Introduction
Colorectal cancer is among the three leading causes
of cancer-related mortality worldwide. Approximately 50% of
patients with colon cancer, the predominant type of colorectal
cancer, develop liver metastasis, which is considered to be the
main cause of death from advanced-stage colon cancer (1). Therefore, it is crucial to elucidate
the biological mechanisms underlying liver metastasis of colon
cancer and accelerate the development of new treatment
strategies.
The liver is the most common site for metastasis
from colon and pancreatic cancer (2). Hepatectomy is a potentially curative
treatment option for liver metastasis from colon cancer; however,
liver metastasis from pancreatic cancer is not considered an
indication for surgical treatment (3). Similarities or differences in the
biology of liver metastasis between colon and pancreatic cancer
remain to be elucidated. We previously established and investigated
the highly liver-metastatic human colorectal cancer cell sublines
SW48LM2 and LM-BxPC-3, through the serial intrasplenic transfer of
hepatic tumor foci formed by parental SW48 colon cancer cells and
BxPC-3 pancreatic cancer cells in
NOD/Shi-scid/IL-2Rγnull mice (4–6). We
then performed a quantitative proteome analysis utilizing these
established cell lines by our original method (7). The comparison of cellular protein
abundance between a pair of ‘highly liver-metastatic’ cells and its
parental cells revealed a series of metastasis-related proteins. In
order to identify more universal metastasis-related proteins, we
subsequently selected 11 proteins commonly detected among two
pairs, i.e., the BxPC-3 and the SW48 subline pairs (unpublished
data). These proteins are expected to be good biomarker candidates
and/or plausible causal factors for cancer metastasis. Zinc finger
protein 185 (ZNF185) is one of these 11 proteins thus selected.
ZNF185 belongs to the family of LIM domain proteins
and contains one LIM zinc-binding domain at the COOH-terminus and
an actin-targeting domain (ATD) at the NH2-terminus. The
LIM domain is a cysteine- and histidine-rich double zinc-finger
motif named after the three homeodomain proteins: Lin-l 1, Isl-1
and Mec-3 (8–10). The LIM domain is present in a wide
range of proteins whose functions include a number of fundamental
biological processes, such as cell lineage specification,
cytoskeleton organization and organ development (11). Whereas the Zinc finger motif
contains the typical DNA binding structures, there is little
evidence to support the observation that LIM domains may directly
bind DNA (12). LIM domain
proteins were found to be distributed in the cytoplasm or nucleus
of cells and perform regulatory functions through protein:protein
interactions rather than direct interactions with DNA (13,14).
ZNF185 is located on chromosome Xq28 and is expressed in the
kidney, prostate, pancreas, blood, placenta and ovary, but not in
the liver (15). ZNF185 may be
involved in regulating cellular differentiation and/or
proliferation (16,17). Certain LIM domain-containing
proteins were previously shown to be involved in carcinogenic
processes (18–27). In basic research on prostate
cancer, craniocervical squamous cell carcinoma and non-small-cell
lung cancer, the underexpression of ZNF185 mRNA in tumoral tissue
was compared to that in matched normal tissue (16,28,29).
However, the detailed properties and functions of ZNF185 in the
multistep process of tumor invasion have not been investigated in
detail (12). The clinical
behavior of ZNF185 also remains unknown in relation to the
prognosis or treatment of various cancers.
In this study, we investigated the expression level
of ZNF185 using immunohistochemistry in 87 cases of colon cancer
obtained by complete surgical resection. We also discussed the
association between prognosis and the clinical significance of
ZNF185 expression.
Materials and methods
Patients
A total of 87 colon cancer specimens were obtained
from the surgical specimens of patients with informed consent
between April, 2002 and May, 2005. This study has been approved by
the Institutional Research Review Board of Tokai University. The
tissues were immediately fixed in 40% formaldehyde. The surgical
specimens were also processed for routine histopathological
analysis.
The patient sample included 48 men and 39 women,
with a mean age of 68.30±9.26 years. Well-differentiated
adenocarcinomas were found in 60 patients, moderately
differentiated adenocarcinomas in 21, poorly differentiated
adenocarcinomas in 2 and mucinous adenocarcinomas in 4 patients.
The tumors were clinically staged according to the Union for
International Cancer Control TNM system. The tumor status was T1 in
5 patients, T2 in 11, T3 in 55 and T4 in 16 patients. A total of 46
patients had lymph node metastasis (N1) and 18 patients had distant
metastasis (M1). Lymphatic and venous involvement was found in 74
and 46 patients, respectively. A total of 19 patients had liver
metastasis, including 10 synchronous liver metastasis patients. The
pathological stages were as follows: stage I, 9 patients; stage II,
30 patients; stage III, 30 patients; and stage IV, 18 patients.
Immunohistochemical (IHC) analysis
Formalin-fixed, paraffin-embedded tissue sections of
the tumor samples were analyzed. The paraffin-embedded sections
were deparaffinized and stained using the
streptavidin-biotin-peroxidase complex method. Rabbit antibodies
specific to ZNF185, activated RNA polymerase II transcriptional
coactivator p15 (SUB1), β-N-acetylhexosaminidase A (HEXA),
general transcription initiation factor IIF α subunit (GTF2F1),
actinin α 4 (ACTN4) and interleukin enhancer-binding factor 3
(ILF3) were purchased from Sigma-Aldrich Co. (St. Louis, MO, USA).
Rabbit anti-glucosidase 2 subunit β (Gluco) and clathrin heavy
chain (CLTC) antibodies were purchased from Abcam®
(Cambridge, UK). Briefly, the sections were incubated in 0.3%
H2O2 in methanol, washed in
phosphate-buffered saline (PBS) and non-specific protein binding
was blocked with normal rabbit serum (Nichirei, Tokyo, Japan). The
sections were then incubated overnight in a humid chamber at 4°C,
with affinity purified antibodies diluted in PBS, as recommended by
the manufacturers. Following three PBS washes, the sections were
incubated with peroxidase-labeled polymer conjugated rabbit
anti-goat antibody (Histofine Simple Stain Max PO; Nichirei). The
amplified immune products were visualized using a
3,3′-diaminobenzidine tetrahydrochloride reaction.
Statistical analysis
Statistical comparisons of data sets were performed
by non-parametric analysis using the Mann-Whitney U test. The
G-test (likelihood ratio Chi-square test) was applied for
comparisons between group frequencies. On multivariate analyses of
the cause-specific survival rate, the Cox proportional hazards
model was used. Data are presented as means ± standard deviation.
The analyses were performed using JMP version 8 software (SAS
Institute Inc., Cary, NC, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Identification of ZNF185 as a liver
metastasis-associated factor
We selected 8 proteins for the IHC staining
experiment among the 11 liver metastasis-associated proteins
identified and selected by quantitative proteome studies
(unpublished data). The expression of each protein in surgically
resected specimens from colon cancer was evaluated by IHC staining.
Statistical analyses were performed between expression of the
ZNF185, SUB1 and HEXA proteins and liver metastasis in 87 colon
cancer cases. A significant correlation was only observed for
ZNF185 expression, whereas the correlations were not significant
for the expression of SUB1 and HEXA (P=0.030, G-test) (Table I). Specific expression of the
GTF2F1, ACTN4, ILF3, CLTC and Gluco proteins could not be detected
using standard IHC procedures.
 | Table IExpression of candidate molecules in
liver metastasis from colon cancer. |
Table I
Expression of candidate molecules in
liver metastasis from colon cancer.
| Total liver
metastasis | |
|---|
|
| |
|---|
| Candidate molecules
(n) | Positive (19) | Negative (68) | P-value |
|---|
| ZNF185 |
| Positive (78) | 19 | 59 | 0.030a |
| Negative (9) | 0 | 9 | |
| HEXA |
| Positive (69) | 14 | 55 | 0.347 |
| Negative (18) | 5 | 13 | |
| SUB1 |
| Positive (84) | 19 | 65 | 0.473 |
| Negative (3) | 0 | 3 | |
ZNF185 expression and clinicopathological
characteristics
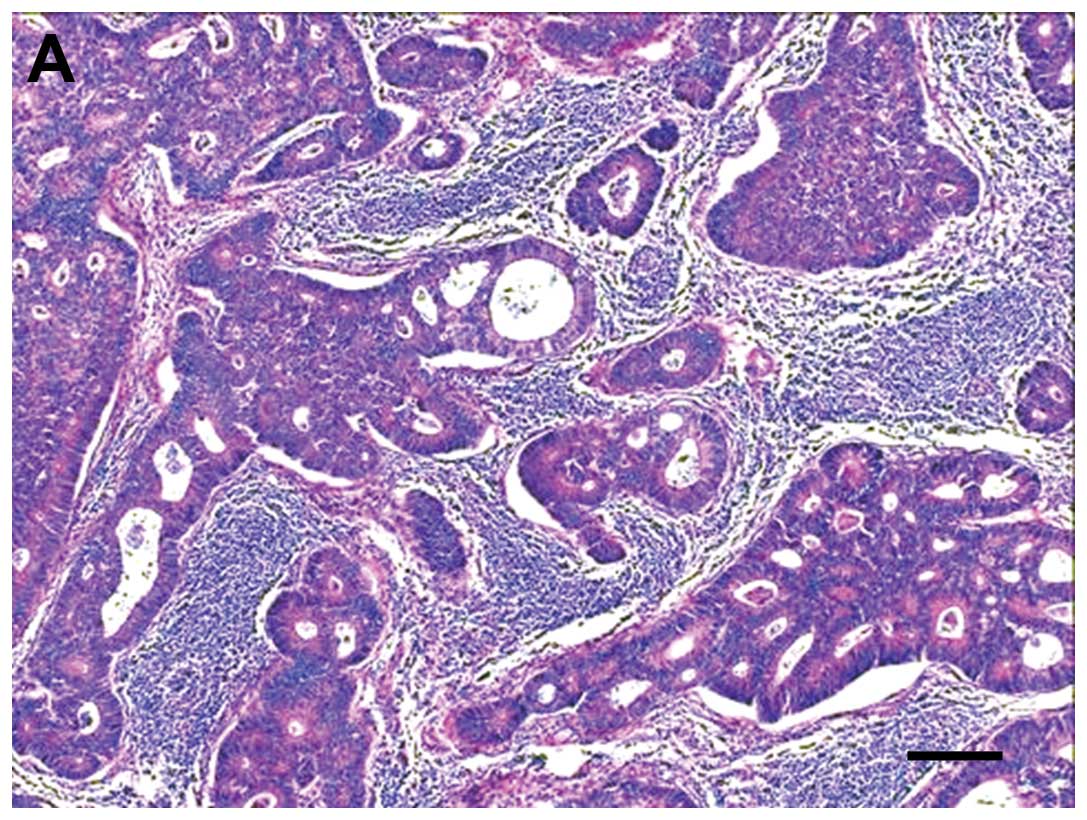
ZNF185 expression was observed in 78 of the 87 colon
cancer cases (Fig. 1). A
significant difference was observed between histological type and
ZNF185 expression (P=0.010, G-test). Other clinicopathological
correlations, including synchronous liver metastasis, were not
significant (Table II). The mean
age of ZNF185-positive and -negative patients was 67.67±9.07 and
73.78±9.61 years, respectively (P=0.071, Mann-Whitney U test).
 | Table IIZNF185 expression in colon
cancer. |
Table II
ZNF185 expression in colon
cancer.
| ZNF185
expression | |
|---|
|
| |
|---|
| Clinicopathological
characteristics (n) | (+) | (−) | P-value |
|---|
| Gender |
| Male (48) | 44 | 4 | 0.496 |
| Female (39) | 34 | 5 | |
| Histological
type |
| Well
differentiated adenocarcinoma (60) | 56 | 4 | 0.010a |
| Moderately
differentiated adenocarcinoma (21) | 18 | 3 | |
| Poorly
differentiated adenocarcinoma (2) | 0 | 2 | |
| Mucinous
adenocarcinoma (4) | 4 | 0 | |
| T status |
| T1 (5) | 5 | 0 | 0.599 |
| T2 (11) | 9 | 2 | |
| T3 (55) | 50 | 5 | |
| T4 (16) | 14 | 2 | |
| N status |
| N0 (41) | 35 | 6 | 0.213 |
| N1 (46) | 43 | 3 | |
| M status |
| M0 (69) | 61 | 8 | 0.424 |
| M1 (18) | 17 | 1 | |
| Lymphatic
involvement |
| Positive (74) | 67 | 7 | 0.538 |
| Negative (13) | 11 | 2 | |
| Venous
involvement |
| Positive (46) | 41 | 5 | 0.835 |
| Negative (41) | 37 | 4 | |
| Synchronous liver
metastasis |
| Positive (10) | 10 | 0 | 0.127 |
| Negative (77) | 68 | 9 | |
| Stage |
| I (9) | 7 | 2 | 0.502 |
| II (30) | 26 | 4 | |
| III (30) | 28 | 2 | |
| IV (18) | 17 | 1 | |
Correlations between prognosis and ZNF185
expression in colon cancer
We analyzed the correlations among cause-specific
survival rate, ZNF185 expression and clinicopathological
characteristics, such as patient age and gender, histological type,
lymphatic and venous involvement, T and N status, synchronous liver
metastasis and stage, using the Cox proportional hazards model. The
multivariate analyses identified histological type, synchronous
liver metastasis and ZNF185 expression as independent prognostic
indicators (P=0.029, P<0.0001 and P=0.020, respectively)
(Table III).
 | Table IIIMultivariate analyses using the Cox
proportional hazards model. |
Table III
Multivariate analyses using the Cox
proportional hazards model.
| Variable | Strata | P-value |
|---|
| Age (years) | 68.30±9.26 | 0.108 |
| Gender | Male, female | 0.967 |
| Histological
type | Well, moderate,
(adenocarcinoma) poorly differentiated, mucinous | 0.029a |
| T status | T1, T2, T3, T4 | 0.087 |
| N status | N0, N1 | 0.268 |
| Synchronous liver
metastasis | Positive,
negative | <0.0001b |
| Lymphatic
involvement | Positive,
negative | 0.216 |
| Venous
involvement | Positive,
negative | 0.319 |
| ZNF185
expression | Positive,
negative | 0.020a |
Discussion
In this study, we identified ZNF185 as a significant
liver metastasis-associated factor in colon cancer. To the best of
our knowledge, this is the first study to investigate the
association between ZNF185 and the clinical characteristics of
cancer. ZNF185 expression in colon cancer was found to be an
indicator of liver metastasis, as well as an independent prognostic
indicator. The histological type and synchronous liver metastasis
were found to significantly affect the prognosis of colon cancer
patients.
ZNF185 belongs to the LIM domain protein family
containing two zinc-finger motifs in the C-terminus, classified as
group 3 (12,30). ZNF185 is located on chromosome Xq28
and is expressed in the kidney, prostate, pancreas, blood, placenta
and ovary, but not in the liver (15). The complete ZNF185 gene was
originally cloned from normal human prostate tissue by Zhang et
al (17). The expression and
localization of ZNF185 in prostate cancer cells and fibroblasts
revealed that, in addition to F-actin stress fibers, ZNF185
localized to several other cytoskeleton-related areas, including
focal adhesions and filopodia/lamellipodia. ZNF185 was also shown
to contain an ATD in the N-terminal region and binds to F-actin
directly through the ATD, but not the LIM domains (17). Thus, ZNF185 interacts with F-actin
and focal adhesion components. Further studies, focused on
identifying proteins interacting with the other domains of ZNF185,
may help clarify the mechanism underlying its diverse subcellular
localization and function (12).
The LIM domains are generally cysteine- and histidine-rich domains,
50–60 amino acids in size, sharing double characteristic zinc
finger motifs. A diverse group of proteins containing LIM domains
has been identified, which displays various functions, including
gene regulation, cell fate determination, tumoral formation and
cytoskeleton organization. LIM domain proteins were previously
shown to be distributed in the nucleus as well as the cytoplasm and
exert their functions through interactions with various protein
partners (12). Certain LIM domain
proteins are known to play a role in the carcinogenic processes.
Epithelial protein lost in neoplasm and testin were found to be
downregulated in various cancer cell lines (18,19),
whereas LIM domain-only protein 4 is considered to be a negative
regulator of breast cancer susceptibility gene 1 and promotes
breast tumorigenesis (21,23). LIM and SH3 protein 1 was also
identified as a promoter of breast cancer, ovarian cancer and
hepatocellular carcinoma and is suggested to be the transcriptional
target of p53 (25–27).
ZNF185 gene expression was only shown to be
downregulated in matched normal tissues from prostate cancer,
craniocervical squamous cell carcinoma and non-small-cell lung
cancer (16,17,28,29).
Vanaja et al (16) reported
that the gene expression levels in high-grade (Gleason score 9)
prostate cancer cells were suppressed more compared to intermediate
grade (Gleason score 6) prostate cancer cells (16). Thus, the dysregulation of ZNF185
gene expression appears to be a frequent event in several cancer
types, which suggests its potential role in cancer development.
However, there are currently no published reports on ZNF185 in
colon cancer. ZNF185 expression was significantly high in
well-differentiated adenocarcinoma. In this study, we demonstrated
that ZNF185 is a liver metastasis-associated factor, as well as an
independent prognosis-deteriorating factor in colon cancer.
Adjuvant chemotherapy is commonly performed to
reduce the risk of recurrence and improve the prognosis in patients
with colon cancer. According to the National Comprehensive Cancer
Network guidelines 2012, all patients with stage III disease should
undergo adjuvant chemotherapy. However, stage II patients should
also undergo adjuvant chemotherapy when they have high-risk
factors, such as T4 lesions, lymphovascular involvement, or poorly
differentiated histology (31–33).
Liver metastasis is one of the most critical events in the clinical
treatment of advanced colon cancer (34). Due to the recent development of
clinical studies, certain patients with advanced colon cancer and
liver metastasis may become operable. However, the chemotherapeutic
regimens for metastatic colon cancer have also improved (35). In our results, ZNF185 indicated
liver metastasis with a sensitivity of 100% (19/19) and a
specificity of 13% (9/68). These high-sensitivity and
low-specificity properties are appropriate for a screening test.
There is a possibility that adjuvant chemotherapy may be omitted in
ZNF185-negative patients. Therefore, ZNF185 may represent a
potential prognostic biomarker of colon cancer.
Cancer cell invasion is a multistep process that
includes cell attachment, proteolysis of matrix components and cell
migration. Hematogenous liver metastasis, in particular, occurs as
a consequence of a well-characterized set of sequential events. The
detailed properties and functions of ZNF185 in cancerous invasion
have not been fully elucidated. We investigated the clinical
significance of ZNF185 in the prognosis and treatment of patients
with various types of cancer. The results of the present study,
which investigated the behavior of ZNF185 in cancerous invasion,
may contribute to the development of novel treatment strategies for
advanced colon cancer.
Acknowledgements
The authors would like to thank Tomohisa Machida
(Tokai University Hachioji Hospital, Tokyo, Japan) for his
technical assistance and helpful discussions.
References
|
1
|
Jemal A, Siegel R, Ward E, et al: Cancer
statistics, 2008. CA Cancer J Clin. 58:71–96. 2008. View Article : Google Scholar
|
|
2
|
Sadahiro S, Suzuki T, Ishikawa K, et al:
Recurrence patterns after curative resection of colorectal cancer
in patients followed for a minimum of ten years.
Hepatogastroenterology. 50:1362–1366. 2003.PubMed/NCBI
|
|
3
|
Yamada H, Hirano S, Tanaka E, Shichinohe T
and Kondo S: Surgical treatment of liver metastases from pancreatic
cancer. HPB (Oxford). 8:85–88. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Suemizu H, Monnai M, Ohnishi Y, Ito M,
Tamaoki N and Nakamura M: Identification of a key molecular
regulator of liver metastasis in human pancreatic carcinoma using a
novel quantitative model of metastasis in NOD/SCID/gammacnull (NOG)
mice. Int J Oncol. 31:741–751. 2007.PubMed/NCBI
|
|
5
|
Hamada K, Monnai M, Kawai K, et al: Liver
metastasis models of colon cancer for evaluation of drug efficacy
using NOD/Shi-scid IL2Rgammanull (NOG) mice. Int J Oncol.
32:153–159. 2008.PubMed/NCBI
|
|
6
|
Matsuyama M, Wakui M, Monnai M, et al:
Reduced CD73 expression and its association with altered purine
nucleotide metabolism in colorectal cancer cells robustly causing
liver metastases. Oncol Lett. 1:431–436. 2010.
|
|
7
|
Matsuo E, Watanabe M, Kuyama H and
Nishimura O: A new strategy for protein biomarker discovery
utilizing 2-nitrobenzenesulfenyl (NBS) reagent and its applications
to clinical samples. J Chromatogr B Analyt Technol Biomed Life Sci.
877:2607–2614. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Way JC and Chalfie M: mec-3, a
homeobox-containing gene that specifies differentiation of the
touch receptor neurons in C. elegans. Cell. 54:5–16. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Freyd G, Kim SK and Horvitz HR: Novel
cysteine-rich motif and homeodomain in the product of the
Caenorhabditis elegans cell lineage gene lin-11. Nature.
344:876–879. 1990. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Karlsson O, Thor S, Norberg T, Ohlsson H
and Edlund T: Insulin gene enhancer binding protein Isl-1 is a
member of a novel class of proteins containing both a homeo- and a
Cys-His domain. Nature. 344:879–882. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dawid IB, Breen JJ and Toyama R: LIM
domains: multiple roles as adapters and functional modifiers in
protein interactions. Trends Genet. 14:156–162. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zheng Q and Zhao Y: The diverse
biofunctions of LIM domain proteins: determined by subcellular
localization and protein-protein interaction. Biol Cell.
99:489–502. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pérez-Alvarado GC, Miles C, Michelsen JW,
et al: Structure of the carboxy-terminal LIM domain from the
cysteine rich protein CRP. Nat Struct Biol. 1:388–398.
1994.PubMed/NCBI
|
|
14
|
Schmeichel KL and Beckerle MC: The LIM
domain is a modular protein-binding interface. Cell. 79:211–219.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Heiss NS, Gloeckner G, Bächner D, et al:
Genomic structure of a novel LIM domain gene (ZNF185) in Xq28 and
comparisons with the orthologous murine transcript. Genomics.
43:329–338. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Vanaja DK, Cheville JC, Iturria SJ and
Young CY: Transcriptional silencing of zinc finger protein 185
identified by expression profiling is associated with prostate
cancer progression. Cancer Res. 63:3877–3882. 2003.PubMed/NCBI
|
|
17
|
Zhang JS, Gong A and Young CY: ZNF185, an
actin-cytoskeleton-associated growth inhibitory LIM protein in
prostate cancer. Oncogene. 26:111–122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maul RS and Chang DD: EPLIN, epithelial
protein lost in neoplasm. Oncogene. 18:7838–7841. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tatarelli C, Linnenbach A, Mimori K and
Croce CM: Characterization of the human TESTIN gene localized in
the FRA7G region at 7q31.2. Genomics. 68:1–12. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tobias ES, Hurlstone AF, MacKenzie E,
McFarlane R and Black DM: The TES gene at 7q31.1 is methylated in
tumours and encodes a novel growth-suppressing LIM domain protein.
Oncogene. 20:2844–2853. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Visvader JE, Venter D, Hahm K, et al: The
LIM domain gene LMO4 inhibits differentiation of mammary epithelial
cells in vitro and is overexpressed in breast cancer. Proc Natl
Acad Sci USA. 98:14452–14457. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Song Y, Maul RS, Gerbin CS and Chang DD:
Inhibition of anchorage-independent growth of transformed NIH3T3
cells by epithelial protein lost in neoplasm (EPLIN) requires
localization of EPLIN to actin cytoskeleton. Mol Biol Cell.
13:1408–1416. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sum EY, Peng B, Yu X, et al: The LIM
domain protein LMO4 interacts with the cofactor CtIP and the tumor
suppressor BRCA1 and inhibits BRCA1 activity. J Biol Chem.
277:7849–7856. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Garvalov BK, Higgins TE, Sutherland JD, et
al: The conformational state of Tes regulates its zyxin-dependent
recruitment to focal adhesions. J Cell Biol. 161:33–39. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang B, Feng P, Xiao Z and Ren EC: LIM and
SH3 protein 1 (Lasp1) is a novel p53 transcriptional target
involved in hepatocellular carcinoma. J Hepatol. 50:528–537. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Grunewald TG, Kammerer U, Schulze E, et
al: Silencing of LASP-1 influences zyxin localization, inhibits
proliferation and reduces migration in breast cancer cells. Exp
Cell Res. 312:974–982. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Grunewald TG, Kammerer U, Winkler C, et
al: Overexpression of LASP-1 mediates migration and proliferation
of human ovarian cancer cells and influences zyxin localisation. Br
J Cancer. 96:296–305. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gonzalez HE, Gujrati M, Frederick M, et
al: Identification of 9 genes differentially expressed in head and
neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg.
129:754–759. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Medina PP, Carretero J, Ballestar E, et
al: Transcriptional targets of the chromatin-remodelling factor
SMARCA4/BRG1 in lung cancer cells. Hum Mol Genet. 14:973–982. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Taira M, Evrard JL, Steinmetz A and Dawid
IB: Classification of LIM proteins. Trends Genet. 11:431–432. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Benson AB III, Schrag D, Somerfield MR, et
al: American Society of Clinical Oncology recommendations on
adjuvant chemotherapy for stage II colon cancer. J Clin Oncol.
22:3408–3419. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Staib L, Link KH, Blatz A and Beger HG:
Surgery of colorectal cancer: surgical morbidity and five- and
ten-year results in 2400 patients - monoinstitutional experience.
World J Surg. 26:59–66. 2002.PubMed/NCBI
|
|
33
|
Schiffmann L, Eiken AK, Gock M and Klar E:
Is the lymph node ratio superior to the Union for International
Cancer Control (UICC) TNM system in prognosis of colon cancer?
World J Surg Oncol. 11:792013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sadahiro S, Suzuki T, Tanaka A, Okada K
and Kamata H: Hematogenous metastatic patterns of curatively
resected colon cancer were different from those of stage IV and
autopsy cases. Jpn J Clin Oncol. 43:444–447. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Baba H, Watanabe M, Okabe H, et al:
Upregulation of ERCC1 and DPD expressions after oxaliplatin-based
first-line chemotherapy for metastatic colorectal cancer. Br J
Cancer. 107:1950–1955. 2012. View Article : Google Scholar : PubMed/NCBI
|