+ BCs where it was
inversely associated with survival and sensitivity to CDK
inhibitor. Mechanistically, ERINA interacts with the E2F
transcription factor 1 (E2F1), which prevents the binding of E2F1
to the tumor suppressor retinoblastoma protein 1 (RB1), thereby
promoting cell-cycle progression. ERINA acts as an ER-responsive
gene, and an intronic ER-binding site was identified as an enhancer
that mediates the transactivation of ERINA (62). With a similar mechanism of action,
Yu et al (63) described
the lncRNA actin gamma 1 pseudogene 25 (AGPG) as a regulator of
E2F1 activity in endocrine resistant BC. AGPG physically interacted
with purine rich element binding protein A, thus releasing E2F1 and
promoting its signaling activation in ERα+ BC cells.
Horie et al (64) revealed that the BC Natural
antisense transcript 1 (BNAT1) can induce tamoxifen-resistant in
ER+ BC cells and its silencing significantly reduced the
in vitro and in vivo growth of cancer cells. This
mechanism is mediated by binding of BNAT1 binding with ERα and its
knockdown reduced ERα expression and its transactivation.
More recently, Chen et al (65) indicated that LINC02568 regulates
estrogen/ERα-induced gene transcriptional activation in
trans by stabilizing ERα mRNA through sponging miR-1233-5p in
the cytoplasm and by regulating carbonic anhydrase CA12 in
cis in the nucleus. This crosstalk modulates BC cell growth
and tumorigenesis as well as endocrine therapy drug resistance
(65).
In conclusion, the ability of numerous lncRNAs
related to Erα activity to modulate resistance to therapies makes
them important predictive biomarkers, and the evaluation of their
aberrant expression could provide an additional tool for the
therapeutic stratification of patients with BC.
Prostate cancer
Some lncRNAs are involved in estrogen signaling in
prostate cancer. In particular, HOTAIR and MALAT1 can interact with
both ERα/ERβ at the chromatin level (35). HOTAIR and MALAT1 silencing
determine the abrogation of estrogen sensitivity of the estrogen
target gene pS2. Nanni et al (66) described that MALAT1 has a role in
the transcription repression of hormone receptors target genes such
as pS2 and PSA in prostate cancer cells and orthotopic models
(35). These observations make
HOTAIR and MALAT1 important biomarkers and potential therapeutic
targets in prostate cancer.
The nuclear enriched abundant transcript 1 (NEAT1)
is a specific transcriptional target of ERα in prostate cancer and
it is significantly overexpressed in cancer as compared with benign
prostate. In prostate cancer cell lines, ERα overexpression and E2
treatment upregulated NEAT1 transcript levels in a time-dependent
manner. NEAT1 acts as a transcriptional activator of different PCa
genes and its silencing modified ERα target genes, suggesting that
NEAT1 is not only a downstream target but also a mediator of ERα
signaling in prostate cancer cells. Moreover, in vitro and
in vivo NEAT1 expression is associated with aggressive
phenotype and drug resistance (67). Additionally, NEAT1 lncRNA could
therefore represent an important prognostic biomarker and a
predictive marker of therapeutic response in patients with prostate
cancer.
As previously described, lncRNA H19 is more
sensitive to estrogens, a stimulus with a pro-tumoral role in
prostate cancer. Bacci et al (68) described H19-dependent mechanisms
by which estrogen and hypoxia signaling promote the acquisition of
an aggressive phenotype in prostate cancer cells. H19 can be
independently upregulated by estrogen or hypoxia. Moreover, the
same authors revealed that H19/estrogen signaling has a potential
impact on the TME by the switch from EMT to a β integrin-mediated
invasion (68).
Other tumors
Numerous other lncRNAs are capable of modulating and
being modulated by ERα also in other neoplastic pathologies. The
lncRNA LINC00312, also known as nasopharyngeal carcinoma-associated
gene 7, a novel putative tumor suppressor, is essential for ERα
binding and suppression activity in nasopharyngeal carcinoma (NPC).
It can promote NPC cell invasion by regulation of ERα and the
HRAS/p-c-RAF and JNK2/AP-1/MMP1 signaling pathways (69).
Estrogen signaling has long been implicated in
epithelial ovarian cancer progression. The lncRNA ElncRNA1 is
transcriptionally regulated by E2 through ERα-EREs and it was
described as able to promote epithelial ovarian cancer cell
proliferation (70). Moreover,
Qiu et al (71) reported
that E2 significantly dysregulated 115 lncRNAs in ERα-positive
SKOV3 ovarian cells compared with E2-untreated controls. Among
these, both TC0101441 and TC0101686 were dysregulated by E2 also in
other ERα-positive PEO1 ovarian cells and dysregulated in ovarian
cancer patients. In particular, high TC0101441 expression was
associated with lymph node metastasis, exhibiting a potential
involvement in the metastatic progression of ovarian cancer
(71).
The lncRNA DSCAM-AS1 overexpression in endometrial
cancer (EC) has a significant association with shorter overall
survival of patients with EC. In these cells, it is associated with
the endometrial tumor promotor prolactin gene and with the
expression of ERα together with its target genes trefoil factor 1
and progesterone receptor (PgR). DSCAM-AS1 silencing in EC cell
lines reduced cell growth and proliferation by inhibition of
NOTCH1, Protein Tyrosine Kinase 2 as well as early growth response
factor 1 (EGR1) expression (45).
The lncRNA LINC00899 overexpression is able to
inhibit the viability, proliferation, migration and invasion of
cervical cancer cells. miR-944 could competitively bind with
LINC00899 and its upregulation can inhibit ER expression, thereby
promoting the migration and invasion of cervical cells. These data
highlight the role of LINC00899/miR-944/ESR1 axis in the regulation
of cervical cancer progression (72).
More recently, Cheng et al (73) revealed that lncRNA
maternally-expressed 3 (MEG3) promoter binds to ERα in liver cancer
cells. ERα upregulation repressed the proliferation, migration as
well as invasion and promoted apoptosis of HepG2 cells under high
glucose conditions. ERα silencing decreased MEG3 expression and in
turn enhanced proliferation, migration and invasion. These data
highlighted that ERα has a protective role in hepatocellular
carcinoma (HCC) being able to inhibit cell progression through
positive regulation of lncRNA MEG3 (73).
3. Crosstalk between ERβ and lncRNAs in
cancer
ERβ is expressed in different types of tissues and
cells, to a higher degree in females as compared with males
(74). ERβ has at least five
different isoforms, four shorter ERβ isoforms, and a full-length
ERβ isoform (75). The five ERβ
variants, which result from alternative splicing of the last coding
exon, and deletion of coding exons, are named ERβ1-5. They can be
detected in multiple normal tissues, especially in BC tissues and
cell lines (76).
In the same manner as other superfamily members of
hormone NRs, ERβ binds EREs located on target genes promoter
regions to modulate their expression levels (75).
MALAT1 and HOTAIR are all, important protagonists in
ERβ signaling especially in prostate cancer (35). In normal conditions, MALAT1 acts
as a suppressor of gene transcription. After estrogen treatment,
ERβ binds to EREs and leads on the one hand the reduction of MALAT1
transcript and on the other hand increases HOTAIR expression.
Mechanistically, MALAT1 acts as a suppressor of estrogen-related
genes transcription. In turn, HOTAIR is involved in the epigenetic
modification associated with transcriptional activation of
estrogen-related genes (77).
Although numerous studies have verified that ERβ is
an independent prognostic and/or predictive factor in BC, very
little information is available regarding the crosstalk between ERβ
and lncRNA in BC. ERβ could play a crucial role in BC cell
progression, particularly EMT and metastasis (78). This suggests a potential
association with HOTAIR as well as ERβ in BC and subsequently
downstream molecules, miR-138/204/217 and miR-200c, in BC,
although, it remains to be investigated (79).
Crosstalk between HOTAIR and ERβ has instead been
abundantly demonstrated in renal cell carcinoma (RCC) cells. Ding
et al (80) showed that
ERβ can increase HOTAIR expression by transcriptional upregulation
in multiple RCC cell lines. In RCC tissues, the ERβ expression can
be aberrant too (80). Both ERβ
activation by 17β-estradiol treatment and its knockdown are able to
modulate HOTAIR expression. Crosstalk between ERβ and HOTAIR, by
miR-138, miR-200c, miR-204, as well as miR-217, can modulate
different oncogenes, such as Cyclin D2, VEGFA and VIM, to induce
RCC proliferation as well as invasion (80).
More recently, He et al (81) demonstrated that the antiangiogenic
drug sunitinib, widely used for RCC therapy, can induce the
expression of lncRNA-ECVSR to influence the RNA stability of ERβ.
This leads to an alteration of vasculogenic mimicry formation in
RCC cells (81).
The crosstalk between SRA and ERβ signaling has been
reported as involved in the pathogenesis from ovarian endometriosis
to ovarian clear cell carcinoma (OCCC). A preliminary study
revealed that SRA interacts preferentially with ERβ rather than
with Erα (82). Lin et al
(83) described that SRA
expression level gradually enhanced from endometriosis (EM) to
EM-associated ovarian clear cell carcinoma (EAOCCC) to OCCC,
showing an inverse relationship with ERβ expression. During
malignant transformation from EM to OCCC, SRAP, as a co-suppressor,
may play a role in hypermethylation of the promoter of ERβ and
silence the latter. Consequentially, overexpression of SRA
accompanied by ERβ reduction may be a crucial step in the EAOCCC
development (83). Therefore,
interfering with the crosstalk mechanisms between SRA and ERβ and
the molecular pathways involved could represent a new potential
strategy to prevent the progression of ovarian cancer.
In papillary thyroid carcinoma, H19 and ERβ
crosstalk are able to promote stem-like phenotype in cancer cells.
E2 significantly promotes H19 transcription via Erβ (84). Both exhibit an aberrant expression
in papillary thyroid cancer stem cells (PTCSCs). Knockdown of ERβ
decreases tumor growth, diminishes ALDH+ cell
populations and suppresses the sphere formation capability of PTC
cells. In the same way, silencing of H19 inhibits E2-induced sphere
formation ability. In papillary thyroid cancer, H19 can act as
competitive endogenous RNA that sequesters miRNA-3126-5p to
reciprocally release ERβ expression. Additionally, H19 appears
highly expressed in PTC tissue specimens, which is associated with
poor overall survival (84).
Thus, as ERβ plays a critical role in regulating PTCSC maintenance,
targeting ERβ could provide a novel therapeutic avenue for patients
with advanced PTC.
4. Crosstalk between ARs and lncRNAs in
cancer
AR is a member of the steroid-hormone family
involved in the regulation of normal growth and development of
different organs (85). AR
transcription is modulated by circulating androgens and it is both
cell-type and age-dependent (86). To perform its function AR needs
ligand-dependent binding and interaction with co-activators as well
as chaperone proteins (87). The
AR inactive form, without a ligand, is located in the cytoplasm,
bound to heat shock proteins (hsp90). Upon exposure to androgen
[testosterone or dihydrotestosterone (DHT)], it results in a
conformational change, dissociation of chaperone proteins and
exposure of the nuclear localization signal which allows it to move
into the nucleus to bind to the genomic androgen response elements
(85). As a result, transcription
of AR target genes, mainly involved in the regulation of cell
growth, proliferation and survival, is activated (85).
Prostate cancer
Androgen as well as ARs play a crucial role in the
initiation and progression of prostate cancer (88). Numerous lncRNAs associated with
prostate cancer play oncogenic roles by modulating AR-mediated
activity through different mechanisms (89).
Two lncRNAs, prostate-specific transcript 1
(PCGEM1), and prostate cancer associated ncRNA 1 have been
described to directly interact with AR under ligand-stimulated
conditions, but the data regarding the two lncRNAs are
contradictory (90,91), questioning their real involvement
in prostate cancer progression and their ability to interact and
modulate the AR signaling pathway.
HOTAIR is involved in cancer progression especially
in androgen-sensitive to castration-resistant prostate cancer
(CRPC) cells (92). HOTAIR can
modulate androgen-independent AR activation and AR-mediated gene
transcription. It interacts with AR thereby inhibiting its
ubiquitination and degradation (92). Moreover, HOTAIR forms a repressive
complex with polycomb proteins and decreases AR transcription which
results in increased cell invasion and MMP9 expression (93). These data evidence HOTAIR as a
novel target gene playing a critical role in invasiveness of
prostate cancer cells.
Ozgur et al (94) revealed that cellular H19
expression strongly decreased after DHT stimulation in
hormone-sensitive prostate cancer cells, and AR inhibition by
enzalutamide restored the DHT effect as well as increased H19
expression. The present study provided evidence of the involvement
of H19 lncRNA in androgen/receptor pathway-dependent prostate
cancer development (94).
Prostate cancer gene 3 (PCA3) lncRNA is
overexpressed in prostate cancer tissues compared with adjacent
non-neoplastic tissues and it can be involved in AR signaling,
since its silencing promotes cell growth and viability (95,96). Moreover, PCA3 knockdown modified
the expression of PSA and PCGEM1, AR-inducible genes, as well as
sensitized PCa cells to enzalutamide (97). PCA3 silencing also highlights its
role in EMT, leading to the overactivation of epithelial markers
E-cadherin, claudin-3 and cytokeratin-18 and the dysregulation of
vimentin (96). Due to its
aberrant oncogenic activity, PCA3 could represent a potential
therapeutic target in patients with prostate cancer.
Growth arrest-specific 5 (GAS5) lncRNA plays a
crucial role in prostate cancer carcinogenesis and its knockdown in
prostate cancer cells leads to the inhibition of tumor progression
(98). The interaction between
GAS5 and AR could perturb cell proliferation of CRPC. GAS5
silencing in CRPC would upregulate the AR axis, while the high
activation of AR could further inhibit the transcription of GAS5
(98). GAS5 can interact with
both GR and AR regulating GR/AR-mediated transcriptional activity
(99).
The lncRNA BCAR4 is overexpressed in prostate cancer
tissues and strongly associated with metastatic progression as well
as poor prognosis (100). In
AR+ PCa cells, upregulation of BCAR4 increases cell
growth as well as migration and induce resistance to androgen
(101). On the contrary, in
AR− prostate cancer cells BCAR4 silencing decreased cell
growth (101). Therefore, as
well as in BC, BCAR4 represents a potential prognostic and
predictive biomarker in patients with prostate cancer.
Instead, some lncRNAs behave like tumor suppressor
genes. For example, downregulated RNA in androgen independent cells
(DRAIC) lncRNA can inhibit the migration of prostate cancer cells.
AR activity induction leads to DRAIC transcription reduction and
promotes prostate cancer progression (102). Moreover, DRAIC downregulation
increased cell invasion and soft agar colony formation by NF-κB
activation (103).
Another tumor suppressor lncRNA, prostate cancer
associated transcript 29 (PCAT29) suppresses androgen signaling in
prostate cancer. PCAT29 silencing promoted prostate cancer growth
and migration while its overexpression reduced these processes.
PCAT29 downregulation results were associated with recurrence in
patients with prostate cancer (104). Several other lncRNAs are
involved in reciprocal modulation with AR and in the progression of
prostate cancer.
AR-regulated lncRNA 1 lncRNA plays a crucial role in
AR-dependent prostate cancer development as well as CRPC. It is
able to interact with AR mRNA transcript and stabilizes it for
protein translation (105).
The androgen-dependent lncRNA suppressor of cytokine
signaling 2 antisense (SOCS2-AS1) promotes androgen-dependent
prostate cancer growth and migration (87). In CRPC cells increased AR
signaling has been strongly associated with SOCS2-AS1 upregulation
compared with androgen-dependent LNCaP cells. Moreover, SOCS2-AS1
can modulate the AR target TNF Superfamily Member 10 gene (106).
The prostate cancer upregulated lncRNA 1 is
overexpressed in prostate cancer cells and its silencing inhibited
AR and NKX3-1 expression, whereas AR silencing decreased PlncRNA-1
expression (107).
TMPO antisense RNA 1 (TMPO-AS1) lncRNA is
overexpressed in prostate cancer cell lines and tissues associated
with a worse prognosis of patients with prostate cancer. AR can
directly regulate the TMPO-AS1 levels in the cytoplasm promoting
growth, progression, cell migration and inhibition of apoptosis in
prostate cancer cells (108).
The AR-regulated lncRNAs, namely CRPC-lncs, are
highly expressed in CRPC tissues. Their silencing reduces the
expression of AR and AR variants which as a consequence inhibit
CRPC tumor growth (109).
MALAT1 is increased in prostate cancer cells after
androgen stimulation, as well as its receptor. MALAT1 knockdown
inhibited the DHT administration-induced increase in the cell
cycle, induced AR expression in these cells and blocked the
tumorigenesis of prostate cancer cells in nude mice. Moreover,
MALAT1 can bind miR-320b, negatively regulating its expression, and
AR is a target of miR-320b. Consequently, changes induced by MALAT1
silencing can be abolished by miR-320b inhibition or AR
overexpression (110).
Recently, a large number of lncRNAs have been
associated with AR activity and AR inhibitors sensibility: i)
LINC00675, overexpressed in androgen-insensitive prostate cancer
cell lines and CRPC, patients can directly modulate AR interaction
with mouse double minute-2 (MDM2) and block the ubiquitination of
AR by binding to it (111), ii)
prostate cancer associated transcript 7 (112) and prostate cancer lncRNA at
chromosome 16 (113) lncRNAs are
directly related with AR signaling and overexpressed in prostate
cancer tissues and AR-dependent cell lines. Their silencing
suppressed AR signaling, tumor growth and proliferation. iii)
Growth hormone secretagogue receptor (GHSR) opposite strand
(GHSROS) lncRNA is overexpressed in prostate cancer cell lines and
modulates the expression of protein phosphatase 2 regulatory
subunit b gamma gene. The loss of GHSROS may drive AR
pathway-independent prostate tumor progression (114). iv) FAM83H-AS1 is upregulated in
prostate adenocarcinoma and its silencing in prostate cancer cells
modulated cell proliferation, cell cycle and migration. AR
signaling is involved in upregulating FAM83H-AS1 expression in
prostate cancer cells (115). v)
Poly(RC) binding protein 1 antisense RNA 1 is involved in CRPC
enzalutamide resistance by enhancing the deubiquitination of
AR/AR-V7 (116). vi) Negative
expression of AR Regulating LncRNA (NXTAR) bound upstream of the AR
promoter inhibiting AR/AR-V7 expression, which in turn upregulates
NXTAR levels, and compromising enzalutamide-resistant prostate
cancer (117). vii) AC016745.3
overexpression inhibits the proliferation and migration of prostate
cancer cells and suppresses the expression of AR target genes by
binding non-POU domain containing octamer binding protein (118). viii) RP11-1023L17.1 expression
can be directly suppressed by the AR as well as promotes
proliferation, migration, cell cycle progression and inhibition of
apoptosis. RP11-1023L17.1 can suppress F-Box Protein 32 expression,
thereby enhancing c-Myc protein stability (119).
BC
AR is overexpressed in normal breast epithelium as
well as in a large proportion of BC tissues and cell models. AR is
co-localized with ER together with PgR in epithelial breast cells,
indicating a combined role in the modulation of mammary epithelial
cell proliferation, and is silent in myoepithelium as well as
stromal cells (120,121). However, in a normal breast,
androgens are involved in inhibiting breast development, whereas,
in BC cell lines, androgens and AR signaling can promote breast
cell proliferation and growth (122).
A strong relation between AR signaling and lncRNA
has been highlighted in BC in particular in triple-negative BC
(TNBC). One of the TNBC subtypes called luminal AR (LAR) was
characterized by AR expression (123). A strong association between the
lncRNA HOTAIR and AR expression was demonstrated in TNBC patient
samples (124). HOTAIR
overexpression appeared associated with lymph node metastases and
with AR expression indicating a role of HOTAIR in TNBC progression
by modulation of the AR signaling pathway (124).
Yang et al (125) studied three TNBC cell lines,
treated with DHT, to investigate the role of AR-related lncRNAs.
They highlighted the downregulation of AR negatively regulated
lncRNA (ARNILA), and its role as a negative prognostic marker in
patients with TNBC. In vitro and in vivo studies
revealed that ARNILA can induce invasion as well as metastatic
progression (125). Moreover, it
can promote EMT by upregulating SOX4 expression (125).
SRA1-mediated AR signaling pathway regulation may be
also involved in BC development, especially in TNBC. Leygue
(126) reported that silencing
of SRA1 could reduce the invasiveness and motility of TNBC cells
(126).
Currently, Huang et al (127) conducted a study to determine the
role of the AR signaling pathways (ARSP)-related lncRNAs in the
incidence and progression of BC and their relationship with the
TME. A total of 340 ARSP-related lncRNAs were detected, of these
203 were upregulated and 137 were downregulated (127). The understanding of the
molecular mechanisms beneath the crosstalk between AR and lncRNAs
in BC could provide new tools for the prognostic stratification of
patients including the development of new therapeutic
strategies.
Additional tumors
The effects of lncRNAs on AR signaling have also
been assessed in other types of cancers.
In bladder cancer, combined overexpression of X
inactive specific transcript (XIST) and AR have been associated
with the advanced TNM stage. XIST knockdown leads to a decrease in
proliferation, invasion and migratory potential through modulation
of AR signaling. Moreover, XIST suppresses the expression of
miR-124 which target 3'-UTR of AR through direct interaction
(128).
Nevertheless, in bladder cancer, another lncRNA,
LINC00460, appeared aberrantly overexpressed and its silencing
decreased the proliferation of bladder cancer cells through
modulation of AR expression. Moreover, LINC00460 overexpression has
been associated with poor prognosis for patients with bladder
cancer (129).
In esophageal squamous cell carcinoma progression,
Wu et al (130)
discovered a micro peptide (YY1BM) encoded by Y-linked lncRNA
LINC00278, able to block YY1 binding to AR to activate the
expression of eEF2K. Recently, He et al (131) showed that AR-mediated
transcription activation is responsible for the overexpression of
LINC01503, which promoted NPC cell proliferation, migration and
invasion in vitro, and facilitated tumor growth as well as
metastasis in vivo.
Schmidt et al (132) demonstrated that in melanoma
cells lncRNA SRA-like ncRNA (SLNCR) binds to AR and complexes with
different transcription factors to mediate invasion or
proliferation. More recently, they revealed that AR and EGR1, a
transcription factor mainly involved in the processes of tissue
injury, immune responses and cancer, bind to the lncRNA SLNCR and
increase melanoma proliferation by p21 (133). The same authors later proved
that point mutations or oligonucleotides that abrogate AR binding
to SLNCR block melanoma invasion, suggesting that targeting
lncRNA-protein complexes holds therapeutic promise (134).
Zhai et al (135) reported that suppressing AR in
RCC (SARCC) lncRNA establishes a physical interaction with AR
acting as a tumor suppressor in RCC cells. SARCC expression appears
downregulated in RCC tissues and metastatic samples compared with
non-neoplastic adjacent tissues. In addition, its dysregulation is
strongly associated with the prognosis of patients with RCC. In
vitro, low expression of SARCC associated with cell invasion,
migration, proliferation and drug resistance to sunitinib.
Mechanistically, SARCC/AR crosstalk inhibited miR-143-3p expression
by modulating K-RAS, MMP-13, AKT and P-ERK expression (135). HOTAIR is also capable of
interacting with AR in RCC cells, enhancing the expression of VEGFA
and platelet derived growth factor receptor alpha (PDGFA) with the
promotion of tumor angiogenesis, and inducing tumor stemness
phenotype (136).
Recently, You et al (137) showed that AR can increase TWIST1
associated lncRNA regulated via AR expression through binding to
the AREs promoting vasculogenesis and metastasis in clear cell RCC
via altering TWIST1.
The lncRNA prostate androgen-regulated transcript 1
(PART1) interacts with AR promoting promyelocytic leukemia zinc
finger (PLZF) expression, followed by recruitment of EZH2, to
mediate epigenetic PDGFB silencing and downstream PI3K/Akt
inhibition, and modulating gastric cancer progression (138).
Finally, Qin et al (139) described the upregulation of
LINC00667 in HCC tissues and cell lines associated with an
unfavourable prognosis of patients with HCC. LINC00667 acts as a
molecular sponge in the miR-130s-3p/AR signaling pathway thereby
promoting HCC tumor progression (139).
5. Crosstalk between other NRs and lncRNAs
in cancer
RAR
All-trans retinoic acid (RA) is the main active
metabolite of vitamin A (140).
RA signaling is involved in different biological processes,
including embryonic development along with organogenesis, cell
growth, proliferation, cell differentiation, together with
metabolism regulation (141). RA
translocates in the nucleus by binding to an intracytoplasmic
transporter, cellular RA binding protein type II. Retinoids bind to
two different NRs, RARs (RAR-α and RAR-β) and retinoid X receptors
(RXRs). RARs function as heterodimers with RXRs. In turn, RXRs can
be involved in the formation of heterodimers with other different
NRs, such as vitamin D3, thyroid hormone and PPARs
(142).
RAR/RXR and RXR/RXR are localized in the nucleus
bound to retinoid hormone response elements (RAREs) in the promoter
regions of retinoid-responsive genes thereby inducing the
transcription of numerous target genes (143).
There are few examples in the literature regarding
crosstalk of RAR signaling and lncRNAs in cancer. In acute
promyelocytic leukemia (APL) RA/RARs signaling can induce the
expression of the lncRNA HOXA-AS2 antisense, which suppresses the
TRAIL pathway and reduces expression of caspases (144). Similarly, RA/RARs signaling
induces H19 expression in APL cells and its upregulation perturbs
telomerase activity, mediated by hTERC-hTR interaction, during
tumor evolution (145).
Conversely, the lncRNA RA early transcript 1K pseudogene (RAET1K)
can reduce RA/RARs signaling. In lung cancer RAET1K interferes with
the protective role of RA, modulating miR-135a-5p, which in turn
inhibits Cyclin E1 (CCNE1) expression, a cyclin required for G1-S
transition. RA promotes the expression of this miRNA, by blocking
lung cancer cells in the G1 phase and thereby tumor cells
proliferation and progression. RAET1K silencing can repress CCNE1
expression and inhibit cell cycle progression from the G1 to the S
phase (146).
A recent study also highlighted crosstalk between
HOTAIR and RA/RARs signaling in acute myeloid leukemia. Different
acute myeloid leukemia cell lines (HL-60, NB4, U937 and THP-1) have
induced myeloid differentiation by all-trans RA (ATRA). All four
cell lines exhibited a higher expression level of HOTAIR after the
ATRA treatment suggesting its role in the regulation of RA/RARs
pathway and consequently in myeloid cell differentiation (147). The lncRNA RNA HOTAIR myeloid 1
(HOTAIRM1) can mediate ATRA-induced differentiation of ALK cells
(148). Upregulation of HOTAIRM1
expression is able to enhance ATRA-induced PML-RARA degradation by
affecting autophagic flux and thereby controlling myeloid cell
differentiation in APL cells (149). lncRNA NEAT1 is also involved in
APL cell differentiation induced by ATRA. C/EBPα binds and
transactivates NEAT1 whereas PML/RARα suppresses this process
suggesting that PML/RARα could contribute to the pathogenesis of
APL by suppressing C/EBPα targets (150).
In glioblastoma multiforme cells ATRA treatment
dysregulated different lncRNAs' expression. In particular, ATRA
dose-dependently decreased the expression of GAS5 within the
microenvironment of the U87MG cell line (151).
RAET1K is necessary for retinoid-induced
differentiation in different cells by binding RXRs. RAET1K is
upregulated in tumor tissues and is associated with a poor
prognosis in patients with lung adenocarcinoma. Aberrant RAET1K
expression upregulated cyclin E1 by targeting miR-135, thereby
promoting tumor progression (146).
Currently, Wang et al (152) asserted that lncHOXA10 is
significantly upregulated in gastric cancer tissues and cell lines,
and can promote proliferation, migration as well as invasion of
cells. lncHOXA10 can suppress RAR-β expression, involved in the
modulation of apoptosis, while ATRA can rescue the expression of
RAR-β, inhibiting lncHOXA10 in gastric cancer cells (152). RAR-β is also modulated by lncRNA
HAND2-AS1 in human bladder cancer. HAND2-AS1 inhibits cell
proliferation and promotes apoptosis in bladder cancer cells by
sponging miR-146. The last of these can promote cell proliferation
by targeting RAR-β and, in turn, HAND2-AS1 can suppress cell
proliferation via releasing RAR-β from miR-146 (153). More recently, Fu et al
(154) described that another
lncRNA, lymphocytic leukemia 2 (DLEU2), can modulate RAR-β in
colorectal cancer cells. DLEU2 induced promoter methylation of
RAR-β to downregulate its expression, and its upregulation induced
the MAPK signaling pathway promoting colorectal cancer progression
(154).
VDR
VDR is a transcription factor that besides
interacting with hormonally active vitamin D3, regulates the
expression of more than 900 genes involved in numerous different
cellular processes. Both vitamin D/VDR signaling and lncRNAs affect
numerous genomic and non-genomic processes, the dysregulation of
which can be associated with a wide range of diseases including
cancer. The principal evidence is associated with skin cancers.
Several studies confirmed the protective effect of VDR in cancer
initiation and progression (155), especially in keratinocyte
carcinoma (156). Jiang and
Bikle (157) profiled 90
well-annotated mouse lncRNAs from cultured mouse keratinocytes
after deleting VDR. They detected that H19, HOTTIP, mHOTAIR,
Malat1, SRA and Nespas were significantly increased, whereas H19
as, Kcnq1ot1 and lincRNA-p21 were decreased (157). Subsequently, other
VDR-associated lncRNAs in BC (158) as well as in lung cancer
(159) have been identified.
VDR, MALAT1 and LINC00511 were significantly upregulated in tumor
cells in BC compared with non-cancerous tissues. In addition, VDR
and another lncRNA SNHG16 were associated in both tumor and
non-tumor breast tissues (158).
In lung cancer, the coordinate expression of VDR and the lncRNAs
MALAT1, SNHG16, SNHG6, LINC00346 and LINC00511 has also been
validated in lung cancer tissues (159).
In oral squamous cell carcinoma (OSCC) Jin et
al (160) evaluated
crosstalk between vitamin D/VDR signaling and lncRNAs selecting 46
pairs of tumor tissue and adjacent non-tumor tissue as well as two
OSCC cell lines. In the latter, they observed 1045 lncRNAs
differentially expressed after the 1,25(OH)2D treatment.
In particular lung cancer-associated transcript 1 (LUCAT1)
expression was reduced by the 1,25(OH)2D treatment in
vitro and it appeared strongly overexpressed in the OSCC tumor
tissues. LUCAT1 silencing in the OSCC cell lines reduced cell
growth in association with a reduction in ERK1/2 phosphorylation
(160).
The oncogenic lncRNA colon cancer-associated
transcript 2 (CCAT2) has been described as overexpressed in ovarian
cancer. Wang et al (161)
described that 1,25(OH)2D inhibited CCAT2 expression
leading to decreased binding of transcription factor 4 (TCF4) to
the MYC promoter in ovarian cancer cell lines. This produced
inhibition of proliferation, migration and invasive features of
ovarian cells (161). In
addition, the lncRNA TOPORS-AS1, overexpressed in ovarian cancer
cells and involved in suppression of cell proliferation, migration
and invasion, is upregulated by VDR. Crosstalk is essential to
interrupt the Wnt/β-catenin signaling thereby defining an improved
prognosis in patients with ovarian cancer (162).
Finally, the relation between the lncRNA H19 and
VDR has been described in colon cancer. VDR can inhibit H19
expression through the regulation of the c-Myc/Mad-1 network. In
turn, H19 upregulation blocked VDR expression by upregulating miRNA
675-5p (163).
Gonadotropin receptor
Human GR (h-GR) belongs to the nuclear hormone
receptor superfamily and is encoded by nuclear receptor subfamily
3C1 gene on chromosome 5q31.3 (164). In the absence of ligands, the GR
is sequestered in the cytoplasm by chaperone proteins.
Glucocorticoids, such as cortisol, prednisolone and dexamethasone
bind to GR leading to both its dimerization and translocation into
the nucleus, where it performs its activity as a transcription
regulator (164). DNA-bound GR
recruits coregulator complexes forming transcription regulatory
complexes that can function in both activation and suppression of
transcription. Some lncRNAs have been described as regular and can
be regulated by glucocorticoid/GR signaling.
GAS5 lncRNA is able to directly interact with the
DNA binding domain of GR acting as a molecular decoy by competing
with the glucocorticoid-response element for GR binding, thereby
inhibiting GR transcription (165). GAS5 is also able to accumulate
in cells that have been starved of growth factors by suppressing GR
activity, and GAS5 sensitizes cells to apoptosis (165). In the same way, overexpression
of GAS5 leads to growth arrest and apoptosis in human breast cell
lines (165).
THRs
Thyroid hormone action is predominantly mediated by
THRs, which are encoded by the THRA and THRB genes (166). THRs act as ligand-activated
nuclear transcription factors to regulate different physiologic
processes through direct gene regulation and with ncRNAs relation
(167,168).
Current evidence indicates a crosstalk between the
thyroid hormone/THR pathway and lncRNAs in liver cancer. Brain
cytoplasmic RNA 1 (BCYRN1/BC200) is abnormally overexpressed in
several tumor types, with a significant overexpression in HCC
tissues. Using the Disease-Related Human lncRNA Profiler to
identify lncRNAs regulated by thyroid hormone/THR signaling in
liver cells, BC200 was identified as a lncRNA downregulated by the
thyroid hormone (169).
In HCC cells, the taurine upregulated gene 1 (TUG1)
is overexpressed and can induce cell proliferation, invasion,
metastatic progression as well as apoptosis by distal-less homeobox
2 (DLX2) activation (170).
Alpha-fetoprotein (AFP) is increased in the majority of patients
with HCC and TUG1 is involved in THR/AFP signaling. Thyroid
hormones suppress TUG1 expression, leading to the downregulation of
AFP (171).
6. Conclusions
NRs function as a transcriptional signaling network
that mediates gene regulatory actions to maintain cellular
homeostasis in response to hormonal and environmental factors. The
dysregulation of NR signaling is known to contribute to the
evolution of hormone-dependent tumors, such as breast and prostate
cancer, which are also therapeutic targets. However, NRs signaling
affects cancer also because the therapeutic response associated
with their target drugs is often altered by changes in the
expression and function of a large number of coregulators. They
could modulate receptor sensitivity, and modify their
protein-protein interaction, thus altering transcription, as well
as regulation of chromatin accessibility. Coregulators include
coactivators that generally associate with agonist-bound NRs to
stimulate gene expression and corepressors that are usually bound
to unliganded or antagonist-bound NRs to suppress gene expression.
lncRNAs are emerging as new genetic/epigenetic coregulators of NRs.
In the last 20 years, lncRNAs have been extensively studied due to
their potential role in cancer development and progression.
However, the functional characterization of lncRNAs remains
complex, interacting with different molecules involved in the
regulation of key processes during cell development and diseases.
lncRNAs can modulate NRs activity, and in turn, they can be
regulated by NRs. Therefore, understanding the mechanisms by which
such coregulators interact and modulate NR activity and vice versa
could offer new opportunities to develop improved prognostic in
addition to diagnostic approaches, along with new therapeutic
targets.
Availability of data and materials
Not applicable.
Authors' contributions
MCa, AB and GF were responsible for the conception
and design of the present review. MCe and PM collected and
assembled the data. MCa, MB and MT contributed to the drafting and
revision of the review. All authors have read and approved the
final version of the manuscript. Data authentication is not
applicable.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the Italian Ministry of
Health (Ricerca Corrente IRCCS; grant. no. 2778553; year 2023).
References
|
1
|
Mangelsdorf DJ, Thummel C, Beato M,
Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M,
Chambon P and Evans RM: The nuclear receptor superfamily: the
second decade. Cell. 83:835–839. 1995.
|
|
2
|
Cheung E and Kraus WL: Genomic analyses of
hormone signaling and gene regulation. Annu Rev Physiol.
72:191–218. 2010.
|
|
3
|
Gadaleta RM and Magnani L: Nuclear
receptors and chromatin: an inducible couple. J Mol Endocrinol.
52:R137–R149. 2014.
|
|
4
|
Lonard DM, Lanz RB and O'Malley BW:
Nuclear receptor coregulators and human disease. Endocr Rev.
28:575–587. 2007.
|
|
5
|
Fatica A and Bozzoni I: Long non-coding
RNAs: New players in cell differentiation and development. Nat Rev
Genet. 15:7–21. 2014.
|
|
6
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021.
|
|
7
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long Non-coding RNAs in Cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016.
|
|
8
|
Foulds CE, Panigrahi AK, Coarfa C, Lanz RB
and O'Malley BW: Long Noncoding RNAs as targets and regulators of
nuclear receptors. Curr Top Microbiol Immunol. 394:143–176.
2016.
|
|
9
|
Wu J, Nagy LE, Liangpunsakul S and Wang L:
Non-coding RNA crosstalk with nuclear receptors in liver disease.
Biochim Biophys Acta Mol Basis Dis. 1867:1660832021.
|
|
10
|
Lanz RB, McKenna NJ, Onate SA, Albrecht U,
Wong J, Tsai SY, Tsai MJ and O'Malley BW: A steroid receptor
coactivator, SRA, functions as an RNA and is present in an SRC-1
complex. Cell. 97:17–27. 1999.
|
|
11
|
Emberley E, Huang GJ, Hamedani MK, Czosnek
A, Ali D, Grolla A, Lu B, Watson PH, Murphy LC and Leygue E:
Identification of new human coding steroid receptor RNA activator
isoforms. Biochem Biophys Res Commun. 301:509–515. 2003.
|
|
12
|
Sheng L, Ye L, Zhang D, Cawthorn WP and Xu
B: New Insights Into the Long Non-coding RNA SRA: Physiological
functions and mechanisms of action. Front Med (Lausanne).
5:2442018.
|
|
13
|
Szwarc MM, Kommagani R, Lessey BA and
Lydon JP: The p160/steroid receptor coactivator family: potent
arbiters of uterine physiology and dysfunction. Biol Reprod.
91:1222014.
|
|
14
|
Tenga A, Beard JA, Takwi A, Wang YM and
Chen T: Regulation of Nuclear Receptor Nur77 by miR-124. PLoS One.
11:e01484332016.
|
|
15
|
Zhang CJ, Liu C, Wang YX, Zhu N, Hu ZY,
Liao DF and Qin L: Long non-coding RNA-SRA promotes neointimal
hyperplasia and vascular smooth muscle cells proliferation via
MEK-ERK-CREB pathway. Vascul Pharmacol. 116:16–23. 2019.
|
|
16
|
Ho JC, Lee CH and Hong CH: Targeting
steroid receptor RNA activator (SRA), a long non-coding RNA,
enhances melanogenesis through activation of TRP1 and inhibition of
p38 phosphorylation. PLoS One. 15:e02375772020.
|
|
17
|
Tello-Flores VA, Beltrán-Anaya FO,
Ramírez-Vargas MA, Esteban-Casales BE, Navarro-Tito N,
Alarcón-Romero LDC, Luciano-Villa CA, Ramírez M, Del
Moral-Hernández Ó and Flores-Alfaro E: Role of Long Non-Coding RNAs
and the molecular mechanisms involved in insulin resistance. Int J
Mol Sci. 22:72562021.
|
|
18
|
Eoh KJ, Paek J, Kim SW, Kim HJ, Lee HY,
Lee SK and Kim YT: Long non-coding RNA, steroid receptor RNA
activator (SRA), induces tumor proliferation and invasion through
the NOTCH pathway in cervical cancer cell lines. Oncol Rep.
38:3481–3488. 2017.
|
|
19
|
Park SA, Kim LK, Kim YT, Heo TH and Kim
HJ: Long non-coding RNA steroid receptor activator promotes the
progression of endometrial cancer via Wnt/β-catenin signaling
pathway. Int J Biol Sci. 6:99–115. 2020.
|
|
20
|
Hong CH, Ho JC and Lee CH: Steroid
Receptor RNA Activator, a Long Noncoding RNA, Activates p38,
facilitates epithelial-mesenchymal transformation, and mediates
experimental melanoma metastasis. J Invest Dermatol.
140:1355–1363.e1. 2020.
|
|
21
|
Mahdevar M, Vatandoost J, Seyed Forootan
F, Kiani-Esfahani A, Esmaeili M, Peymani M, Tavakkoli H, Osmay Gure
A, Nasr Esfahani MH and Ghaedi K: Steroid receptor RNA activator
gene footprint in the progression and drug resistance of colorectal
cancer through oxidative phosphorylation pathway. Life Sci.
285:1199502021.
|
|
22
|
Lanz RB, Chua SS, Barron N, Söder BM,
DeMayo F and O'Malley BW: Steroid receptor RNA activator stimulates
proliferation as well as apoptosis in vivo. Mol Cell Biol.
23:7163–7176. 2003.
|
|
23
|
Kurisu T, Tanaka T, Ishii J, Matsumura K,
Sugimura K, Nakatani T and Kawashima H: Expression and function of
human steroid receptor RNA activator in prostate cancer cells: Role
of endogenous hSRA protein in androgen receptor-mediated
transcription. Prostate Cancer Prostatic Dis. 9:173–178. 2006.
|
|
24
|
Kim LK, Park SA, Yang Y, Kim YT, Heo TH
and Kim HJ: LncRNA SRA mediates cell migration, invasion, and
progression of ovarian cancer via NOTCH signaling and
epithelial-mesenchymal transition. Biosci Rep.
41:BSR202105652021.
|
|
25
|
Nilsson S, Mäkelä S, Treuter E, Tujague M,
Thomsen J, Andersson G, Enmark E, Pettersson K, Warner M and
Gustafsson JA: Mechanisms of estrogen action. Physiol Rev.
81:1535–1565. 2001.
|
|
26
|
Eyster KM: The estrogen receptors: An
overview from different perspectives. Methods Mol Biol. 1366:1–10.
2016.
|
|
27
|
Moggs JG and Orphanides G: Estrogen
receptors: Orchestrators of pleiotropic cellular responses. EMBO
Rep. 2:775–781. 2001.
|
|
28
|
Carroll JS, Meyer CA, Song J, Li W,
Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC,
Hall GF, et al: Genome-wide analysis of estrogen receptor binding
sites. Nat Genet. 38:1289–1297. 2006.
|
|
29
|
Stein RA, Chang CY, Kazmin DA, Way J,
Schroeder T, Wergin M, Dewhirst MW and McDonnell DP:
Estrogen-related receptor alpha is critical for the growth of
estrogen receptor-negative breast cancer. Cancer Res. 68:8805–8812.
2008.
|
|
30
|
Bhan A, Hussain I, Ansari KI, Kasiri S,
Bashyal A and Mandal SS: Antisense transcript long noncoding RNA
(lncRNA) HOTAIR is transcriptionally induced by estradiol. J Mol
Biol. 425:3707–3722. 2013.
|
|
31
|
Sørensen KP, Thomassen M, Tan Q, Bak M,
Cold S, Burton M, Larsen MJ and Kruse TA: Long non-coding RNA
HOTAIR is an independent prognostic marker of metastasis in
estrogen receptor-positive primary breast cancer. Breast Cancer Res
Treat. 142:529–536. 2013.
|
|
32
|
Cantile M, Di Bonito M, Cerrone M, Collina
F, De Laurentiis M and Botti G: Long Non-Coding RNA HOTAIR in
breast cancer therapy. Cancers (Basel). 12:11972020.
|
|
33
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010.
|
|
34
|
Tao S, He H and Chen Q: Estradiol induces
HOTAIR levels via GPER-mediated miR-148a inhibition in breast
cancer. J Transl Med. 13:1312015.
|
|
35
|
Aiello A, Bacci L, Re A, Ripoli C,
Pierconti F, Pinto F, Masetti R, Grassi C, Gaetano C, Bassi PF, et
al: MALAT1 and HOTAIR Long Non-Coding RNAs play opposite role in
estrogen-mediated transcriptional regulation in prostate cancer
cells. Sci Rep. 6:384142016.
|
|
36
|
Zhao Z, Chen C, Liu Y and Wu C:
17β-Estradiol treatment inhibits breast cell proliferation,
migration and invasion by decreas-815 ing MALAT-1 RNA level.
Biochem Biophys Res Commun. 445:388–393. 2014.
|
|
37
|
Wang Z, Katsaros D, Biglia N, Shen Y, Fu
Y, Loo LWM, Jia W, Obata Y and Yu H: High expression of long
non-coding RNA MALAT1 in breast cancer is associated with poor
relapse-free survival. Breast Cancer Res Treat. 171:261–271.
2018.
|
|
38
|
Huang Z, Qin Q, Xia L, Lian B, Tan Q, Yu Y
and Mo Q: Significance of Oncotype DX 21-gene test and expression
of long Non-Coding RNA MALAT1 in early and estrogen
receptor-positive breast cancer patients. Cancer Manag Res.
13:587–593. 2021.
|
|
39
|
Alipoor FJ, Asadi MH and Torkzadeh-Mahani
M: MIAT lncRNA is overexpressed in breast cancer and its inhibition
triggers senescence and G1 arrest in MCF7 cell line. J Cell
Biochem. 119:6470–6481. 2018.
|
|
40
|
Luan T, Zhang X, Wang S, Song Y, Zhou S,
Lin J, An W, Yuan W, Yang Y, Cai H, et al: Long non-coding RNA MIAT
promotes breast cancer progression and functions as ceRNA to
regulate DUSP7 expression by sponging miR-155-5p. Oncotarget.
8:76153–76164. 2017.
|
|
41
|
Li Y, Jiang B, Wu X, Huang Q, Chen W, Zhu
H, Qu X, Xie L, Ma X and Huang G: Long non-coding RNA MIAT is
estrogen-responsive and promotes estrogen-induced proliferation in
ER-positive breast cancer cells. Biochem Biophys Res Commun.
503:45–50. 2018.
|
|
42
|
Wang Z, Katsaros D, Biglia N, Shen Y, Loo
L, Yu X, Lin H, Fu Y, Chu WM, Fei P, et al: ERα upregulates the
expression of long non-coding RNA LINC00472 which suppresses the
phosphorylation of NF-κB in breast cancer. Breast Cancer Res Treat.
175:353–368. 2019.
|
|
43
|
Jonsson P, Coarfa C, Mesmar F, Raz T,
Rajapakshe K, Thompson JF, Gunaratne PH and Williams C:
Single-Molecule sequencing reveals estrogen-regulated clinically
relevant lncRNAs in Breast Cancer. Mol Endocrinol. 29:1634–1645.
2015.
|
|
44
|
Liu D, Rudland PS, Sibson DR and
Barraclough R: Identification of mRNAs differentially-expressed
between benign and malignant breast tumour cells. Br J Cancer.
87:423–431. 2002.
|
|
45
|
Miano V, Ferrero G, Reineri S, Caizzi L,
Annaratone L, Ricci L, Cutrupi S, Castellano I, Cordero F and De
Bortoli M: Luminal long non-coding RNAs regulated by estrogen
receptor alpha in a ligand-independent manner show functional roles
in breast cancer. Oncotarget. 7:3201–3216. 2016.
|
|
46
|
Niknafs YS, Han S, Ma T, Speers C, Zhang
C, Wilder-Romans K, Iyer MK, Pitchiaya S, Malik R, Hosono Y, et al:
The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1
in breast cancer progression. Nat Commun. 7:127912016.
|
|
47
|
Hu C: LncRNA DSCAM-AS1: A pivotal
therapeutic target in cancer. Mini Rev Med Chem. 23:530–536.
2023.
|
|
48
|
Sun W, Li AQ, Zhou P, Jiang YZ, Jin X, Liu
YR, Guo YJ, Yang WT, Shao ZM and Xu XE: DSCAM-AS1 regulates the
G1/S cell cycle transition and is an independent prognostic factor
of poor survival in luminal breast cancer patients treated with
endocrine therapy. Cancer Med. 7:6137–6146. 2018.
|
|
49
|
Treeck O, Weber F, Fritsch J, Skrzypczak
M, Schüler-Toprak S, Buechler C and Ortmann O: DSCAM-AS1 Long
Non-Coding RNA exerts oncogenic functions in endometrial
adenocarcinoma via activation of a tumor-promoting transcriptome
profile. Biomedicines. 10:17272022.
|
|
50
|
Yadav N, Sunder R, Desai S, Dharavath B,
Chandrani P, Godbole M and Dutt A: Progesterone modulates the
DSCAM-AS1/miR-130a/ESR1 axis to suppress cell invasion and
migration in breast cancer. Breast Cancer Res. 24:972022.
|
|
51
|
Xu SP, Zhang JF, Sui SY, Bai NX, Gao S,
Zhang GW, Shi QY, You ZL, Zhan C and Pang D: Downregulation of the
long noncoding RNA EGOT correlates with malignant status and poor
prognosis in breast cancer. Tumour Biol. 36:9807–9812. 2015.
|
|
52
|
Sun M, Gadad SS, Kim DS and Kraus WL:
Discovery, Annotation, and functional analysis of long noncoding
RNAs controlling cell-cycle gene expression and proliferation in
breast cancer cells. Mol Cell. 59:698–711. 2015.
|
|
53
|
Zhang X, Gao S, Li Z, Wang W and Liu G:
Identification and analysis of estrogen receptor α promoting
tamoxifen resistance-related lncRNAs. Biomed Res Int.
20:90317232020.
|
|
54
|
Basak P, Chatterjee S, Bhat V, Su A, Jin
H, Lee-Wing V, Liu Q, Hu P, Murphy LC and Raouf A: Long Non-Coding
RNA H19 acts as an estrogen receptor modulator that is required for
endocrine therapy resistance in ER+ Breast cancer cells. Cell
Physiol Biochem. 51:1518–1532. 2018.
|
|
55
|
Sun H, Wang G, Peng Y, Zeng Y, Zhu QN, Li
TL, Cai JQ, Zhou HH and Zhu YS: H19 lncRNA mediates
17β-estradiol-induced cell proliferation in MCF-7 breast cancer
cells. Oncol Rep. 33:3045–3052. 2015.
|
|
56
|
Ma T, Liang Y, Li Y, Song X, Zhang N, Li
X, Chen B, Zhao W, Wang L and Yang Q: LncRNA LINP1 confers
tamoxifen resistance and negatively regulated by ER signaling in
breast cancer. Cell Signal. 68:1095362020.
|
|
57
|
Si X, Zang R, Zhang E, Liu Y, Shi X, Zhang
E, Shao L, Li A, Yang N, Han X, et al: LncRNA H19 confers
chemoresistance in ERα-positive breast cancer through epigenetic
silencing of the pro-apoptotic gene BIK. Oncotarget. 7:81452–81462.
2016.
|
|
58
|
Tu C, Ren X, He J, Zhang C, Chen R, Wang W
and Li Z: The Value of LncRNA BCAR4 as a prognostic biomarker on
clinical outcomes in human cancers. J Cancer. 10:5992–6002.
2019.
|
|
59
|
Peng R, Cao J, Guo Q, Sun Q, Xu L, Xie X
and Song C: Variant in BCAR4 gene correlated with the breast cancer
susceptibility and mRNA expression of lncRNA BCAR4 in Chinese Han
population. Breast Cancer. 28:424–433. 2021.
|
|
60
|
Liang X, Zhao Y, Fang Z, Shao N, Zhai D,
Zhang M, Yu L and Shi Y: DLGAP1-AS2 promotes estrogen receptor
signalling and confers tamoxifen resistance in breast cancer. Mol
Biol Rep. 49:3939–3947. 2022.
|
|
61
|
Yuan H, Yan L, Wu M, Shang Y, Guo Q, Ma X,
Zhang X, Zhu Y, Wu Z, Lobie PE and Zhu T: Analysis of the Estrogen
Receptor-Associated LncRNA Landscape Identifies a Role for ERLC1 in
breast cancer progression. Cancer Res. 82:391–405. 2022.
|
|
62
|
Fang Z, Wang Y, Wang Z, Xu M, Ren S, Yang
D, Hong M and Xie W: ERINA Is an Estrogen-Responsive LncRNA That
Drives Breast Cancer through the E2F1/RB1 Pathway. Cancer Res.
80:4399–4413. 2020.
|
|
63
|
Yu S, Wang Y, Gong X, Fan Z, Wang Z, Liang
Z, Wu R, Cao B, Wang N, Bi C, et al: LncRNA AGPG confers endocrine
resistance in breast cancer by promoting E2F1 activity. Cancer Res.
Jul 28–2023.Epub ahead of print.
|
|
64
|
Horie K, Takagi K, Takeiwa T, Mitobe Y,
Kawabata H, Suzuki T, Ikeda K and Inoue S: Estrogen-Inducible
LncRNA BNAT1 functions as a modulator for estrogen receptor
signaling in endocrine-resistant breast cancer cells. Cells.
11:36102022.
|
|
65
|
Chen X, Ding JC, Hu GS, Shu XY, Liu Y, Du
J, Wen ZJ, Liu JY, Huang HH, Tang GH and Liu W: Estrogen-Induced
LncRNA, LINC02568, promotes estrogen receptor-positive breast
cancer development and drug resistance through both In Trans and In
Cis Mechanisms. Adv Sci (Weinh). 10:e22066632023.
|
|
66
|
Nanni S, Aiello A, Salis C, Re A, Cencioni
C, Bacci L, Pierconti F, Pinto F, Ripoli C, Ostano P, et al:
Metabolic reprogramming by malat1 depletion in prostate cancer.
Cancers (Basel). 13:152020.
|
|
67
|
Chakravarty D, Sboner A, Nair SS,
Giannopoulou E, Li R, Hennig S, Mosquera JM, Pauwels J, Park K,
Kossai M, et al: The oestrogen receptor alpha-regulated lncRNA
NEAT1 is a critical modulator of prostate cancer. Nat Commun.
5:53832014.
|
|
68
|
Bacci L, Aiello A, Ripoli C, Loria R,
Pugliese D, Pierconti F, Rotili D, Strigari L, Pinto F, Bassi PF,
et al: H19-Dependent transcriptional regulation of β3 and β4
Integrins Upon estrogen and hypoxia favors metastatic potential in
prostate cancer. Int J Mol Sci. 20:40122019.
|
|
69
|
Huang C, Wu M, Tang Y, Li X, Ouyang J,
Xiao L, Li D and Li G: NAG7 promotes human nasopharyngeal carcinoma
invasion through inhibition of estrogen receptor alpha and
up-regulation of JNK2/AP-1/MMP1 pathways. J Cell Physiol.
221:394–401. 2009.
|
|
70
|
Qiu JJ, Zhang XD, Tang XY, Zheng TT, Zhang
Y and Hua KQ: ElncRNA1, a long non-coding RNA that is
transcriptionally induced by oestrogen, promotes epithelial ovarian
cancer cell proliferation. Int J Oncol. 51:507–514. 2017.
|
|
71
|
Qiu J, Ye L, Ding J, Feng W, Zhang Y, Lv
T, Wang J and Hua K: Effects of oestrogen on long noncoding RNA
expression in oestrogen receptor alpha-positive ovarian cancer
cells. J Steroid Biochem Mol Biol. 141:60–70. 2014.
|
|
72
|
Chen Y, Gu Y, Gu Y and Wu J: Long
Noncoding RNA LINC00899/miR-944/ESR1 Axis regulates cervical cancer
cell proliferation, migration, and invasion. J Interferon Cytokine
Res. 41:220–233. 2021.
|
|
73
|
Cheng T, Bai Y, Huang S, Wang Y, Zhou S,
Liu H, Zhang R, Luo X and Yu P: Estrogen receptor 1 inhibits the
progression of hepatocellular carcinoma via positively regulating
lncRNA maternally expressed gene 3 under high glucose conditions. J
Gastrointest Oncol. 13:2485–2496. 2022.
|
|
74
|
Rochira V, Granata AR, Madeo B, Zirilli L,
Rossi G and Carani C: Estrogens in males: What have we learned in
the last 10 years? Asian J Androl. 7:3–20. 2005.
|
|
75
|
Girault I, Andrieu C, Tozlu S, Spyratos F,
Bièche I and Lidereau R: Altered expression pattern of
alternatively spliced estrogen receptor beta transcripts in breast
carcinoma. Cancer Lett. 215:101–112. 2004.
|
|
76
|
Tong D, Schuster E, Seifert M, Czerwenka
K, Leodolte S and Zeillinger R: Expression of estrogen receptor
beta isoforms in human breast cancer tissues and cell lines. Breast
Cancer Res Treat. 71:249–255. 2002.
|
|
77
|
Nanni S, Bacci L, Aiello A, Re A, Salis C,
Grassi C, Pontecorvi A, Gaetano C and Farsetti A: Signaling through
estrogen receptors modulates long non-coding RNAs in prostate
cancer. Mol Cell Endocrinol. 511:1108642020.
|
|
78
|
Piperigkou Z, Bouris P, Onisto M, Franchi
M, Kletsas D, Theocharis AD and Karamanos NK: Estrogen receptor
beta modulates breast cancer cells functional properties, signaling
and expression of matrix molecules. Matrix Biol. 56:4–23. 2016.
|
|
79
|
Mozdarani H, Ezzatizadeh V and Rahbar
Parvaneh R: The emerging role of the long non-coding RNA HOTAIR in
breast cancer development and treatment. J Transl Med.
18:1522020.
|
|
80
|
Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou
Z, Chang C, Qi J and Yeh S: Estrogen receptor β promotes renal cell
carcinoma progression via regulating LncRNA
HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene.
37:5037–5053. 2018.
|
|
81
|
He M, Yang H, Shi H, Hu Y, Chang C, Liu S
and Yeh S: Sunitinib increases the cancer stem cells and
vasculogenic mimicry formation via modulating the
lncRNA-ECVSR/ERβ/Hif2-α signaling. Cancer Lett. 524:15–28.
2022.
|
|
82
|
Chooniedass-Kothari S, Vincett D, Yan Y,
Cooper C, Hamedani MK, Myal Y and Leygue E: The protein encoded by
the functional steroid receptor RNA activator is a new modulator of
ER alpha transcriptional activity. FEBS Lett. 584:1174–1180.
2010.
|
|
83
|
Lin K, Zhan H, Ma J, Xu K, Wu R, Zhou C
and Lin J: Increased steroid receptor RNA activator protein (SRAP)
accompanied by decreased estrogen receptor-beta (ER-β) levels
during the malignant transformation of endometriosis associated
ovarian clear cell carcinoma. Acta Histochem. 116:878–882.
2014.
|
|
84
|
Li M, Chai HF, Peng F, Meng YT, Zhang LZ,
Zhang L, Zou H, Liang QL, Li MM, Mao KG, et al: Estrogen receptor β
upregulated by lncRNA-H19 to promote cancer stem-like properties in
papillary thyroid carcinoma. Cell Death Dis. 9:11202018.
|
|
85
|
Davey RA and Grossmann M: Androgen
receptor structure, function and biology: From bench to bedside.
Clin Biochem Rev. 37:3–15. 2016.
|
|
86
|
Wilson EM: Analysis of interdomain
interactions of the androgen receptor. Methods Mol Biol.
776:113–129. 2011.
|
|
87
|
van de Wijngaart DJ, Dubbink HJ, van Royen
ME, Trapman J and Jenster G: Androgen receptor coregulators:
Recruitment via the coactivator binding groove. Mol Cell
Endocrinol. 352:57–69. 2012.
|
|
88
|
Culig Z and Santer FR: Androgen receptor
signaling in prostate cancer. Cancer Metastasis Rev. 33:413–427.
2014.
|
|
89
|
Kumar S, Prajapati KS, Singh AK, Kushwaha
PP, Shuaib M and Gupta S: Long non-coding RNA regulating androgen
receptor signaling in breast and prostate cancer. Cancer Lett.
504:15–22. 2021.
|
|
90
|
Yang L, Lin C, Jin C, Yang JC, Tanasa B,
Li W, Merkurjev D, Ohgi KA, Meng D, Zhang J, et al:
lncRNA-dependent mechanisms of androgen-receptor-regulated gene
activation programs. Nature. 500:598–602. 2013.
|
|
91
|
Prensner JR, Sahu A, Iyer MK, Malik R,
Chandler B, Asangani IA, Poliakov A, Vergara IA, Alshalalfa M,
Jenkins RB, et al: The IncRNAs PCGEM1 and PRNCR1 are not implicated
in castration resistant prostate cancer. Oncotarget. 5:1434–1438.
2014.
|
|
92
|
Zhang A, Zhao JC, Kim J, Fong KW, Yang YA,
Chakravarti D, Mo YY and Yu J: LncRNA HOTAIR enhances the
androgen-receptor-mediated transcriptional program and drives
castration-resistant prostate cancer. Cell Rep. 13:209–221.
2015.
|
|
93
|
Li L, Dang Q, Xie H, Yang Z, He D, Liang
L, Song W, Yeh S and Chang C: Infiltrating mast cells enhance
prostate cancer invasion via altering LncRNA-HOTAIR/PRC2-androgen
receptor (AR)-MMP9 signals and increased stem/progenitor cell
population. Oncotarget. 6:14179–14190. 2015.
|
|
94
|
Ozgur E and Gezer U: Investigation of
lncRNA H19 in prostate cancer cells and secreted exosomes upon
androgen stimulation or androgen receptor blockage. Bratisl Lek
Listy. 121:362–365. 2020.
|
|
95
|
Warrick JI, Tomlins SA, Carskadon SL,
Young AM, Siddiqui J, Wei JT, Chinnaiyan AM, Kunju LP and
Palanisamy N: Evaluation of tissue PCA3 expression in prostate
cancer by RNA in situ hybridization-a correlative study with urine
PCA3 and TMPRSS2-ERG. Mod Pathol. 27:609–620. 2014.
|
|
96
|
Lemos AE, Ferreira LB, Batoreu NM, de
Freitas PP, Bonamino MH and Gimba ER: PCA3 long noncoding RNA
modulates the expression of key cancer-related genes in LNCaP
prostate cancer cells. Tumour Biol. 37:11339–11348. 2016.
|
|
97
|
Özgür E, Celik AI, Darendeliler E and
Gezer U: PCA3 silencing sensitizes prostate cancer cells to
enzalutamide-mediated androgen receptor blockade. Anticancer Res.
37:3631–3637. 2017.
|
|
98
|
Lv S, Pu X, Luo M, Wen H, Xu Z, Wei Q and
Dang Q: Long noncoding RNA GAS5 interacts and suppresses androgen
receptor activity in prostate cancer cells. Prostate. 81:893–901.
2021.
|
|
99
|
Zhou Y and Chen B: GAS5-mediated
regulation of cell signaling (Review). Mol Med Rep. 22:3049–3056.
2020.
|
|
100
|
Wang B, Xu W, Cai Y, Liu K, Wu J, Guo C
and Yuan C: The functional role of oncogenic LncRNA BCAR4 for
cancer outcome. Curr Pharm Des. 27:4107–4113. 2021.
|
|
101
|
Cai Z, Wu Y, Li Y, Ren J and Wang L: BCAR4
activates GLI2 signaling in prostate cancer to contribute to
castration resistance. Aging (Albany NY). 10:3702–3712. 2018.
|
|
102
|
Sakurai K, Reon BJ, Anaya J and Dutta A:
The lncRNA DRAIC/PCAT29 locus constitutes a tumor-suppressive
nexus. Mol Cancer Res. 13:828–838. 2015.
|
|
103
|
Saha S, Kiran M, Kuscu C, Chatrath A,
Wotton D, Mayo MW and Dutta A: Long Noncoding RNA DRAIC inhibits
prostate cancer progression by interacting with IKK to Inhibit
NF-κB Activation. Cancer Res. 80:950–963. 2020.
|
|
104
|
Malik R, Patel L, Prensner JR, Shi Y, Iyer
MK, Subramaniyan S, Carley A, Niknafs YS, Sahu A, Han S, et al: The
lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol
Cancer Res. 12:1081–1087. 2014.
|
|
105
|
Zhang Y, Pitchiaya S, Cieślik M, Niknafs
YS, Tien JC, Hosono Y, Iyer MK, Yazdani S, Subramaniam S, Shukla
SK, et al: Analysis of the androgen receptor-regulated lncRNA
landscape identifies a role for ARLNC1 in prostate cancer
progression. Nat Genet. 50:814–824. 2018.
|
|
106
|
Misawa A, Takayama K, Urano T and Inoue S:
Androgen-induced Long Noncoding RNA (lncRNA) SOCS2-AS1 promotes
cell growth and inhibits apoptosis in prostate cancer cells. J Biol
Chem. 291:17861–17880. 2016.
|
|
107
|
Fang Z, Xu C, Li Y, Cai X, Ren S, Liu H,
Wang Y, Wang F, Chen R, Qu M, et al: A feed-forward regulatory loop
between androgen receptor and PlncRNA-1 promotes prostate cancer
progression. Cancer Lett. 374:62–74. 2016.
|
|
108
|
Huang W, Su X, Yan W, Kong Z, Wang D,
Huang Y, Zhai Q, Zhang X, Wu H, Li Y, et al: Overexpression of
AR-regulated lncRNA TMPO-AS1 correlates with tumor progression and
poor prognosis in prostate cancer. Prostate. 78:1248–1261.
2018.
|
|
109
|
Takayama KI, Fujimura T, Suzuki Y and
Inoue S: Identification of long non-coding RNAs in advanced
prostate cancer associated with androgen receptor splicing factors.
Commun Biol. 3:3932020.
|
|
110
|
Dai X, Liu L, Liang Z, Guo K, Xu S and
Wang H: Silencing of lncRNA MALAT1 inhibits cell cycle progression
via androgen receptor signaling in prostate cancer cells. Pathol
Res Pract. 215:712–721. 2019.
|
|
111
|
Yao M, Shi X, Li Y, Xiao Y, Butler W,
Huang Y, Du L, Wu T, Bian X, Shi G, et al: LINC00675 activates
androgen receptor axis signaling pathway to promote
castration-resistant prostate cancer progression. Cell Death Dis.
11:6382020.
|
|
112
|
Li Z, Teng J, Jia Z, Zhang G and Ai X: The
long non-coding RNA PCAL7 promotes prostate cancer by strengthening
androgen receptor signaling. J Clin Lab Anal. 35:e236452021.
|
|
113
|
Shi Z, Chen J, Wumaner A and Li M, Liang C
and Li M: A novel long non-coding RNA PCLN16 facilitates androgen
receptor signaling in prostate cancer. Biochem Biophys Res Commun.
537:78–84. 2021.
|
|
114
|
Thomas PB, Jeffery P, Gahete MD, Whiteside
E, Walpole C, Maugham M, Jovanovic L, Gunter J, Williams E, Nelson
C, et al: The long non-coding RNA GHSROS reprograms prostate cancer
cell lines toward a more aggressive phenotype. Peer J.
9:e102802021.
|
|
115
|
Liu B, Qian D, Zhou W, Jiang H, Xiang Z
and Wu D: A Novel Androgen-Induced lncRNA FAM83H-AS1 Promotes
Prostate Cancer Progression via the miR-15a/CCNE2 Axis. Front
Oncol. 10:6203062021.
|
|
116
|
Zhang B, Zhang M, Shen C, Liu G, Zhang F,
Hou J and Yao W: LncRNA PCBP1-AS1-mediated AR/AR-V7
deubiquitination enhances prostate cancer enzalutamide resistance.
Cell Death Dis. 12:8562021.
|
|
117
|
Ghildiyal R, Sawant M, Renganathan A,
Mahajan K, Kim EH, Luo J, Dang HX, Maher CA, Feng FY and Mahajan
NP: Loss of Long Noncoding RNA NXTAR in prostate cancer augments
androgen receptor expression and enzalutamide resistance. Cancer
Res. 82:155–168. 2022.
|
|
118
|
Lu Y, Wan X, Huang W, Zhang L, Luo J, Li
D, Huang Y, Li Y and Xu Y: AC016745.3 regulates the transcription
of AR target genes by antagonizing NONO. Life (Basel).
11:12082021.
|
|
119
|
Huang W, Chen Q, Lu Y, Kong Z, Wan X,
Huang Y, Qiu M and Li Y: Androgen-Responsive Oncogenic lncRNA
RP11-1023L17.1 Enhances c-Myc protein stability in prostate cancer.
Int J Mol Sci. 23:122192022.
|
|
120
|
Zhou J, Anderson K, Bievre M, Ng S and
Bondy CA: Primate mammary gland insulin-like growth factor system:
Cellular localization and regulation by sex steroids. J Investig
Med. 49:47–55. 2001.
|
|
121
|
Dimitrakakis C and Bondy C: Androgens and
the breast. Breast Cancer Res. 11:2122009.
|
|
122
|
Hickey TE, Robinson JL, Carroll JS and
Tilley WD: Minireview: The androgen receptor in breast tissues:
Growth inhibitor, tumor suppressor, oncogene? Mol Endocrinol.
26:1252–1267. 2012.
|
|
123
|
Burstein MD, Tsimelzon A, Poage GM,
Covington KR, Contreras A, Fuqua SA, Savage MI, Osborne CK,
Hilsenbeck SG, Chang JC, et al: Comprehensive genomic analysis
identifies novel subtypes and targets of triple-negative breast
cancer. Clin Cancer Res. 21:1688–1698. 2015.
|
|
124
|
Collina F, Aquino G, Brogna M, Cipolletta
S, Buonfanti G, De Laurentiis M, Di Bonito M, Cantile M and Botti
G: LncRNA HOTAIR up-regulation is strongly related with lymph nodes
metastasis and LAR subtype of Triple Negative Breast Cancer. J
Cancer. 10:2018–2024. 2019.
|
|
125
|
Yang F, Shen Y, Zhang W, Jin J, Huang D,
Fang H, Ji W, Shi Y, Tang L, Chen W, et al: An androgen receptor
negatively induced long non-coding RNA ARNILA binding to miR-204
promotes the invasion and metastasis of triple-negative breast
cancer. Cell Death Differ. 25:2209–2220. 2018.
|
|
126
|
Leygue E: Steroid receptor RNA activator
(SRA1): Unusual bifaceted gene products with suspected relevance to
breast cancer. Nucl Recept Signal. 5:e0062007.
|
|
127
|
Huang G, Cao H, Liu G and Chen J: Role of
androgen receptor signaling pathway-related lncRNAs in the
prognosis and immune infiltration of breast cancer. Sci Rep.
12:206312022.
|
|
128
|
Xiong Y, Wang L, Li Y, Chen M, He W and Qi
L: The Long Non-Coding RNA XIST Interacted with MiR-124 to modulate
bladder cancer growth, invasion and migration by targeting androgen
receptor (AR). Cell Physiol Biochem. 43:405–418. 2017.
|
|
129
|
Wen L, Zhang X, Bian J, Han L, Huang H, He
M, Wei M and Wang P: The long non-coding RNA LINC00460 predicts the
prognosis and promotes the proliferation and migration of cells in
bladder urothelial carcinoma. Oncol Lett. 17:3874–3880. 2019.
|
|
130
|
Wu S, Zhang L, Deng J, Guo B, Li F, Wang
Y, Wu R, Zhang S, Lu J and Zhou Y: A novel micropeptide encoded by
Y-Linked LINC00278 links cigarette smoking and AR signaling in male
esophageal squamous cell carcinoma. Cancer Res. 80:2790–2803.
2020.
|
|
131
|
He SW, Xu C, Li YQ, Li YQ, Zhao Y, Zhang
PP, Lei Y, Liang YL, Li JY, Li Q, et al: AR-induced long non-coding
RNA LINC01503 facilitates proliferation and metastasis via the
SFPQ-FOSL1 axis in nasopharyngeal carcinoma. Oncogene.
39:5616–5632. 2020.
|
|
132
|
Schmidt K, Joyce CE, Buquicchio F, Brown
A, Ritz J, Distel RJ, Yoon CH and Novina CD: The lncRNA SLNCR1
Mediates Melanoma Invasion through a Conserved SRA1-like Region.
Cell Rep. 15:2025–2037. 2016.
|
|
133
|
Schmidt K, Carroll JS, Yee E, Thomas DD,
Wert-Lamas L, Neier SC, Sheynkman G, Ritz J and Novina CD: The
lncRNA SLNCR recruits the androgen receptor to EGR1-Bound genes in
melanoma and inhibits expression of tumor suppressor p21. Cell Rep.
27:2493–2507.e4. 2019.
|
|
134
|
Schmidt K, Weidmann CA, Hilimire TA, Yee
E, Hatfield BM, Schneekloth JS Jr, Weeks KM and Novina CD:
Targeting the Oncogenic Long Non-coding RNA SLNCR1 by blocking its
sequence-specific binding to the androgen receptor. Cell Rep.
30:541–554.e5. 2020.
|
|
135
|
Zhai W, Sun Y, Guo C, Hu G, Wang M, Zheng
J, Lin W, Huang Q, Li G, Zheng J and Chang C: LncRNA-SARCC
suppresses renal cell carcinoma (RCC) progression via altering the
androgen receptor(AR)/miRNA-143-3p signals. Cell Death Differ.
24:1502–1517. 2017.
|
|
136
|
Bai JY, Jin B, Ma JB, Liu TJ, Yang C,
Chong Y, Wang X, He D and Guo P: HOTAIR and androgen receptor
synergistically increase GLI2 transcription to promote tumor
angiogenesis and cancer stemness in renal cell carcinoma. Cancer
Lett. 498:70–79. 2021.
|
|
137
|
You B, Sun Y, Luo J, Wang K, Liu Q, Fang
R, Liu B, Chou F, Wang R, Meng J, et al: Androgen receptor promotes
renal cell carcinoma (RCC) vasculogenic mimicry (VM) via altering
TWIST1 nonsense-mediated decay through lncRNA-TANAR. Oncogene.
40:1674–1689. 2021.
|
|
138
|
Han H, Wang S, Meng J, Lyu G, Ding G, Hu
Y, Wang L, Wu L, Yang W, Lv Y, et al: Long noncoding RNA PART1
restrains aggressive gastric cancer through the epigenetic
silencing of PDGFB via the PLZF-mediated recruitment of EZH2.
Oncogene. 39:6513–6528. 2020.
|
|
139
|
Qin Z, Liu X, Li Z, Wang G, Feng Z, Liu Y,
Yang H, Tan C, Zhang Z and Li K: LncRNA LINC00667 aggravates the
progression of hepatocellular carcinoma by regulating androgen
receptor expression as a miRNA-130a-3p sponge. Cell Death Discov.
7:3872021.
|
|
140
|
D'Ambrosio DN, Clugston RD and Blaner WS:
Vitamin A metabolism: An update. Nutrients. 3:63–103. 2011.
|
|
141
|
Siddikuzzaman, Guruvayoorappan C and
Berlin Grace VM: All trans retinoic acid and cancer.
Immunopharmacol Immunotoxicol. 33:241–249. 2011.
|
|
142
|
Dawson MI and Xia Z: The retinoid X
receptors and their ligands. Biochim Biophys Acta. 1821:21–56.
2012.
|
|
143
|
Ghyselinck NB and Duester G: Retinoic acid
signaling pathways. Development. 146:dev1675022019.
|
|
144
|
Zhao H, Zhang X, Frazão JB, Condino-Neto A
and Newburger PE: HOX antisense lincRNA HOXA-AS2 is an apoptosis
repressor in all trans retinoic acid treated NB4 promyelocytic
leukemia cells. J Cell Biochem. 114:2375–2383. 2013.
|
|
145
|
El Hajj J, Nguyen E, Liu Q, Bouyer C,
Adriaenssens E, Hilal G and Ségal-Bendirdjian E: Telomerase
regulation by the long non-coding RNA H19 in human acute
promyelocytic leukemia cells. Mol Cancer. 17:852018.
|
|
146
|
Zheng C, Li X, Ren Y, Yin Z and Zhou B:
Long Noncoding RNA RAET1K Enhances CCNE1 expression and cell cycle
arrest of lung adenocarcinoma cell by sponging miRNA-135a-5p. Front
Genet. 10:13482020.
|
|
147
|
Hu L, Liu J, Meng Y, Zheng H, Ding C, Wang
H, Charwudzi A, Li M, Li J, Zhai Z and Xiong S: Long non-coding RNA
HOTAIR regulates myeloid differentiation through the upregulation
of p21 via miR-17-5p in acute myeloid leukaemia. RNA Biol.
18:1434–1444. 2021.
|
|
148
|
Wei S, Zhao M, Wang X, Li Y and Wang K:
PU.1 controls the expression of long noncoding RNA HOTAIRM1 during
granulocytic differentiation. J Hematol Oncol. 9:442016.
|
|
149
|
Chen ZH, Wang WT, Huang W, Fang K, Sun YM,
Liu SR, Luo XQ and Chen YQ: The lncRNA HOTAIRM1 regulates the
degradation of PML-RARA oncoprotein and myeloid cell
differentiation by enhancing the autophagy pathway. Cell Death
Differ. 24:212–224. 2017.
|
|
150
|
Tang D, Hu P, Zhu D, Luo Y, Chen M, Zhang
G and Wang Y: C/EBPα is indispensable for PML/RARα-mediated
suppression of long non-coding RNA NEAT1 in acute promyelocytic
leukemia cells. Aging (Albany NY). 13:13179–13194. 2021.
|
|
151
|
Pokorná M, Hudec M, Juříčková I, Vácha M,
Polívková Z, Kútna V, Pala J, Ovsepian SV, Černá M and O'Leary VB:
All-Trans retinoic acid fosters the multifarious U87MG cell line as
a model of glioblastoma. Brain Sci. 11:8122021.
|
|
152
|
Wang C, Zhao D, Wang K, Gao L, He Y, Wu H,
Ruan L, Chen W, Zhang D, Xia T, et al: All-Trans retinoic acid
rescues the tumor suppressive role of RAR-β by Inhibiting LncHOXA10
expression in gastric tumorigenesis. Nutr Cancer. 73:2065–2077.
2021.
|
|
153
|
Shan L, Liu W and Zhan Y: LncRNA HAND2-AS1
exerts anti-oncogenic effects on bladder cancer via restoration of
RARB as a sponge of microRNA-146. Cancer Cell Int. 21:3612021.
|
|
154
|
Fu L, Shi Z and Chen B: Deleted in
lymphocytic leukemia 2 induces retinoic acid receptor beta promoter
methylation and mitogen activated kinase-like protein activation to
enhance viability and mobility of colorectal cancer cells.
Bioengineered. 13:12847–12862. 2022.
|
|
155
|
Bikle DD: Vitamin D metabolism, mechanism
of action, and clinical applications. Chem Biol. 21:319–329.
2014.
|
|
156
|
Bikle DD, Jiang Y, Nguyen T, Oda Y and Tu
CL: Disruption of Vitamin D and Calcium signaling in keratinocytes
predisposes to skin cancer. Front Physiol. 7:2962016.
|
|
157
|
Jiang YJ and Bikle DD: LncRNA profiling
reveals new mechanism for VDR protection against skin cancer
formation. J Steroid Biochem Mol Biol. 144(Pt A): 87–90. 2014.
|
|
158
|
Kholghi Oskooei V, Geranpayeh L, Omrani MD
and Ghafouri-Fard S: Assessment of functional variants and
expression of long noncoding RNAs in vitamin D receptor signaling
in breast cancer. Cancer Manag Res. 10:3451–3462. 2018.
|
|
159
|
Gheliji T, Oskooei VK, Ashrafi Hafez A,
Taheri M and Ghafouri-Fard S: Evaluation of expression of vitamin D
receptor related lncRNAs in lung cancer. Noncoding RNA Res.
5:83–87. 2020.
|
|
160
|
Jin T, Guo Y and Huang Z, Zhang Q and
Huang Z, Zhang Y and Huang Z: Vitamin D inhibits the proliferation
of oral squamous cell carcinoma by suppressing lncRNA LUCAT1
through the MAPK pathway. J Cancer. 11:5971–5981. 2020.
|
|
161
|
Wang L, Zhou S and Guo B: Vitamin D
suppresses ovarian cancer growth and invasion by targeting long
Non-Coding RNA CCAT2. Int J Mol Sci. 21:23342020.
|
|
162
|
Fu Y, Katsaros D, Biglia N, Wang Z, Pagano
I, Tius M, Tiirikainen M, Rosser C, Yang H and Yu H: Vitamin D
receptor upregulates lncRNA TOPORS-AS1 which inhibits the
Wnt/β-catenin pathway and associates with favorable prognosis of
ovarian cancer. Sci Rep. 11:74842021.
|
|
163
|
Chen S, Bu D, Ma Y, Zhu J, Chen G, Sun L,
Zuo S, Li T, Pan Y, Wang X, et al: H19 overexpression induces
resistance to 1,25(OH)2D3 by Targeting VDR Through miR-675-5p in
colon cancer cells. Neoplasia. 19:226–236. 2017.
|
|
164
|
Timmermans S, Souffriau J and Libert C: A
general introduction to glucocorticoid biology. Front Immunol.
10:15452019.
|
|
165
|
Kino T, Hurt DE, Ichijo T, Nader N and
Chrousos GP: Noncoding RNA gas5 is a growth arrest- and
starvation-associated repressor of the glucocorticoid receptor. Sci
Signal. 3:ra82010.
|
|
166
|
Brent GA: Mechanisms of thyroid hormone
action. J Clin Invest. 122:3035–3043. 2012.
|
|
167
|
Aranda A: MicroRNAs and thyroid hormone
action. Mol Cell Endocrinol. 525:1111752021.
|
|
168
|
Huang PS, Chang CC, Wang CS and Lin KH:
Functional roles of non-coding RNAs regulated by thyroid hormones
in liver cancer. Biomed J. 44:272–284. 2021.
|
|
169
|
Lin YH, Wu MH, Huang YH, Yeh CT, Chi HC,
Tsai CY, Chuang WY, Yu CJ, Chung IH, Chen CY and Lin KH: Thyroid
hormone negatively regulates tumorigenesis through suppression of
BC200. Endocr Relat Cancer. 25:967–979. 2018.
|
|
170
|
Dai Q, Deng J, Zhou J, Wang Z, Yuan XF,
Pan S and Zhang HB: Long non-coding RNA TUG1 promotes cell
progression in hepatocellular carcinoma via regulating
miR-216b-5p/DLX2 axis. Cancer Cell Int. 20:82020.
|
|
171
|
Lin YH, Wu MH, Huang YH, Yeh CT and Lin
KH: TUG1 Is a Regulator of AFP and serves as prognostic marker in
Non-Hepatitis B Non-Hepatitis C hepatocellular carcinoma. Cells.
9:2622020.
|















