Introduction
Glioma is the most common malignant tumor of the
nervous system, with high malignancy, fast growth, a short disease
course, ease of recurrence and a high incidence of postoperative
complications (1,2). Glioma is considered to be one of the
most challenging and drug-resistant tumors in neurosurgical
treatment (3). Thus, it is urgent
to identify novel treatments for glioblastoma (GBM).
Copper is an integrant mineral for life and an
important component of numerous vital activities, including
biological oxidation-reduction, iron metabolism, antioxidant
effects and detoxification (4).
Copper has also been reported to promote angiogenesis, which is
important for tumor progression and metastasis. Researchers have
identified a novel method of cell death, cuproptosis, that differs
from known cell death types. Direct copper binding to the fatty
acylation component of the tricarboxylic acid cycle results in
cuproptosis, causing aggregation of fatty acylated proteins and
iron loss (5). The synthesis of
sulfur cluster proteins results in proteotoxic stress, in turn
causing cell death (5). The
potential impact of copper on the development of cancer cells has
been the subject of numerous investigations (5,6).
Copper has a strong affinity for MEK1 and binds to it directly. By
activating ERK1/2 downstream, it stimulates the growth of tumors
(6). Disruption of copper
homeostasis reportedly triggers cuproptosis, thereby synergizing
with regorafenib-mediated lethal inhibition of autophagy in GBM.
Therefore, Cu2+ and regorafenib may serve a role in the
treatment of GBM by regulating autophagy and cuproptosis (7). Ferredoxin 1 (FDX1), a pivotal
determiner of cuproptosis, works by using ferredoxin reductase to
transport electrons from NADPH to cytochrome P450 (8). Unlike the majority of malignancies,
GBM multiforme, gastric adenocarcinoma and endometrial cancer all
exhibit high levels of FDX1 expression (9). High FDX1 expression is inversely
related to the prognosis of patients with GBM (9). Research has reported the possibility
of reducing copper levels in patients with newly diagnosed GBM. The
safety profile is consistent with the known toxicity profile, and
primarily related to hematology, penicillamine and copper
deficiency (10). A phase II
trial of penicillamine for copper depletion and anti-angiogenic
therapy in GBM is underway (10).
Copper has also been found to induce GBM cell senescence by
downregulating B lymphoma Mo-MLV insertion region 1 homolog.
Therefore, by inducing premature senescence in GBM cells, it may
provide a useful in vitro model for the development of novel
tumor therapeutic strategies (11). Disulfiram combined with copper can
improve the efficacy of temozolomide in the treatment of GBM
(12). Cuproptosis-related genes
have also been used in tumor diagnosis. Solute carrier family 31
member 1 has been selected as a potential cuproptosis-related gene
in breast cancer because its expression was upregulated, and
exhibited the ability to predict diagnosis, prognosis and drug
response (13).
Cuproptosis-related signatures help predict prognosis and guide
treatment in patients with hepatocellular carcinoma (14). Cuproptosis serves an important
role in tumors, and the levels of cuproptosis, prognosis, diagnosis
and mechanisms of immune responses in different tumors warrant
further study.
EMD-1204831 was first developed and synthesized by
Merck Serono; Merck KGaA. It could highly selectively inhibit c-Met
receptor tyrosine kinase activity in both a ligand-dependent and
-independent manner (15). In
mice with ligand-dependent Hs746T and hepatocyte growth
factor-dependent U87MG cancer, EMD-1204831 treatment was associated
with potent tumor growth suppression and regression; however, to
the best of our knowledge, the specific mechanism of action is
unknown (15). Since studies of
EMD-1204831 in GBM are not precise enough, a comprehensive
mechanistic analysis of its anti-GBM action was conducted.
Materials and methods
Data collection
The Cancer Genome Atlas (TCGA; https://portal.gdc.cancer.gov/) and Chinese
Glioma Genome Atlas (CGGA; https://www.cgga.org.cn/) were mined to collect data
of patients with GBM and clinical information, which included tumor
tissues and normal tissues from healthy controls (TCGA, TCGA-GBM;
CGGA, mRNAseq_693). The transcript information of patients with GBM
was obtained from the GSE100675 dataset in the Gene Expression
Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), including 19
control individuals and 19 patients with GBM. The transcripts per
kilobase of exon model per million mapped reads method was used for
data normalization. The CGGA dataset was used for the consensus
analysis. TCGA data were used to construct the least absolute
shrinkage and selector operator (LASSO) model. The GSE100675
dataset was used to perform differentially expressed gene (DEG)
analysis. The cuproptosis-related genes, including FDX1, LIPT1,
DLD, LIAS, dihydrolipoamide S-acetyltransferase (DLAT), PDHA1,
PDHB, MTF1, GLS and CDKN2A, were obtained from a previous study
(5).
DEG analysis
The R (R-4.2.0; https://www.r-project.org/) package limma (3.58.1;
https://www.bioconductor.org/packages/release/bioc/html/limma.html)
was used for DEG analysis. Statistically, genes were considered
significantly differentially expressed when log|fold change|≥1 and
P<0.05. Additionally, for cuproptosis-related genes in TCGA-GBM
dataset, consensus clustering was conducted using the
ConsensusClusterPlus (1.70.0; https://bioconductor.org/packages/release/bioc/html/ConsensusClusterPlus.html)
R package, and principal component analysis was performed to
visualize the distribution of subtypes.
Gene set enrichment analysis (GSEA)
After obtaining the DEGs, GSEA was performed to
assess enrichment based on Gene Ontology terms and signaling
pathways related to these genes. This process was performed using
the enrichplot (1.22.0; https://bioconductor.org/packages/release/bioc/html/enrichplot.html)
and clusterprofiler (4.10.0; https://www.bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
packages in R. Enrichment results were considered statistically
significant when P<0.05.
LASSO regression analysis
A risk score model was constructed using LASSO
regression. First, univariate Cox proportional hazard regression
analysis was implemented by applying the R package survival (3.5-7;
https://cran.r-project.org/web/packages/survival/index.html)
and genes related to overall survival (OS) were identified. Next,
to screen the most characteristic genes among them, LASSO
regression analysis was performed, where the dataset from TCGA was
selected as the training set and the dataset from CGGA was selected
as the verification set. The risk score was calculated using the
following formula: Risk score=∑(Xi x Coefi), where Xi is the
normalized expression value of the gene and Coefi is the
coefficient.
Single-cell RNA sequencing (scRNA-seq)
analysis
For scRNA-seq analysis, the Seurat package in R
(5.0.2; https://cran.r-project.org/web/packages/Seurat/index.html)
was used to analyze the GSM6432709 dataset from GEO (16). First, the data were preprocessed
and all batch effects were removed. The data were clustered using
the FindCluster function, and the marker genes of each cell cluster
were obtained using the Findmarkers function. The Monocle3 package
(4.1.0; https://cole-trapnell-lab.github.io/monocle3/docs/installation/)
was also used to perform pseudotemporal trajectory analysis to
explore the relationship between the prognostic genes and disease
development.
Protein level validation of central
genes
The Human Protein Atlas (https://www.proteinatlas.org/) is designed to create
human proteome-wide maps. To validate the protein expression levels
of the nine genes [AE binding protein 1 (AEBP1), NOP2/Sun RNA
methyltransferase 5 (NSUN5), MED10, CCM2 scaffold protein (CCM2),
oncostatin M receptor (OSMR), ribosomal protein L39 like (RPL39L),
EN2, collagen type XXII α1 chain (COL22A1) and DNAJC3] obtained
from the LASSO regression analysis, the protein expression levels
of target genes in the Human Protein Atlas were screened in
patients with colon, breast and lung cancer.
Immune cell infiltration analysis
To analyze immune infiltration in GBM, the dataset
was divided into high-risk and low-risk groups, and the control and
disease groups from the dataset were compared. This analysis
process was performed using the Cibersort (R script v1.04;
https://cibersortx.stanford.edu/)
algorithm. When P<0.05, the analysis results were considered
statistically significant.
DNA methylation analysis
A dataset from TCGA was used in the present study.
First, quality control and normalization analysis were performed on
the dataset (Benjamini-Hochberg correction; P<0.05).
Subsequently, differential methylation sites (Δβ values >0.1)
were obtained using the ChAMP package in R (2.36.0; https://www.bioconductor.org/packages/release/bioc/html/ChAMP.html).
Somatic mutation analysis
The somatic mutation data of GBM samples were
downloaded from TCGA. The mutation data were analyzed and
visualized using the maftools package in R (2.22.0; https://www.bioconductor.org/packages/release/bioc/html/maftools.html).
Drug sensitivity analysis
The CellMiner tool (v2.9; https://discover.nci.nih.gov/cellminer/home.do)
was used for drug susceptibility analysis. Drugs with high
sensitivity [correlation coefficient (cor) >0.3; P<0.05] were
screened. Next, the relationship between drugs and target genes was
visualized through box plots and correlation curves.
Collection of active ingredients of
Ginseng
The Traditional Chinese Medicine Systems
Pharmacology database (TCMSP) was investigated to determine the
ingredients of Ginseng (https://old.tcmsp-e.com/tcmsp.php). This database
includes a set of ingredients, targeted genes and pharmacokinetic
properties of natural compounds. To determine the active
ingredients of Ginseng in the database, the screening criteria
were: Oral bioavailability ≥30% and drug-likeness ≥0.18.
Molecular docking
Molecular docking tools were used to analyze the
interaction between the predicted drugs and the target genes. This
was performed using AutoDock software (v1.2.x; https://autodock.scripps.edu/), and the results were
visualized using Discovery Studio Visualizer (v.19; https://discover.3ds.com/discovery-studio-visualizer-download)
software.
Cell culture
The pharmacological experiments in the present study
were conducted using GBM cells. The 293T, LN229 and A172 cells
obtained from American Type Culture Collection were grown in DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin and streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
at 37°C in an incubator with 5% CO2.
Transfection
To silence COL22A1, short hairpin RNA (shRNA/sh)
specific to COL22A1 (shCOL22A1-1, 5′-GGT CTT GTT TAG AGT CTG A-3′;
shCOL22A1-2, 5′-GTC TGA GTT TGT GAG ATT A-3′) and negative control
shRNA (shNC; 5′-CAA CAA GAT GAA GAG CAC CAA-3′) were designed and
synthesized by Sangon Biotech Co., Ltd. The 2nd generation system
was used to generate lentivirus. The shRNA was integrated into the
pLKO.1 vector (2 µg; 8453; Addgene, Inc.), and transfected
into 293T cells along with psPAX2 (1 µg; 8454; Addgene,
Inc.) and PMD2.G (1 µg; 12260; Addgene, Inc.) using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) for
48 h at 37°. Viruses were collected at 48 and 72 h and filtered
using a 0.45-µm nitrocellulose filter. Polyethylene glycol
6000 was added and the viruses were concentrated by
ultracentrifugation (4°C; 1,500 × g; 30 min). Subsequently, the
virus was added to the cells for 2 days. Target cells were infected
with virus using polybrene (8 mg/ml). The multiplicity of infection
used to infect cells was 10. The next day, the cells were selected
with puromycin for 48 h (2-5 µg/ml; MilliporeSigma). The
transfection efficiencies were detected by western blotting. Next,
the cells were used for subsequent experiments. The time interval
between transduction and subsequent experimentation was ~5 days.
Puromycin (1-2 µg/ml; MilliporeSigma) was used for
maintenance.
Cell Counting Kit-8 (CCK-8) assay
To study cell proliferation following drug
intervention, LN229 and A172 cells were treated with DMSO (<0.1%
v/v; HY-Y0320; MedChemExpress), kaempferol (80 µM; HY-14590;
MedChemExpress), EMD-1204831 (9 µM; HY-164394;
MedChemExpress), the combination of kaempferol (9 µM) and
EMD-1204831 (80 µM) or elesclomol (HY-12040; MedChemExpress)
for 24 h at 37°C, and CCK-8 cell viability experiments (Merck KGaA)
were conducted. First, 5,000 cells per well were seeded in 96-well
plates. Cells were collected and the CCK-8 reagent (10 µl
per well) was added at 72 h for incubation for 2 h in the dark at
37°C with 5% CO2. The VersaMax microplate reader
(Molecular Devices, LLC) was used to measure absorbance at a
wavelength of 450 nm.
Cell colony formation assay
A cell colony formation experiment was conducted to
assess cell proliferation. A total of 2×103 LN229 and
A172 cells were grown for 14 days on 6-well plates and treated with
DMSO, kaempferol (80 µM), EmdMD-1204831 (9 µM) or the
combination of both at 37°C for 24 h. When there were >50 cells
for a single cell clone, cloning was deemed complete. The cells
were then dyed with viola crystallina for 30 min at 37°C after
fixation with 10% paraformaldehyde (Thermo Fisher Scientific, Inc.)
for 30 min at room temperature. Subsequently, the clone numbers
were counted manually. The definition of colonies was >50 cells
for a single cell clone.
Transmission electron microscopy
For electron microscopy analysis, tumors were
dissected (1 mm3) in an ice-cold fixative (2.5%
glutaraldehyde in 0.1 mol/l PIPES buffer at pH 7.4). After 10 h of
fixation at 4°C, samples were washed with 0.1 mol/l PIPES,
post-fixed in 1% OsO4 (30 min at room temperature) and
stained in 2% uranyl acetate (1 h at room temperature). Samples
were dehydrated in an ethanol series (50, 70 and 100%) and embedded
in epoxy (37°C; overnight; 12-16 h). The tissues were divided into
~70 nm slices. The slices were observed and images were captured
with a HT-7700 transmission electron microscope.
Ki67 staining
Tumor tissues were stained using Ki67 antibody
(9027; 1:400; Cell Signaling Technology, Inc.) and Ki67-positive
cells were counted as described previously (17). The results of Ki67 staining were
recorded as the percentage of Ki67-positive cells.
Mice
A total of 20 male BALB/c-nude mice (6 weeks;
weight, 17-21 g) from Shanghai Model Organisms Center, Inc. were
stochastically separated into four groups, with 5 mice in each
group. The housing conditions were as follows: Temperature,
20-26°C; humidity, 50-70%; light/dark cycle, 12 h/12 h; and feeding
food and water in the cage every day. A total of 5×106
LN229 cells were resuspended in 1 ml PBS, 8 µl
(4×104) of which were stereotactically injected into the
right hemisphere of the brain, at 0.5 mm posterior of the bregma, 2
mm to the right of the sagittal suture and 3 mm below the surface
of the skull. Deep anesthesia was used to control and treat pain
(isoflurane; induction, 4-5%; maintenance, 2%). The cells were
injected into the brain through microsyringes. When the volume of
the tumor reached 100 mm3, the four groups of mice were
treated as follows: No treatment in the control group, injection of
EMD-1204831 (10 mg/kg), injection of kaempferol (20 mg/kg) and
simultaneous injection of EMD-1204831 and kaempferol. The drugs
were injected every 2 days. The tumor cells were infected with
lentivirus generating luciferase and tumor growth was monitored by
bioluminescent imaging. Tumor volumes were measured every 4 days.
Tumors were peeled off for testing. Mice were euthanized by
intraperitoneal injection of sodium pentobarbital (100-200 mg/kg)
after 4 weeks. The tumors were examined by H&E staining
according to the protocol in the kit (fixation with 4%
paraformaldehyde for 6-8 h at room temperature; thickness of
sections, 5 µm; staining with haematoxylin for 4 min,
hydrochloric acid alcohol for 3 sec and eosin for 25 sec-1 min; all
staining steps were performed at room temperature; type of
microscope used, light microscope). After euthanasia, the death of
the animals was confirmed based on the disappearance of pain
response, no response when pressing the toes with hands or forceps,
and observation of cardiac and respiratory arrest. The time
interval between injection/implantation and the final tumor growth
measurement was ~28 days.
Reactive oxygen species (ROS) assay
A ROS detection kit (C10445; Invitrogen; Thermo
Fisher Scientific, Inc.) was used to detect the levels of ROS in
LN229 and A172 glioma cell lines treated with CuCl2 and
the cuproptosis inducer elesclomol (STA-4783; MedChemExpress).
Firstly, GBM carcinoma cells in the logarithmic growth phase were
inoculated into a 12-well plate and further cultured. When the cell
density reached 40-50%, 200 nM elesclomol was added to treat the
cells for 24 h at 37°C, followed by the addition of 10 µM
ROS fluorescence probe (S0033S; Beyotime Institute of
Biotechnology) for incubation at 37°C for 20 min. Finally, flow
cytometry (BD LSRFortessa; BD Biosciences) was used to detect the
intensity of the fluorescence signals at an excitation wavelength
of 488 nm, which represented the levels of ROS (fluorescence
emission: Before lipid peroxidation, analyte detector, 581 nm,
analyte reporter, 591 nm; after lipid peroxidation, analyte
detector, 488 nm, analyte reporter, 510 nm). Lipid peroxidation was
determined by quantitating the fluorescence intensities and
calculating the ratio of intensity in the Texas Red®
channel (581/591 nm) to the intensity in the FITC channel (488/519
nm). Flowjo v9 software (BD Biosciences) was used.
Measurement of intracellular copper
A total of 2×105 LN229 and A172 GBM cells
per well in the logarithmic growth phase were inoculated into a
6-well plate and cultured for 12 h. Subsequently, cells were
incubated with different concentrations of CuCl2 (0,
0.5, 1, 2, 2.5, 3, 4 and 5 µmol/l) to generate a standard
curve (data not shown) and the cuproptosis inducer elesclomol (200
nM) for 24 h at 37°C. Cells were collected and resuspended in 120
µl water. Ultrasound was used to break the cells to collect
copper ions inside the cells. Finally, copper ions inside the cells
were detected according to the instructions (E-BC-K775-M;
Elabscience Bionovation Inc.) using a microplate reader.
Western blotting
LN229 and A172 glioma cells in the logarithmic
growth phase were inoculated into a 6-well plate, and the cells
were cultured until they reached a density of 40-50%. Different
concentrations of CuCl2 (0, 0.5, 1, 2, 2.5, 3, 4 and 5
µmol/l) and the cuproptosis inducer elesclomol (200 nM) were
added to treat the cells at 37°C overnight. The cells were washed
with PBS once. Total protein was extracted from the cells with an
appropriate amount of RIPA lysis buffer (P0013C; Beyotime Institute
of Biotechnology) and the protein concentration was measured using
a BCA kit (23225; Thermo Fisher Scientific, Inc.). The mass of
protein loaded per lane was ~100 µg. The percentage of the
SDS-PAGE gel was 10%. The expression of proteins was detected
through western blotting. Proteins were transferred to
nitrocellulose membranes. The membranes were blocked in 5% milk
containing 2% BSA (A8020; Beijing Solarbio Science & Technology
Co., Ltd.) at room temperature for 1 h. Membranes were incubated at
4°C overnight (12-16 h) with the primary antibodies, including
COL22A1 (1:2,000; PA5-70816; Invitrogen; Thermo Fisher Scientific,
Inc.), DLAT (1:3,000; cat. no. 13426-1-AP; Proteintech Group,
Inc.), dihydrolipoamide S-succinyltransferase (1:10,000; cat. no.
ab177934; Abcam), FDX1 (1:2,000; cat. no. 12592-1-AP; Proteintech
Group, Inc.), ACTB (1:3,000; ab8226; Abcam) and GAPDH (1:50,000;
cat. no. 60004-1-Ig; Proteintech Group, Inc.) antibodies, then
washed with PBS with 0.5% Tween-20 three times, and an appropriate
amount of the HRP-conjugated secondary antibody was added for
incubation at room temperature for 1 h (anti-mouse IgG; 1:5,000;
ab7068; Abcam; anti-rabbit IgG; 1:5,000; ab288151; Abcam). An ECL
kit was used for visualization (322209; Thermo Fisher Scientific,
Inc.). Images were captured using the BIO-RAD ChemiDoc MP
multifunctional chemical imaging instrument (Bio-Rad Laboratories,
Inc.).
Statistical analysis
Kaplan-Meier plots were plotted and the log-rank
test was performed to estimate and compare the OS between the two
risk groups in the training set. The Renyi test was performed to
estimate and compare the OS between the two risk groups in the
verification set. The hazard ratio and Cox P-values were generated
using Cox analysis. The correlations between different factors were
analyzed using Spearman's statistical methods. Statistical analysis
was performed using GraphPad Prism 8.0 software (Dotmatics). Data
are presented as the mean ± standard deviation of three
experimental repeats. An unpaired Student's t-test was used for
comparisons between two groups. Differences among multiple groups
were analyzed by one-way ANOVA followed by Tukey's post hoc test.
Differences among multiple groups containing two variables were
analyzed using two-way ANOVA followed by Bonferroni's post hoc
analysis. The tumor volumes of mice in different drug groups were
analyzed using two-way mixed ANOVA with the Bonferroni post hoc
test. The immune cell infiltration based on Cibersort was analyzed
using the Wilcox rank-sum test. P<0.05 was considered to
indicate a statistically significant difference.
Results
Consensus clustering of
cuproptosis-related genes in patients with GBM
Although cuproptosis has been shown to serve an
important role in GBM (9), to the
best of our knowledge, the precise mechanism of its action remains
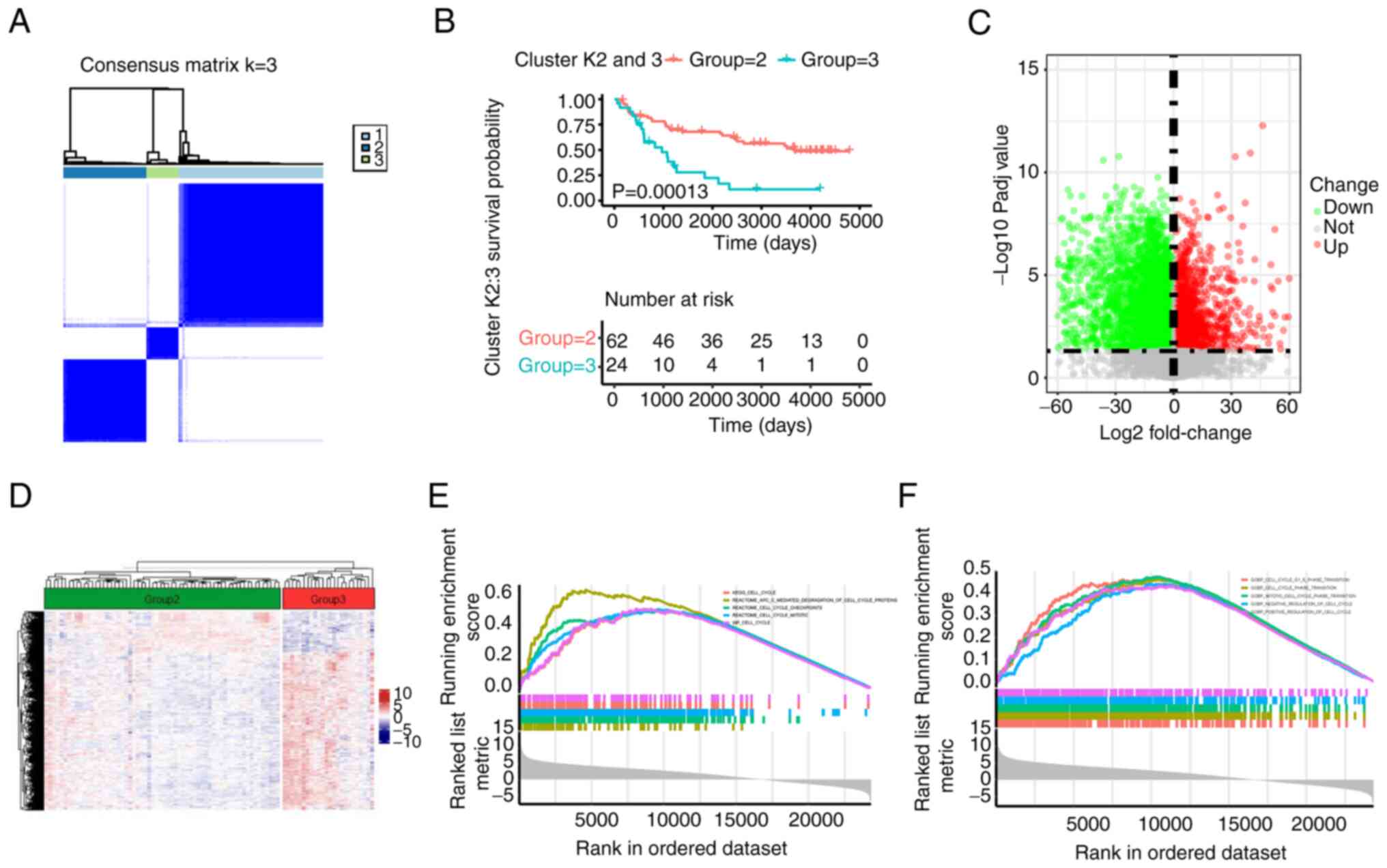
unknown. First, the present study grouped the GBM cohort (k=3) by
consensus clustering analysis according to cuproptosis-related
genes, which means there were three cuproptosis levels (high,
middle and low; Figs. 1A, and
S1A and B). Next, different
cuproptosis levels of patients will be associated with different
outcomes. For example, high cuproptosis levels of patients are
associated with a longer OS than low cuproptosis levels (18). Therefore, to identify the high and
low groups of cuproptosis levels in patients, survival analysis for
the three groups was performed and the results revealed that the
prognosis of cluster 2 and 3 was significantly better than that of
cluster 1, and cluster 2 had a longer OS than cluster 3, thus
cluster 2 was considered as relative high level of cuproptosis and
cluster 3 as cuproptosis low level because the OS in cluster 2 was
longer than that in cluster 3 (18) (data not shown). Therefore, groups
2 and 3 were selected for comparison. The survival performance of
the former was improved according to the Kaplan-Meier analysis
(P=0.00013; Fig. 1B).
Subsequently, DEGs were compared between the two groups. As shown
in Figs. 1C and D, and S1C, a total of 6,935 DEGs were
identified, including 1,885 upregulated genes and 5,050
downregulated genes. GSEA verified that the DEGs were primarily
gathered in 'cell cycle' pathways (Figs. 1E and F, and S1D). These results suggested that the
cuproptosis-related DEGs were associated with malignant progression
of GBM.
LASSO regression analysis for patients
with GBM
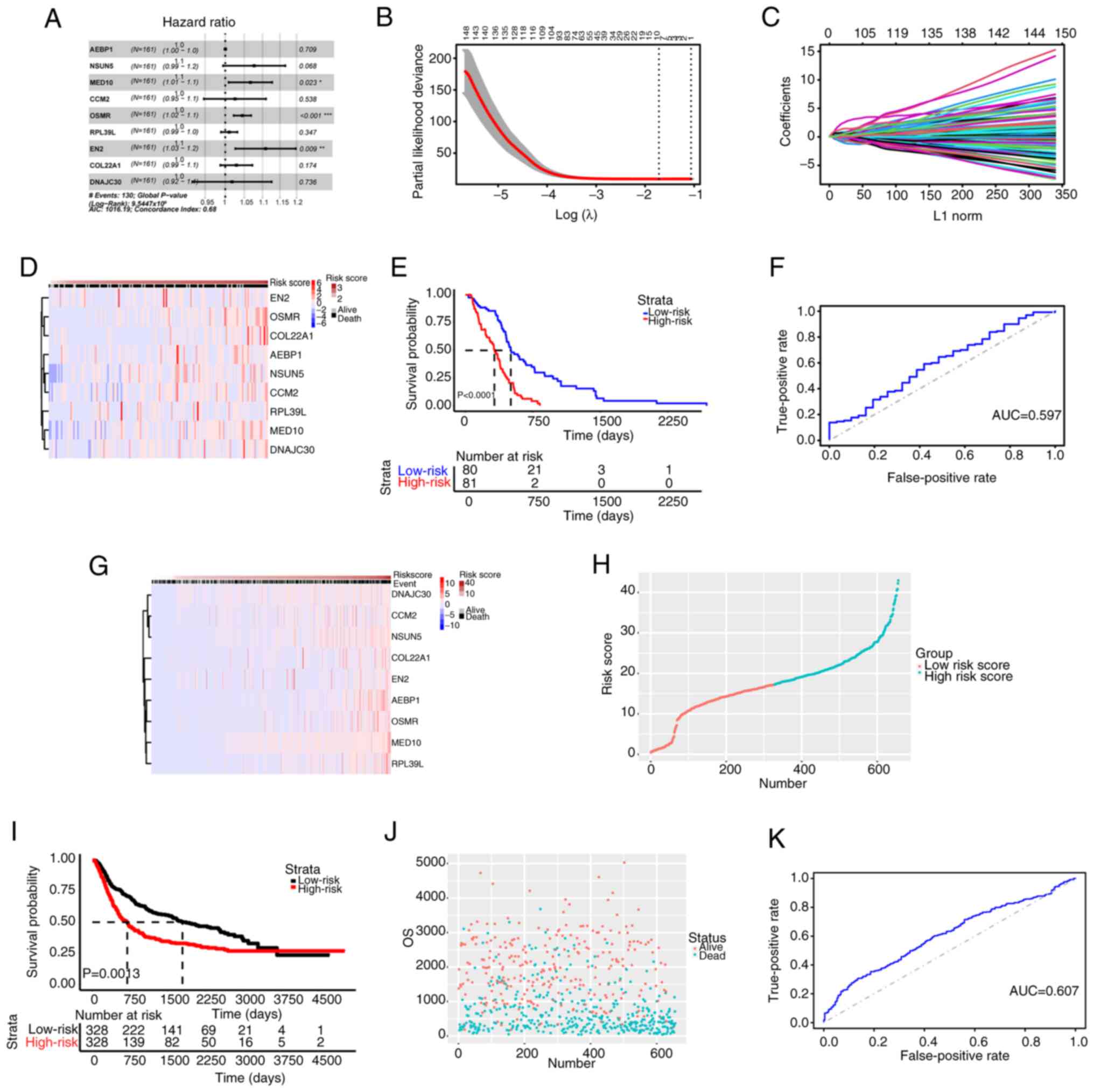
To ascertain the characteristic genes associated
with GBM prognosis, univariate Cox and LASSO regression analyses
were conducted for an in-depth analysis of the identified DEGs. The
candidate genes were screened by univariate Cox analysis (Fig. 2A). To eliminate analysis errors,
the dataset from TCGA was used as the training set and
characteristic prognostic genes were screened by LASSO analysis. As
shown in Fig. 2B-D, nine
prognostic genes were screened out. The risk score was calculated
as follows: Risk score=A EBP1 × 0.007204771 + NSUN5 × 0.200917630 +
MED10 × 0.257867168 + CCM2 × 0.017182529 + OSMR × 0.15343 0963 +
RPL39L × 0.019123077 + EN2 × 0.038036644 + C OL22A1 × 0.048587190 +
DNAJC30 × 0.001146385. Based on the riskscore values in the
patients with GBM from CGGA, the median value was calculated and
the patients were divided into high and low risk groups according
to the median value (Fig.
S2A-C). Furthermore, the low-risk group exhibited improved
survival performance compared with the high-risk group (Fig. 2E), and the 1-year area under the
curve (AUC) value was 0.597 (Fig.
2F). The aforementioned results were confirmed in the
validation set (Fig. 2G-K). The
aforementioned findings showed that the prognostic model in the
present study was closely associated with the prognosis of GBM and
could predict the risk score of GBM based on the assessment of the
risk score gene value of each patient.
Expression profiles of risk score-related
genes
The DNA, mRNA and protein levels of these nine genes
were assessed to examine the risk score-related genes in GBM. The
DNA of COL22A1, OSMR, NSUN5, AEBP1,
CCM2 and EN2 was mutated in GBM (Fig. 3A). Additionally,
post-translational modification of DNA serves an important role in
tumor regulation, and DNA methylation is one of the most common
modification methods (19). Thus,
the differentially expressed methylated genes were compared between
the GBM group and the paracancerous group, and the methylation
levels of numerous genes were significantly different between the
two groups (Fig. 3B and D), which
caused changes in their RNA levels (Figs. 3C and S3A). Next, the Human Protein Atlas
database was queried and the protein expression levels of AEBP1,
NSUN5, CCM2, OSMR, RPL39L and COL22A1 were found to be increased to
varying degrees in tumor tissues of the colon (Fig. 3E). The result was validated in
other cancer tissues (Fig. S3B).
These results indicated that the status of most risk score-related
genes was different at the multiomics level in GBM and normal
tissues.
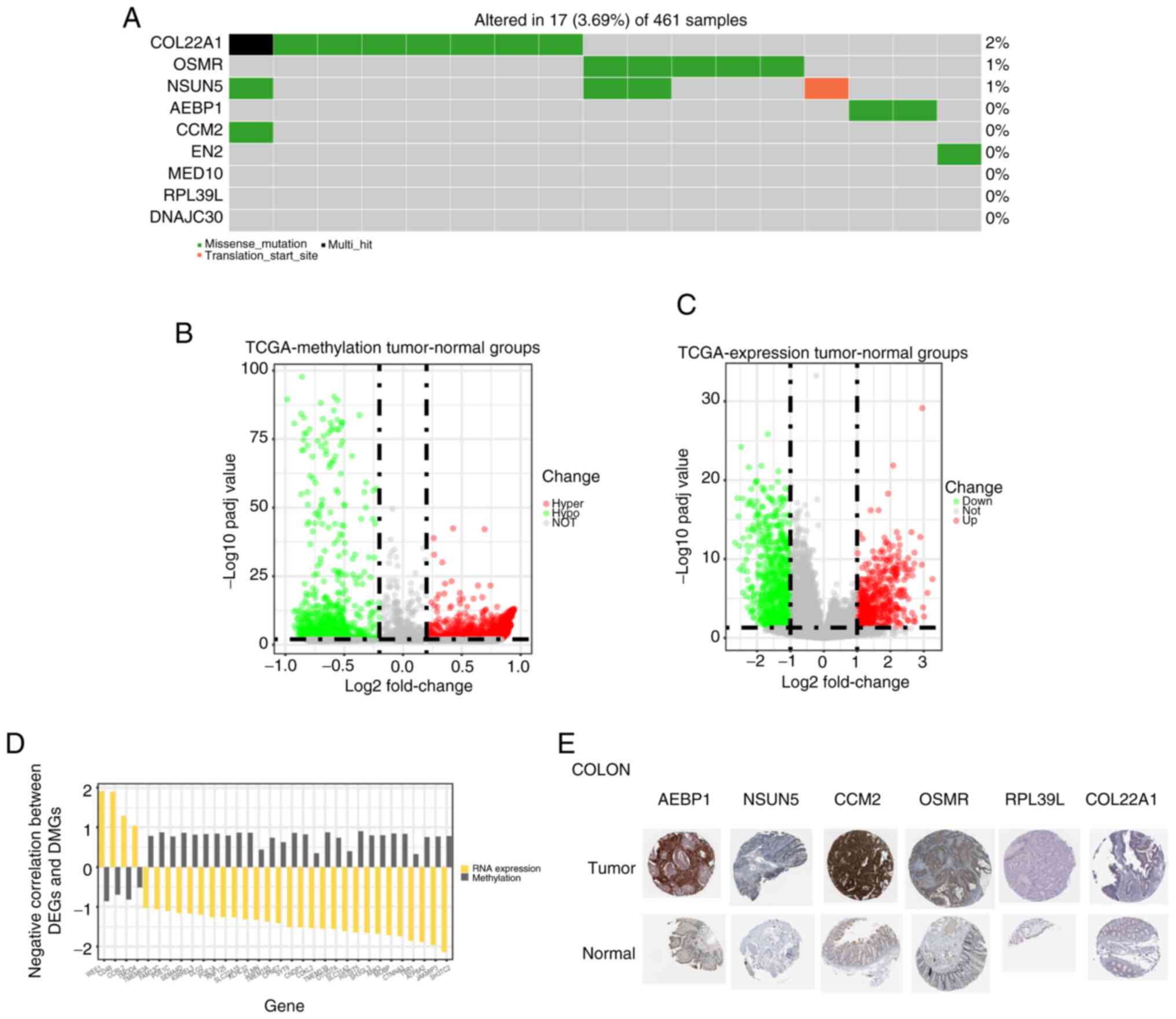 | Figure 3Expression profiles of risk
score-related genes. (A) Somatic mutation of nine risk
score-related genes in the dataset from TCGA. (B) Volcano plot of
differential methylation sites in the tumor and normal groups. (C)
Volcano plot of DEGs in the tumor and normal groups. (D) DEGs and
DMGs with a negative correlation in the tumor and normal groups.
(E) Representative immunohistochemistry images of six prognostic
proteins in tumor and normal tissues of the colon. Magnification,
×200. AEBP1, AE binding protein 1; CCM2, CCM2 scaffold protein;
COL22A1, collagen type XXII α1 chain; DEG, differentially expressed
gene; DMG, differentially methylated gene; NSUN5, NOP2/Sun RNA
methyltransferase 5; OSMR, oncostatin M receptor; Padj value,
adjusted P-value; RPL39L, ribosomal protein L39 like; TCGA, The
Cancer Genome Atlas. |
Somatic mutation analysis in GBM
Tumor occurrence is highly associated with DNA
mutation (9). Mutation analysis
was performed to examine DNA mutations in GBM. As shown in Fig. S4A, in the GBM group, the mutation
frequencies of PTEN, TP53, EGFR and TTN were the highest. The
mutation frequencies of the high-risk group and low-risk group were
different (Fig. S4B and C).
Therefore, the prognostic model in the present study was closely
related to DNA mutation.
Biofunction analysis of risk
score-related genes via scRNA-seq
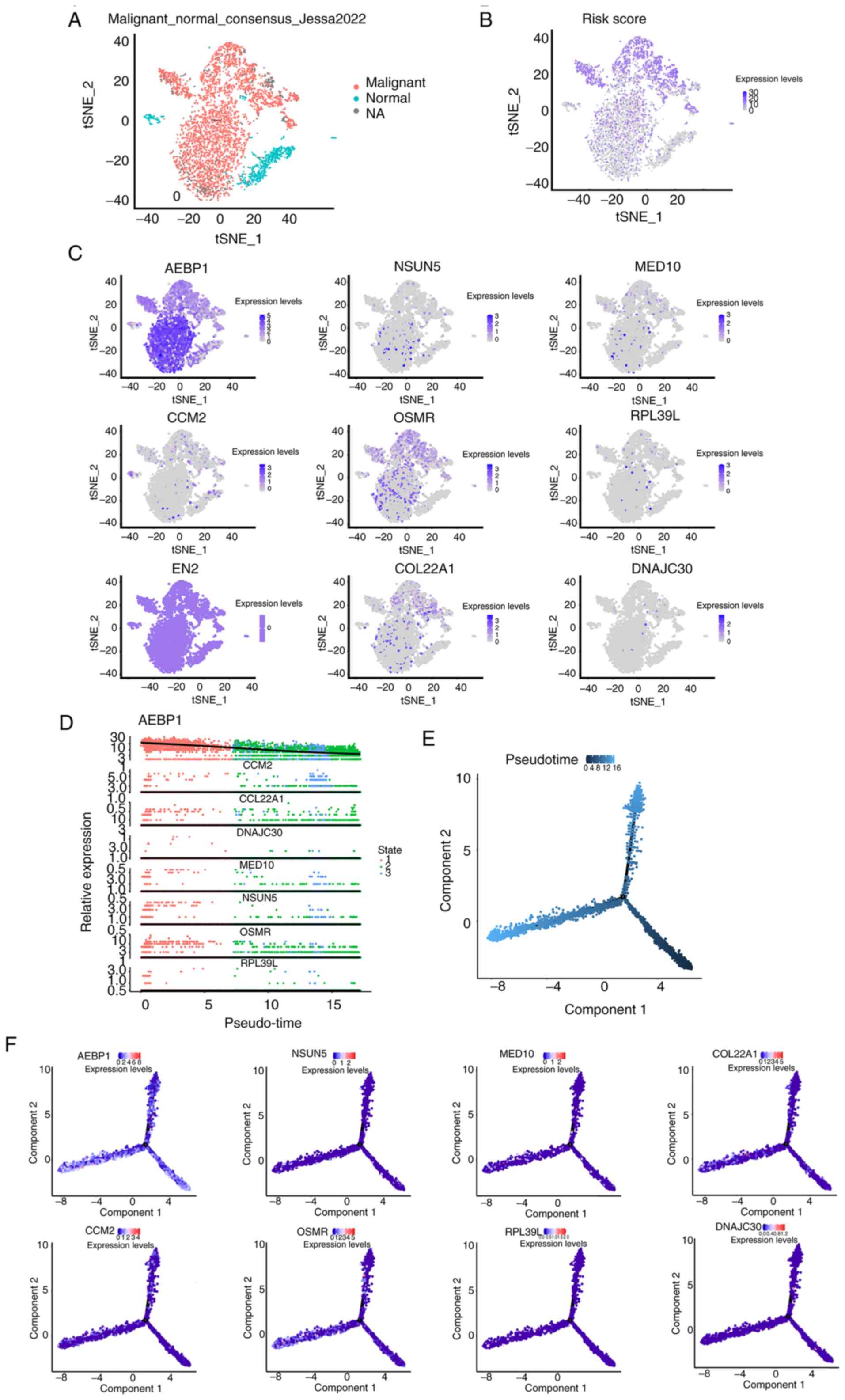
Subsequently, to visualize the expression of risk
score-related genes more accurately in different types of cells in
GBM tissues, scRNA-seq was performed to analyze these genes. First,
three cell populations that made up the GBM tissue were identified
(Fig. 4A). Subsequently, risk
scores of these cell populations were evaluated and risk
score-related genes were highly expressed in malignant cells, which
means that these risk score genes may exert their function mainly
by influencing tumor cells (Fig. 4B
and C). To further examine the dynamic changes in these genes
during tumor development, a pseudo-timeline analysis was performed.
As shown in Fig. 4D-F, the
expression of these genes increased with tumor development.
Simultaneously, the biomarkers based on a previous study (16) in each cell type were displayed
using a heatmap (Fig. S5). The
aforementioned findings of the scRNA-seq analysis further
demonstrated the reliability of the prognostic model.
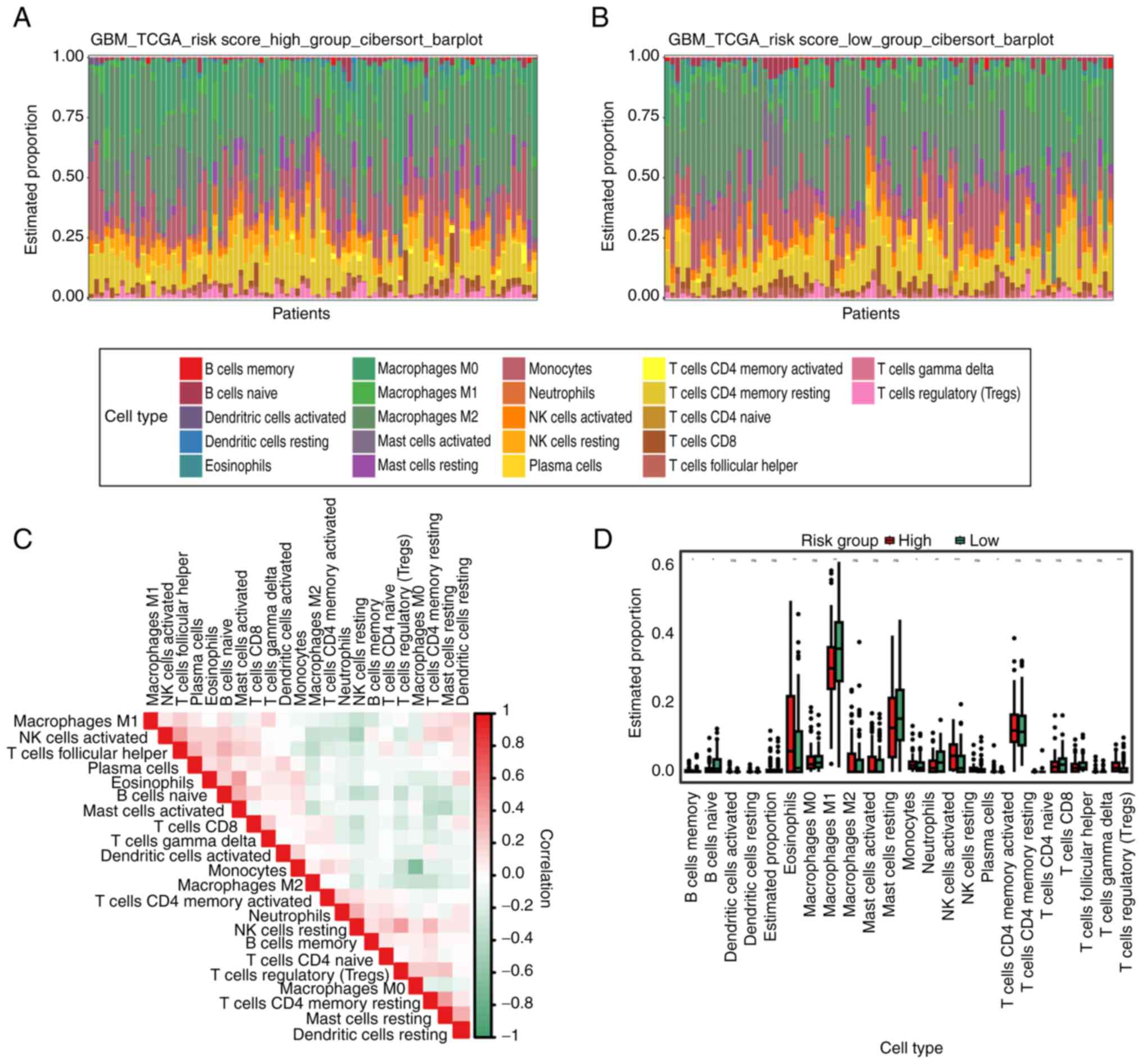
Immune cell infiltration analysis
Understanding the tumor-related immune
microenvironment is crucial for understanding the occurrence and
development of malignancies, since immune infiltration serves an
important role in their formation (13). The present study used the
Cibersort algorithm to examine the immune microenvironment in the
high-risk group, low-risk group, GBM tumor group and paracancerous
group. First, the proportions of different immune cell types in
each group were examined. The proportion of different immune cell
types in each group was analyzed, and there were significant
differences in the degree of immune invasion of some B cells,
macrophages and T cells not only in glioma and normal cells, but
also in the high-risk group and the low-risk group (Figs. 5A-D and S6A-F). Based on the aforementioned
results, it was concluded that the immune microenvironment changed
during the development of GBM, and this change was closely related
to the risk score.
Exploration of compounds and sensitive
drugs targeting risk score-related genes
Integrative traditional Chinese and Western medicine
is a current research hotspot in tumor treatment (20-22). First, traditional Chinese medicine
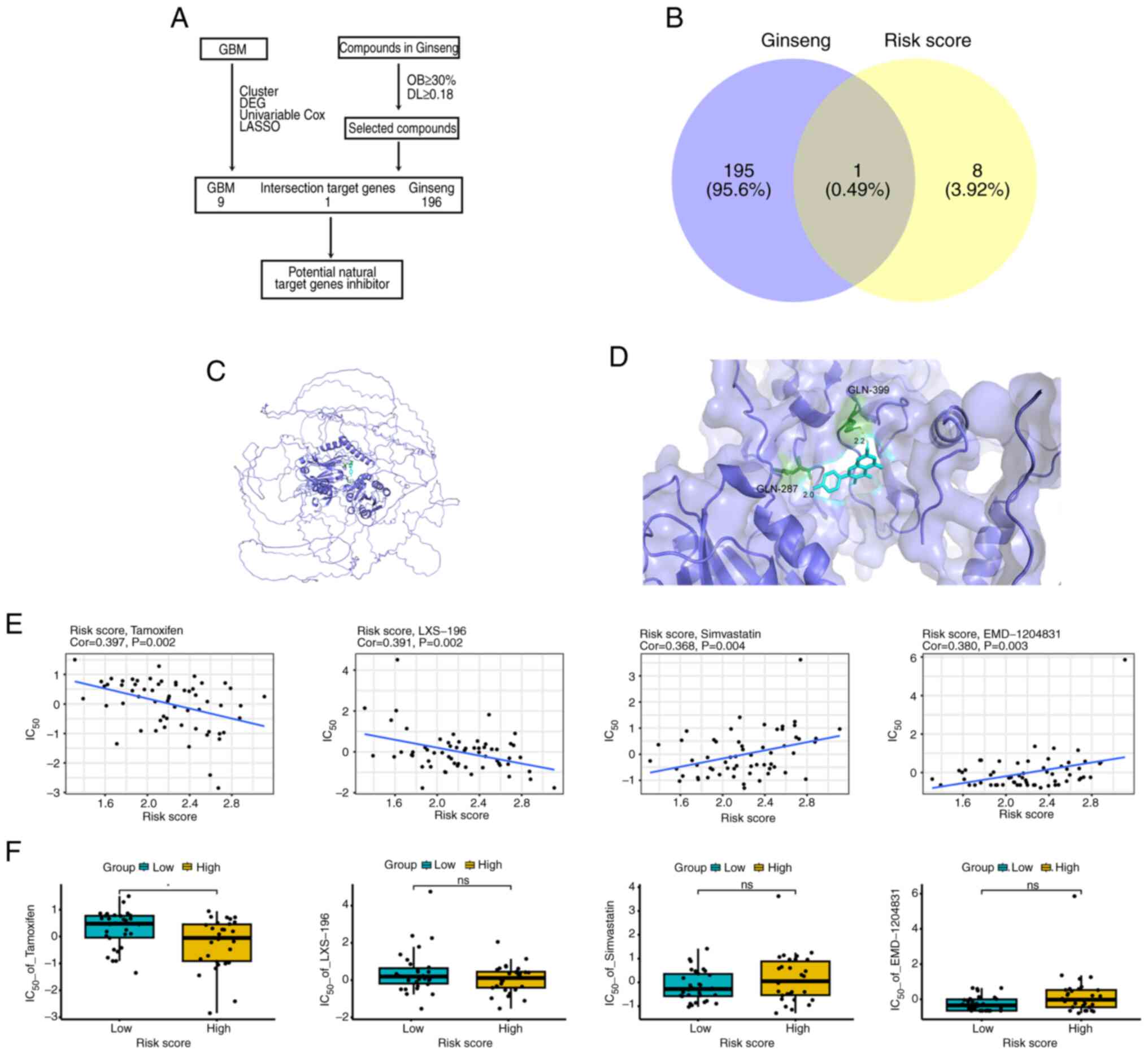
(TCM) compounds targeting GBM were explored (Fig. 6A). Querying the TCMSP database
showed that the Ginseng drug could only target the COL22A1 gene in
the prognostic model (the other genes in the model could not be
targeted by Ginseng), and the active ingredient was kaempferol
(Fig. 6B). Fig. 6C shows the simulated binding
sites. Further molecular docking analysis helped calculate the
combining ability of the active ingredient and the targeted gene.
The results were as follows: Kaempferol and COL22A1 score, -6.5
kcal/mol (Fig. 6D). To explore
the therapeutic implications of the prognostic model, the risk
score-related genes were subjected to a CellMiner drug sensitivity
examination. The outcomes demonstrated that 386 medications were
related to the prediction model risk score (P<0.05;
|cor|>0.3). The drug tamoxifen exhibited a significant
correlation and effect, while other drugs had no significant effect
(Figs. 6E and F, and S7A and B). The results indicated that
the COL22A1 gene in the prognostic model could be targeted by
several drugs, suggesting that it possessed potential therapeutic
significance.
Silencing COL22A1 enhances GBM
cuproptosis
To verify the role of COL22A1 in the development of
GBM, proliferation was detected using CCK-8 and colony formation
assays, and the results demonstrated that inhibiting COL22A1
significantly decreased the proliferation and colony formation of
GBM (Figs. 7A and B, and S8C and D). To further investigate the
effect of COL22A1 on GBM cuproptosis, ROS levels were detected by
flow cytometry. Low concentrations of elesclomol reduced ROS levels
in the COL22A1 knockdown group, while this was not observed in the
shNC group (Figs. 7C, and
S8E and F). Copper
concentrations were measured in the same groups, and the results
were consistent with the aforementioned results; low concentrations
of elesclomol significantly increased copper concentrations in the
COL22A1 knockdown group, while this was not observed in the shNC
group (Fig. 7D). Furthermore, the
protein levels of the representative marker (FDX1) of cuproptosis
were detected after knocking down COL22A1. The results showed that
the levels of the downstream proteins of cuproptosis were decreased
in shCOL22A1 cells treated with low concentrations of elesclomol
compared with the shNC group (Fig.
7E). Similarly, transmission electron microscopy showed that
low concentrations of elesclomol caused mitochondrial shrinkage in
the shCOL22A1 group, which was not observed in shNC group (Fig. 7F). In conclusion, knockdown of
COL22A1 promoted cuproptosis in GBM. Notably, EMD-1204831 and
kaempferol inhibited COL22A1 expression, and the effect was greater
when used in combination (Fig.
7G). Therefore, future studies should focus on the effect of
the combination of EMD-1204831 and kaempferol on GBM development
and cuproptosis.
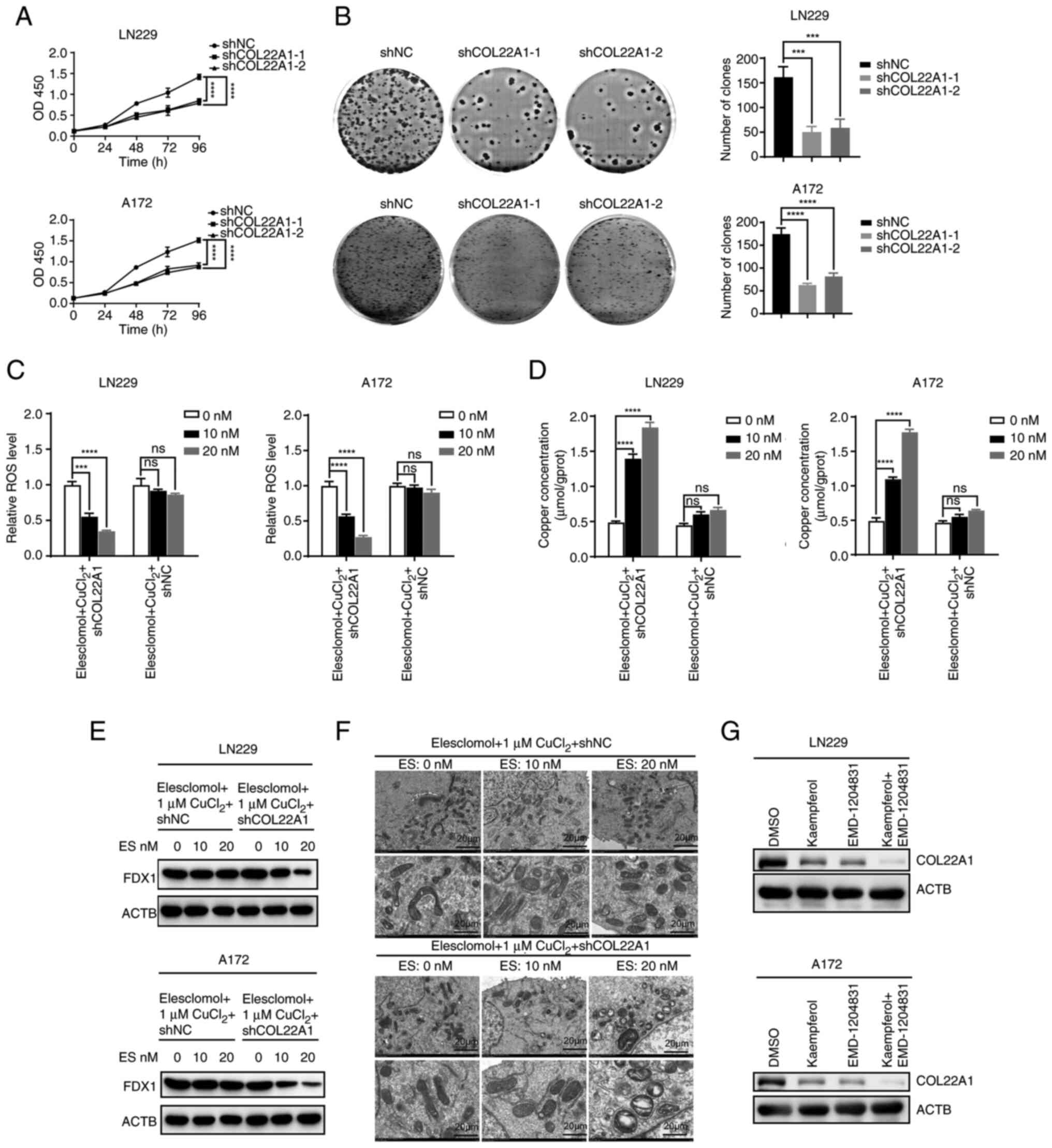 | Figure 7Silencing of COL22A1 enhances
glioblastoma cuproptosis. (A) Cell Counting Kit-8 assays were
performed in shNC and shCOL22A1 LN229 and A172 cell lines. (B)
Colony formation assays were performed in shNC and shCOL22A1 LN229
and A172 cell lines. (C) ROS levels of shNC and shCOL22A1 LN229 and
A172 cells exposed to increasing doses of elesclomol for 24 h. (D)
Concentration of copper following exposure to increasing doses of
elesclomol for 24 h in shNC and shCOL22A1 LN229 and A172 cell
lines. (E) Western blotting detecting the representative marker
(FDX1) of cuproptosis in shNC or shCOL22A1 LN229 and A172 cells
following exposure to increasing doses of elesclomol for 24 h. (F)
Transmission electron microscopy images were captured following
exposure to increasing doses of elesclomol for 24 h in shNC or
shCOL22A1 LN229 cells. Scale bar, 20 µm. (G) Protein levels
of COL22A1 in LN229 and A172 cells treated with DMSO or kaempferol
combined with EMD-1204831 examined by western blotting. Data are
presented as the mean ± SD of three experiments.
***P<0.001, ****P<0.0001 vs. controls.
ACTB, actin β; COL22A1, collagen type XXII α1 chain; ES,
elesclomol; FDX1, ferredoxin 1; NC, negative control; ns, not
significant; OD, optical density; ROS, reactive oxygen species; sh,
short hairpin RNA. |
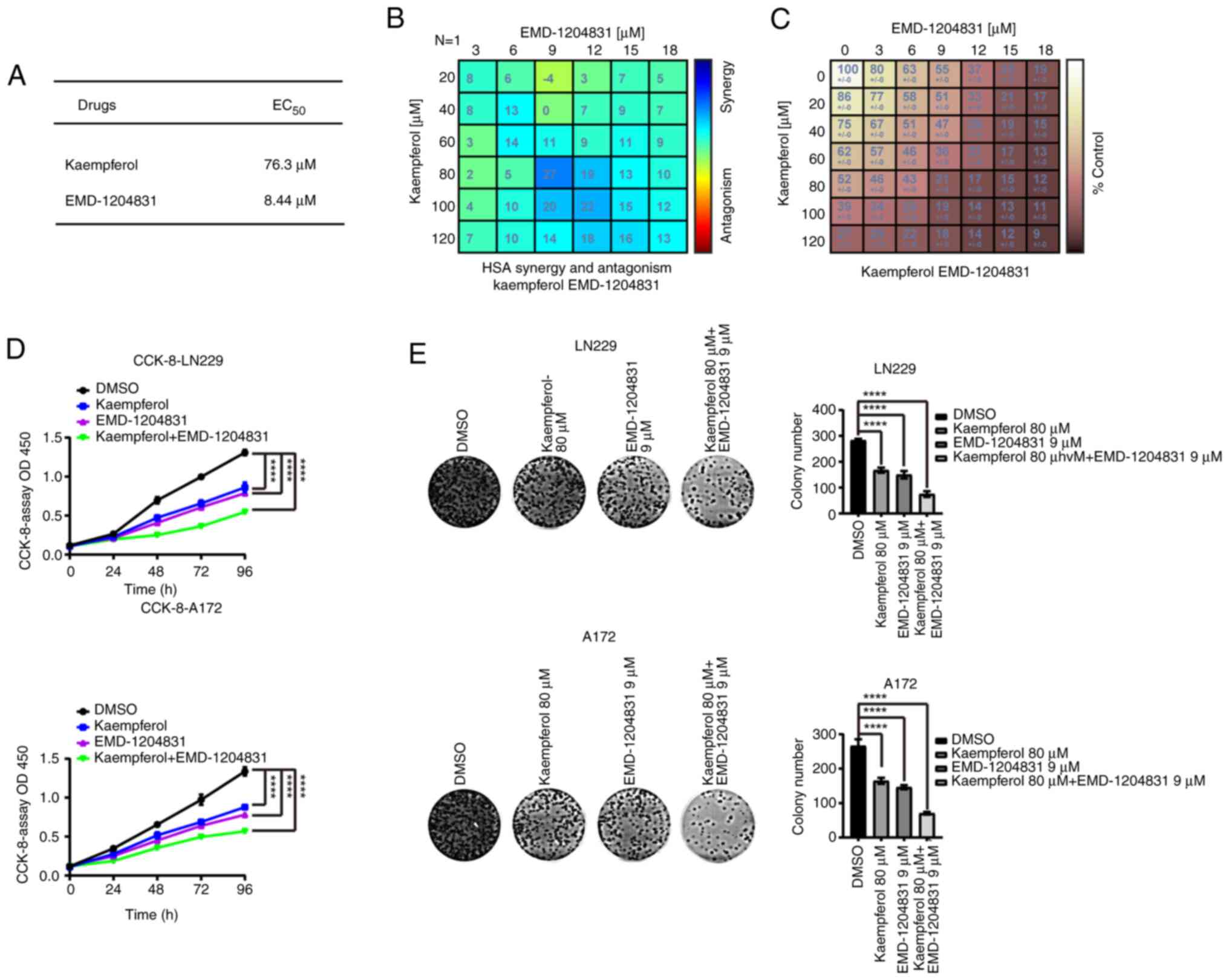
EMD-1204831 and kaempferol
synergistically treat GBM
The median effect concentration values of the two
drugs were obtained through CCK-8 experiments in LN229 cells
(Figs. 8A, and S8A and B). Subsequently, EMD-1204831
and kaempferol were used for treatment intervention in LN229 and
A172 cells. The cell viability assay showed that the combined drugs
reduced the cell activity more effectively than either drug alone,
and their best concentrations of combination were determined
(kaempferol, 80 µM; EMD-1204831, 9 µM) (Fig. 8B-D). Furthermore, the same results
were obtained in colony formation assays (Fig. 8E). In order to further explore the
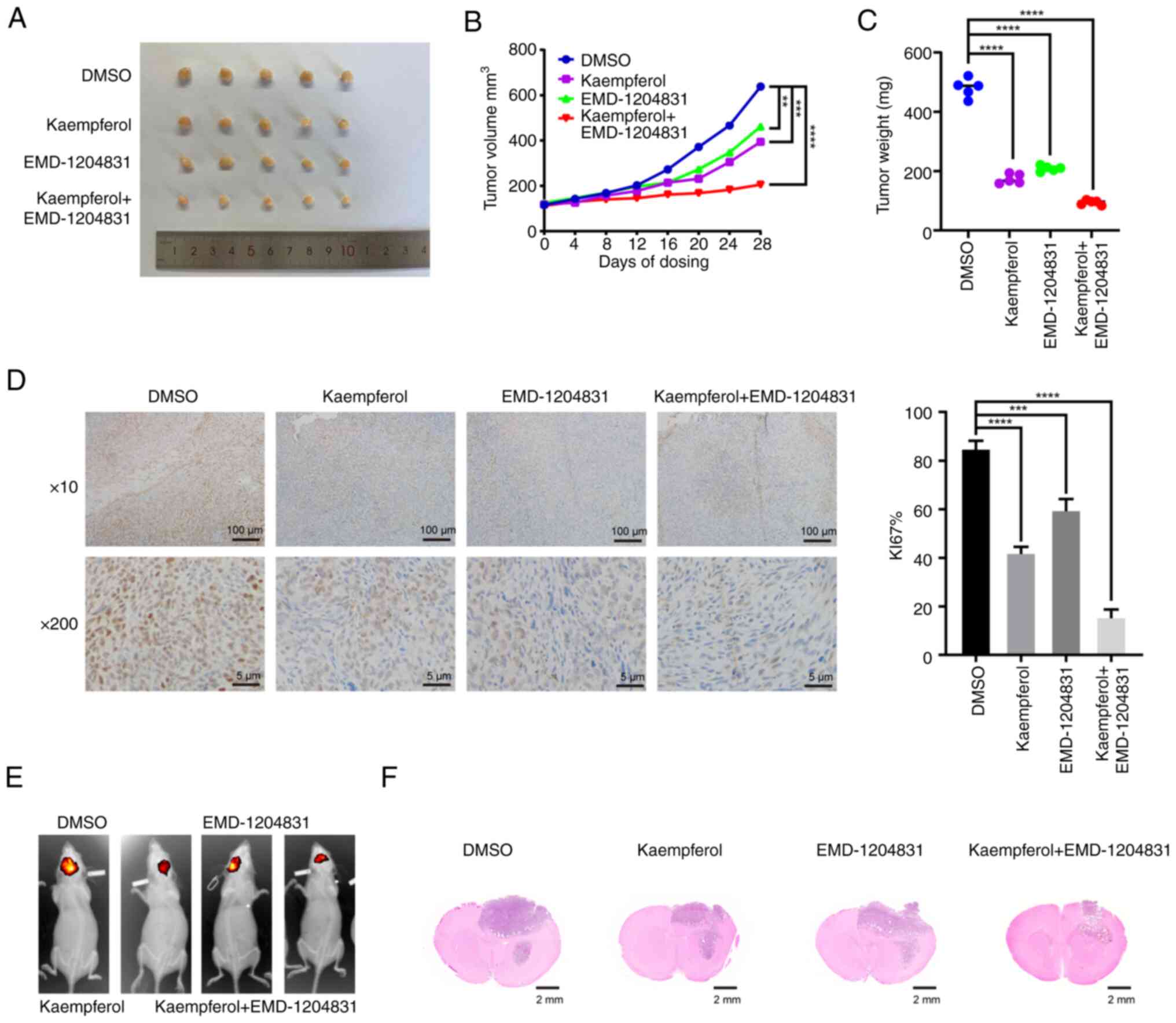
combination effect of EMD-1204831 and kaempferol, LN229 cells were
first injected into nude mice to prepare xenograft tumor models.
The nude mice were divided into four groups, namely the vehicle,
EMD-1204831, kaempferol and combination treatment groups. Compared
with the vehicle group, the tumor volume and mass of the two groups
of mice treated with either kaempferol or EMD-1204831 alone were
reduced, while the tumor inhibition effect of the combined
treatment group was the most obvious (Fig. 9A-C). After 4 weeks of treatment,
tumor tissues were collected and Ki67 was examined. The proportion
of positive tumor cells in the single drug groups was lower than
that in the vehicle group, while the inhibitory effect in the
combined drug group was greater (Fig.
9D). The results of the intracranial tumorigenesis experiment
in mice were consistent with the results of the present in
vitro experiments; the tumor size was smaller in the combined
drug group than in the control and single drug groups (Fig. 9E and F). The aforementioned
results indicated that the combination of EMD-1204831 and
kaempferol had therapeutic effects on GBM both in vivo and
in vitro.
Combination of EMD-1204831 and kaempferol
sensitizes GBM to cuproptosis inducers
To clarify the effect of the combination of the two
drugs on cuproptosis, the IC50 of the
cuproptosis-inducer elesclomol was determined in GBM cell lines
using a CCK-8 assay (Fig. 10A).
Then, low concentrations of elesclomol (10 and 20 nM) were added in
the control group and the combination group. The results of flow
cytometry showed that the changes in ROS levels were not
significant in the control group induced by low concentrations of
elesclomol. Conversely, low concentrations of elesclomol in the
combination group (EMD-1204831 and kaempferol) significantly
reduced ROS levels (Figs. 10B,
and S8G and H). To further
verify that the changes in ROS levels were related to cuproptosis,
copper concentrations and cuproptosis checkpoint proteins were
examined in both groups, and the results were consistent with the
aforementioned results; compared with the DMSO group, the
combination group exhibited increased copper concentrations and
cuproptosis (the expression levels of FDX1 could reflect the levels
of cuproptosis) (Fig. 10C and
D). Furthermore, transmission electron microscopy showed that
low concentrations of elesclomol caused mitochondrial shrinkage in
the combination group, which was not observed in the DMSO group
(Fig. 10E). In conclusion, the
combination of EMD-1204831 and kaempferol increased the sensitivity
of GBM to cuproptosis inducers.
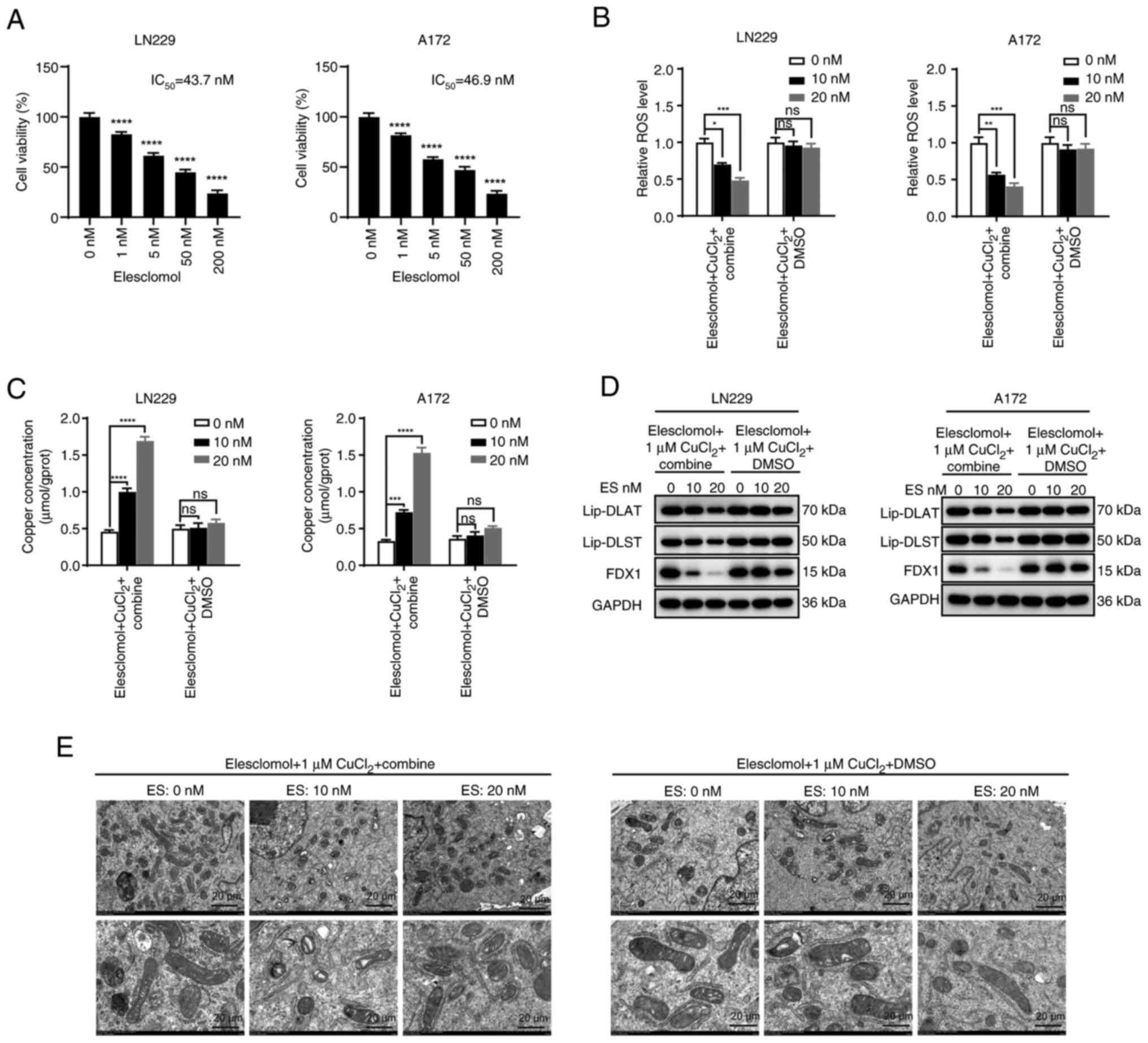 | Figure 10Combination of EMD-1204831 and
kaempferol sensitizes glioblastoma to cuproptosis inducers. (A)
Cell viability of LN229 and A172 cells following exposure to
increasing doses of elesclomol for 24 h. (B) ROS level of LN229 and
A172 cells treated with DMSO or kaempferol combined with
EMD-1204831 while exposed to increasing doses of elesclomol for 24
h. (C) Concentration of copper following exposure to increasing
doses of elesclomol for 24 h in LN229 and A172 cells treated with
DMSO or kaempferol combined with EMD-1204831. (D) Representative
markers of cuproptosis in LN229 and A172 cells treated with DMSO or
kaempferol combined with EMD-1204831 while exposed to increasing
doses of elesclomol for 24 h examined by western blotting. (E)
Transmission electron microscopy images captured following exposure
to increasing doses of elesclomol for 24 h in LN229 cells treated
with DMSO or kaempferol combined with EMD-1204831. Scale bar, 20
µm. Data are presented as the mean ± SD of three
experiments. *P<0.05, **P<0.01,
***P<0.001, ****P<0.0001 vs. control.
DLAT, dihydrolipoamide S-acetyltransferase; DLST, dihydrolipoamide
S-succinyltransferase; ES, elesclomol; FDX1, ferredoxin 1; Lip,
lipoylation; ns, not significant; ROS, reactive oxygen species. |
Discussion
More than half of all gliomas are GBM, the most
common primary malignant central nervous system tumor and the most
aggressive, with the worst clinical prognosis (1). The standard treatment for GBM is
based on surgery, combined with comprehensive treatments, including
radiotherapy and chemotherapy; however, their efficacy is limited,
with a median survival of 14-16 months (23,24). Cuproptosis is a recently reported
novel form of death caused by excessive copper levels; however, the
specific molecular mechanism of copper ion-induced cell death has
not been elucidated (25).
Cuproptosis can induce tumor cell death, including GBM cell death;
however, the underlying mechanisms remain largely unknown and
warrant investigation (4).
Exploring the molecular mechanisms by which copper toxicity leads
to cell death is expected to provide a deeper understanding of
copper imbalance in human diseases and the development of treatment
strategies. Using LASSO regression analysis, a prediction model of
genes linked to cuproptosis in GBM was created. In parallel, the
relationship between the GBM risk score and methylation, DNA
mutation or scRNA-seq data was investigated. The results of the
multiomics analysis showed that the prognostic pattern was
associated with the immune microenvironment, and kaempferol and
EMD-1204831 had a collaborative inhibitory effect on GBM cells. The
present findings provide a novel predictive model for the diagnosis
of GBM based on cuproptosis-related genes, and novel targets and
potential therapeutic options for its treatment.
Cluster analysis was adopted according to the
expression levels of cuproptosis genes in GBM to obtain different
cuproptosis expression level groups. Subsequently, DEG analysis and
Cox proportional hazards model analysis between the two groups of
patients were performed, the acquired genes were examined by LASSO
regression, and the risk score genes were used to establish a
prognostic model related to cuproptosis in GBM. A total of nine
genes, including AEBP1, NSUN5, MED10,
CCM2, OSMR, RPL39L, EN2, COL22A1
and DNAJC30, were related to GBM. The fibril-associated
collagens with interrupted triple-helices protein family is
represented structurally by the collagen encoded by the COL22A1
gene on human chromosome 8q24.2. The extracellular matrix protein
known as collagen XXII, a unique gene product, is exclusively found
at tissue junctions in the skin, muscle, articular cartilage, heart
and tendon. At the tendon junction, collagen XXII is deposited in
the basement membrane region (26). Transcriptional regulation of
COL22A1 suppresses nasopharyngeal carcinoma cell senescence through
G1/S arrest (27).
High proliferation and recurrence rates of squamous cell carcinoma
of the head and neck are positively associated with high COL22A1
expression, which can be used as a prognostic indicator for this
tumor, and COL22A1 is also closely related to the prognosis of
osteosarcoma (28,29). A pooled analysis or Kaplan-Meier
survival analysis was performed for these nine genes in the
training and validation sets to calculate AUC values and it was
verified that the risk scores of the predictive models were
consistent with the condition of patients, which means this
prognostic model could predict the survival status of patients.
Overall, different approaches were taken in both the training set
and validation set to check the usefulness and reliability of the
model.
scRNA-seq technology refers to high-throughput
sequencing of genetic information such as the genome, transcriptome
and epigenome at the single-cell level. scRNA-seq reflects the
heterogeneity and genetic information of rare cells that cannot be
obtained by sequencing mixed samples. It reveals the gene structure
and gene expression status of individual cells and provides more
detailed information on individual cells (30). The status of immune cell
heterogeneity was investigated by scRNA-seq (31). The application and development of
scRNA-seq technology is a novel method to develop treatments for
neurodegenerative diseases and identify targets and biomarkers
(32). GBM samples were sorted
into malignant, normal and no classification groups based on
scRNA-seq data. Among the three groups, the highest risk score was
observed in the malignant group, whereby the risk score could
assess tumor development and progression. Next, a timeline analysis
of risk scores and associated genes was performed, which showed
that risk scores and associated genes gradually increased with
tumor progression. The aforementioned results demonstrated that the
present prognostic model could predict tumor occurrence to some
extent.
Chemosensitivity analysis revealed a strong
association between risk score-related genes and the drug
susceptibility of several widely used chemotherapeutic medications.
The present study also revealed that the risk score exhibited an
obvious correlation with EMD-1204831 indicating that this drug may
be useful in treating GBM based on the prognostic model.
Furthermore, the present experiments also verified this
association. EMD-1204831 treatment is associated with potent tumor
growth suppression and regression; however, to the best of our
knowledge, the specific mechanism of action is unknown (15,33). The present study attempted to
investigate combination therapy based on integrative traditional
Chinese and Western medicine. Therefore, drug sensitivity, network
pharmacology and molecular docking analyses were performed, and
only COL22A1 could be targeted by EMD-1204831 and kaempferol
simultaneously, according to the overlap between the prognostic
genes and the genes targeted by Ginseng. Although COL22A1 was not
the most markedly altered protein compared with in normal tissues
based on immunohistochemistry data, it was selected for further
study. Kaempferol was found to target the COL22A1 protein in
prognostic models based on molecular docking and network
pharmacology analyses. Kaempferol is a natural compound and the
most common flavonoid found widely in vegetables and fruits
(34,35). Kaempferol and its glycosylated
derivatives have been shown to have numerous functions, including
antioxidant, anti-inflammatory, antimicrobial, anticancer,
cardioprotective, neuroprotective, antidiabetic, anti-osteoporotic,
estrogenic/antiestrogenic, anxiolytic, analgesic and antiallergic
activities (36). Several
anticancer effects of kaempferol have been reported, including in
breast cancer, gastric cancer, prostate cancer, bladder cancer,
cervical cancer, colon cancer, liver cancer, lung cancer, ovarian
cancer and leukemia (37-39). Kaempferol can also inhibit the
invasion/migration of GBM cells by inhibiting
12-O-tetradecanoylphorbol-13-acetate-induced protein kinase
Cα/ERK/NF-κB activation, migration and MMP-9 activation (40). Since the studies of kaempferol and
EMD-1204831 in GBM are not precise enough, a comprehensive
mechanistic analysis of their anti-GBM action was conducted. The
two drugs synergistically inhibited the proliferation of GBM cells.
Through cluster analysis and LASSO regression analysis, the
prognostic cuproptosis-related genes in GBM were identified.
Furthermore, the drugs EMD-1214063 and kaempferol could target one
of the aforementioned genes, COL22A1, simultaneously based on the
drug sensitivity analysis and network pharmacology. Therefore, we
hypothesized that these two drugs exert their function through the
same target and validated their synergistic effect in subsequent
experiments. EMD-1204831 is a c-Met inhibitor that has been
reported to inhibit cancer cell proliferation (14); however, to the best of our
knowledge, its mechanism of action has not been studied. As one of
the natural flavonoids, kaempferol is often considered to inhibit
tumors (36,41). However, to the best of our
knowledge, the application of kaempferol in GBM has not yet been
reported. Kaempferol, the main ingredient in natural medicine, has
no clearly-defined purpose (36,37). In summary, the present study has
found a reliable method for the potential treatment of GBM and a
theoretical basis for the treatment of GBM based on integrated
traditional Chinese and Western medicine.
In conclusion, the purpose of the present study was
to investigate the important role of cuproptosis in the diagnosis
and treatment of GBM. First, a cuproptosis-related prediction model
was constructed using bioinformatics analysis. The results of
single-cell sequencing analysis showed that the risk score
calculated by the prognostic model was higher in malignant GBM cell
clusters and increased with the progression of malignant tumors.
The multiomics analysis revealed that the prognostic pattern was
strongly associated with immune system infiltration, DNA
methylation and mutations. The combination of kaempferol and
EMD-1204831 exhibited improved treatment efficiency in inhibiting
GBM progression.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
YC was involved in conceptualization, performed
experiments and wrote the original draft. HX and HP confirm the
authenticity of all the raw data. YZ was involved in
conceptualization and funding acquisition. HY, QL, RS, JS, HL and
JL performed experiments. HX performed experiments, revised and
edited the manuscript, and acquired funding. HP performed
experiments, acquired funding and supervised the study. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was carried out in strict
accordance with the recommendations in the Guide for the Care and
Use of Laboratory Animals of the National Institutes of Health. All
procedures that involved animals were approved by the Experimental
Animal Ethics Committee: China Medical University (approval no.
CMUXN2023010; approval date, March 9, 2023; Shenyang, China). All
surgery was performed under isoflurane anesthesia, and all efforts
were made to minimize suffering.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Abbreviations:
|
GBM
|
glioblastoma
|
|
FDX1
|
ferredoxin 1
|
|
ROS
|
reactive oxygen species
|
|
GSEA
|
gene set enrichment analysis
|
|
TCGA
|
The Cancer Genome Atlas
|
|
AUC
|
area under the curve
|
Acknowledgments
Not applicable.
Funding
The present study was supported by the Key Research and
Development Project of Liaoning Province (grant no. 2018225040),
the Science and Technology Planning Project of Shenyang (grant no.
22-321-33-50), the Special Fund for Clinical Research of Wu Jieping
Medical Foundation (grant no. 320.6750.2023-17-13) and the Natural
Science Foundation of Liaoning Province (grant no.
2023-BS-046).
References
|
1
|
Ostrom QT, Gittleman H, Liao P, Rouse C,
Chen Y, Dowling J, Wolinsky Y, Kruchko C and Barnholtz-Sloan J:
CBTRUS statistical report: Primary brain and central nervous system
tumors diagnosed in the United States in 2007-2011. Neuro Oncol.
16(Suppl 4): iv1–iv63. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Salcman M: Glioblastoma multiforme. Am J
Med Sci. 279:84–94. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dolecek TA, Propp JM, Stroup NE and
Kruchko C: CBTRUS statistical report: Primary brain and central
nervous system tumors diagnosed in the United States in 2005-2009.
Neuro Oncol. 14(Suppl 5): v1–v49. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ala A, Walker AP, Ashkan K, Dooley JS and
Schilsky ML: Wilson's disease. Lancet. 369:397–408. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kahlson MA and Dixon SJ: Copper-induced
cell death. Science. 375:1231–1232. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grasso M, Bond GJ, Kim YJ, Boyd S, Matson
Dzebo M, Valenzuela S, Tsang T, Schibrowsky NA, Alwan KB, Blackburn
NJ, et al: The copper chaperone CCS facilitates copper binding to
MEK1/2 to promote kinase activation. J Biol Chem. 297:1013142021.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jia W, Tian H, Jiang J, Zhou L, Li L, Luo
M, Ding N, Nice EC, Huang C and Zhang H: Brain-targeted HFn-Cu-REGO
nanoplatform for site-specific delivery and manipulation of
autophagy and cuproptosis in glioblastoma. Small. 19:e22053542023.
View Article : Google Scholar
|
|
8
|
Tsvetkov P, Detappe A, Cai K, Keys HR,
Brune Z, Ying W, Thiru P, Reidy M, Kugener G, Rossen J, et al:
Mitochondrial metabolism promotes adaptation to proteotoxic stress.
Nat Chem Biol. 15:681–689. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yang L, Zhang Y, Wang Y, Jiang P, Liu F
and Feng N: Ferredoxin 1 is a cuproptosis-key gene responsible for
tumor immunity and drug sensitivity: A pan-cancer analysis. Front
Pharmacol. 13:9381342022. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Brem S, Grossman SA, Carson KA, New P,
Phuphanich S, Alavi JB, Mikkelsen T and Fisher JD; New Approaches
to Brain Tumor Therapy CNS Consortium: Phase 2 trial of copper
depletion and penicillamine as antiangiogenesis therapy of
glioblastoma. Neuro Oncol. 7:246–253. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Hu J, Guan F, Song L, Fan R, Zhu H,
Hu X, Shen E and Yang B: Copper induces cellular senescence in
human glioblastoma multiforme cells through downregulation of
Bmi-1. Oncol Rep. 29:1805–1810. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lun X, Wells JC, Grinshtein N, King JC,
Hao X, Dang NH, Wang X, Aman A, Uehling D, Datti A, et al:
Disulfiram when combined with copper enhances the therapeutic
effects of temozolomide for the treatment of glioblastoma. Clin
Cancer Res. 22:3860–3875. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Ma Z and Mei L: Cuproptosis-related
gene SLC31A1 is a potential predictor for diagnosis, prognosis and
therapeutic response of breast cancer. Am J Cancer Res.
12:3561–3580. 2022.PubMed/NCBI
|
|
14
|
Zhang Z, Zeng X, Wu Y, Liu Y, Zhang X and
Song Z: Cuproptosis-related risk score predicts prognosis and
characterizes the tumor microenvironment in hepatocellular
carcinoma. Front Immunol. 13:9256182022. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bladt F, Faden B, Friese-Hamim M, Knuehl
C, Wilm C, Fittschen C, Grädler U, Meyring M, Dorsch D, Jaehrling
F, et al: EMD 1214063 and EMD 1204831 constitute a new class of
potent and highly selective c-Met inhibitors. Clin Cancer Res.
19:2941–2951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jessa S, Mohammadnia A, Harutyunyan AS,
Hulswit M, Varadharajan S, Lakkis H, Kabir N, Bashardanesh Z,
Hébert S, Faury D, et al: K27M in canonical and noncanonical H3
variants occurs in distinct oligodendroglial cell lineages in brain
midline gliomas. Nat Genet. 54:1865–1880. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Arnone AA, Tsai YT, Cline JM, Wilson AS,
Westwood B, Seger ME, Chiba A, Howard-McNatt M, Levine EA, Thomas
A, et al: Endocrine-targeting therapies shift the breast microbiome
to reduce estrogen receptor-α breast cancer risk. Cell Rep Med.
6:1018802025. View Article : Google Scholar
|
|
18
|
Wu Z, Li W, Zhu H, Li X, Zhou Y, Chen Q,
Huang H, Zhang W, Jiang X and Ren C: Identification of
cuproptosis-related subtypes and the development of a prognostic
model in glioma. Front Genet. 14:11244392023. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nishiyama A and Nakanishi M: Navigating
the DNA methylation landscape of cancer. Trends Genet.
37:1012–1027. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang H and Bu L: Progress in the
treatment of lung adenocarcinoma by integrated traditional Chinese
and Western medicine. Front Med (Lausanne). 10:13233442024.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen L, Xu YX, Wang YS, Ren YY, Chen YM,
Zheng C, Xie T, Jia YJ and Zhou JL: Integrative Chinese-Western
medicine strategy to overcome docetaxel resistance in prostate
cancer. J Ethnopharmacol. 331:1182652024. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin J, Sun L, Chen H, Chen W, Zhang Z, Cao
Y and Lin L: Chinese and Western integrative medicine for stage
IIIb-IVb non-small cell lung cancer: Design and rationale of a
multi-center, prospective registry (NSCLC-Chinese and Western
integrative medicine cohort). Integr Cancer Ther.
22:153473542311851092023. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Komori T: The 2016 WHO classification of
tumours of the central nervous system: The major points of
revision. Neurol Med Chir (Tokyo). 57:301–311. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu W, Klockow JL, Zhang M, Lafortune F,
Chang E, Jin L, Wu Y and Daldrup-Link HE: Glioblastoma multiforme
(GBM): An overview of current therapies and mechanisms of
resistance. Pharmacol Res. 171:1057802021. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Chen L, Min J and Wang F: Copper
homeostasis and cuproptosis in health and disease. Signal Transduct
Target Ther. 7:3782022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Koch M, Schulze J, Hansen U, Ashwodt T,
Keene DR, Brunken WJ, Burgeson RE, Bruckner P and Bruckner-Tuderman
L: A novel marker of tissue junctions, collagen XXII. J Biol Chem.
279:22514–22521. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang ML and Luo WL: Engrailed homeobox 1
transcriptional regulation of COL22A1 inhibits nasopharyngeal
carcinoma cell senescence through the G1/S phase arrest. J Cell Mol
Med. 26:5473–5485. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Misawa K, Kanazawa T, Imai A, Endo S,
Mochizuki D, Fukushima H, Misawa Y and Mineta H: Prognostic value
of type XXII and XXIV collagen mRNA expression in head and neck
cancer patients. Mol Clin Oncol. 2:285–291. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pan R, Pan F, Zeng Z, Lei S, Yang Y, Yang
Y, Hu C, Chen H and Tian X: A novel immune cell signature for
predicting osteosarcoma prognosis and guiding therapy. Front
Immunol. 13:10171202022. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu
J, Yu X and Shi S: Applications of single-cell sequencing in cancer
research: Progress and perspectives. J Hematol Oncol. 14:912021.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Papalexi E and Satija R: Single-cell RNA
sequencing to explore immune cell heterogeneity. Nat Rev Immunol.
18:35–45. 2018. View Article : Google Scholar
|
|
32
|
Ofengeim D, Giagtzoglou N, Huh D, Zou C
and Yuan J: Single-Cell RNA sequencing: Unraveling the brain one
cell at a time. Trends Mol Med. 23:563–576. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Walker K and Padhiar M:
AACR-NCI-EORTC--21st international symposium. Molecular targets and
cancer therapeutics-Part 2. IDrugs. 13:10–12. 2010.
|
|
34
|
Devi KP, Malar DS, Nabavi SF, Sureda A,
Xiao J, Nabavi SM and Daglia M: Kaempferol and inflammation: From
chemistry to medicine. Pharmacol Res. 99:1–10. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yang RY, Lin S and Kuo G: Content and
distribution of flavonoids among 91 edible plant species. Asia Pac
J Clin Nutr. 17(Suppl 1): S275–S279. 2008.
|
|
36
|
Calderón-Montaño JM, Burgos-Morón E,
Pérez-Guerrero C and López-Lázaro M: A review on the dietary
flavonoid kaempferol. Mini Rev Med Chem. 11:298–344. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kim TW, Lee SY, Kim M, Cheon C and Ko SG:
Kaempferol induces autophagic cell death via IRE1-JNK-CHOP pathway
and inhibition of G9a in gastric cancer cells. Cell Death Dis.
9:8752018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X, Yang Y, An Y and Fang G: The
mechanism of anticancer action and potential clinical use of
kaempferol in the treatment of breast cancer. Biomed Pharmacother.
117:1090862019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tie F, Ding J, Hu N, Dong Q, Chen Z and
Wang H: Kaempferol and kaempferide attenuate oleic acid-induced
lipid accumulation and oxidative stress in HepG2 cells. Int J Mol
Sci. 22:88472021. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lin CW, Shen SC, Chien CC, Yang LY, Shia
LT and Chen YC: 12-O-tetradecanoylphorbol-13-acetate-induced
invasion/migration of glioblastoma cells through activating
PKCalpha/ERK/NF-kappaB-dependent MMP-9 expression. J Cell Physiol.
225:472–481. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Imran M, Salehi B, Sharifi-Rad J, Aslam
Gondal T, Saeed F, Imran A, Shahbaz M, Tsouh Fokou PV, Umair Arshad
M, Khan H, et al: Kaempferol: A key emphasis to its anticancer
potential. Molecules. 24:22772019. View Article : Google Scholar : PubMed/NCBI
|