Introduction
Renal cell carcinoma (RCC) is one of the leading
causes of urological cancer-related mortality and clear cell renal
cell carcinoma (ccRCC) is the most common type of RCC, accounting
for 82% of RCC cases (1).
Early-stage ccRCC is usually asymptomatic and difficult to
accurately diagnose; in addition, ccRCC is resistant to
conventional chemotherapeutic drugs and the overall clinical
outcome is poor (2). The treatment
of advanced ccRCC remains a major challenge for clinicians and
ccRCC is responsible for ∼35% of RCC-related mortality cases
(1). Thus, there is an urgent need
for diagnostic and prognostic biomarkers in ccRCC. However, there
are currently no biomarkers for ccRCC in routine clinical practice.
Over the last few years, prognostic markers for ccRCC, such as
senescence-associated protein p400 (3), glyoxalase-1 (4), transforming growth factor-β-activated
kinase-1 (5) and serum miR-210
(6), have emerged; however, their
large-scale clinical application is not feasible. Therefore, novel
diagnostic and prognostic markers of ccRCC would be valuable in
high-risk individuals and in those with existing disease.
Pituitary tumor-transforming gene-1 (PTTG1) was
first isolated from rat pituitary tumor cells in 1997 and was
identified as an oncogene, as PTTG1 overexpression was found to
induce cellular transformation in vitro and tumor formation
in nude mice (7). As a human
securin, PTTG1 is involved in the mitotic spindle checkpoint
pathway and inhibits sister chromatid separation to ensure
chromosomal stability (7, 8). In contrast to its restricted
expression in normal tissues, PTTG1 is abundantly detected in a
wide variety of tumors and is associated with metastasis and poor
clinical outcome, suggesting that PTTG1 may play a role in
tumorigenesis (9, 10). A higher PTTG1 expression is
observed in cancer cells compared to that in adjacent normal cells,
although the underlying mechanisms has not been fully elucidated.
Moreover, the association between PTTG1 expression and prognosis
remains ambiguous. In this study, we aimed to investigate the
expression and clinical significance of PTTG1 in ccRCC and assess
the association between PTTG1 expression level and prognosis.
Materials and methods
Patients and tissue samples
The 44 paired samples of ccRCC and normal adjacent
tissues (ADTs) were collected from patients who underwent radical
nephrectomy at the Department of Urology, Hefei Hospital Affiliated
to Anhui Medical University, between November, 2007 and December,
2012. The ADT samples were collected at a distance of 2.0 cm from
visible ccRCC lesions and were all properly maintained until
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis. For immunohistochemical analysis of the PTTG1
protein, a total of 192 paraffin-embedded pathologically verified
ccRCC samples were collected. All the patients had undergone
radical nephrectomy performed at the Department of Urology, Hefei
Hospital Affiliated to Anhui Medical University, between 2000 and
2013. The histological and clinical diagnoses of the tumors in all
the patients were performed by the Department of Pathology in the
same hospital. The characteristics of the 192 patients are
summarized in Table I. Patient
survival data were obtained via telephonical communication and data
on clinical characteristics were obtained from the medical records.
Tumor stage was reclassfied based on the 2011 Union for
International Cancer Control TNM classification of malignant tumors
and nuclear grading was performed according to the Fuhrman
classification (11).
 | Table I.Association of pituitary
tumor-transforming gene-1 (PTTG1) expression level with clinic o
pathologic al characteristics in clear cell renal cell carcinoma
patients. |
Table I.
Association of pituitary
tumor-transforming gene-1 (PTTG1) expression level with clinic o
pathologic al characteristics in clear cell renal cell carcinoma
patients.
| | PTTG1 expression | |
|---|
| |
| |
|---|
| Parameters | Total no.
(n=192) | High (+) (n=113) | Low (-) (n=79) | χ2 | P-value |
|---|
| Gender | | | |
1.819 |
0.177 |
| Male | 108 | 59 | 49 | | |
|
Female | 84 | 54 | 30 | | |
| Age (years) | | | |
0.560 |
0.454 |
| ≥50 | 125 | 76 | 49 | | |
|
<50 | 67 | 37 | 30 | | |
| T stage | | | |
10.816 |
0.004 |
| T1 | 97 | 47 | 50 | | |
| T2 | 51 | 32 | 19 | | |
| T3/4 | 44 | 34 | 10 | | |
| N stage | | | |
4.674 |
0.031 |
| N0 | 145 | 79 | 66 | | |
| N+ | 47 | 34 | 13 | | |
| Metastasis | | | |
4. 169 |
0.041 |
| No
(M0) | 130 | 70 | 60 | | |
| Yes
(M1) | 62 | 43 | 19 | | |
| Recurrence | | | |
7.903 |
0.005 |
| No | 157 | 85 | 72 | | |
| Yes | 35 | 28 | 7 | | |
| Fuhrman grade | | | |
14.719 |
0.002 |
| F1 | 111 | 53 | 48 | | |
| F2 | 51 | 26 | 25 | | |
| F3 | 27 | 22 | 5 | | |
| F4 | 13 | 12 | 1 | | |
This study was approved by the Ethics Committee of
Anhui Medical University and all the patients provided written
informed consent.
RT-qPCR
RT-qPCR was performed as previously described
(12). The corresponding primer
sequences were as follows: PTTG1: sense primer, 5′-AAAGCTCTGTTCCTG
CCTCA-3′; and reverse primer, 5′-GAGAGGCACTCC ACTCAAGG-3′. GAPDH :
sense primer, 5′-GGAGTCCAC TGGCGTCTTCACC-3′; and reverse primer,
5′-GAGGAG TGGG TGTCGCTGTTG-3′. The relative expression levels of
PTTG1 were normalized to the geometric mean of GAPDH (internal
control gene). The data were analysed via the comparative threshold
cycle method (13).
Immunohistochemical analysis
Immunohistochemistry was performed to investigate
PTTG1 expression in the 44 paired ccRCC and normal tissue samples.
This was also implemented in the 192 ccRCC samples. All the
procedures were performed as previously described (12). The sections were incubated with the
monoclonal rabbit anti-human PTTG1 antibody (cat. no. sc-5843,
dilution 1:400; Abcam, Cambridge, MA, USA).
Staining evalvation
The stained sections were reviewed by two
independent pathologists. Scoring was based mainly on color
intensity and extent. The proportion of cells expressing PTTG1
varied between 0 and 100% and the intensity of staining ranged from
weak to strong. The proportion of PTTG1-expressing tumour cells was
scored at low magnification on a scale of 0–5 (0, no positive
cells; 1, 0–5%; 2, 6–25%; 3, 26–50%; 4, 51–75%; and 5, 76–100%
positive cells). The intensity score was determined at high
magnification on a scale of 0–3 (0, negative; 1, weakly positive;
2, moderately positive; and 3, strongly positive staining) and the
final score was calculated by multiplying the two parameters, with
scores of 0, 1, 2, 3, 4, 5, 6, 8, 9, 10, 12 and 15. The optimal
cut-off values for PTTG1 levels were set by measuring the
heterogeneity in overall survival rates using the log-rank test
method. Low expression was defined as a total score of <5 and
high expression as a total score of ≥5. Thus, the stained sections
were divided into two different groups by PTTG1 expression
level.
Statistical analysis
Paired-sample t-tests were used in the RT-qPCR and
immunohistochemical assays to analyze the significance of the
differences in mRNA and protein expression level between ccRCCs and
the ADTs. The correlation of PTTG1 expression with
clinicopathological characteristics was assessed by the
χ2 test. Survival curves were plotted according to the
Kaplan-Meier method and compared by the log-rank test. A
multivariate analysis according to Cox's proportional hazards
regression model adjusted for clinicopathological factors (age,
gender, tumor size, Fuhrman grade, TNM stage and PTTG1 expression)
was performed to identify the clinical variables that were
independently correlated with overall survival. The statistical
analysis was performed using the SPSS 17.0 package (SPSS Inc.,
Chicago, IL, USA) and P<0.05 was considered to indicate a
statistically significant difference.
Results
RT-qPCR analysis of PTTG1 mRNA in 44
ccRCC tumor samples
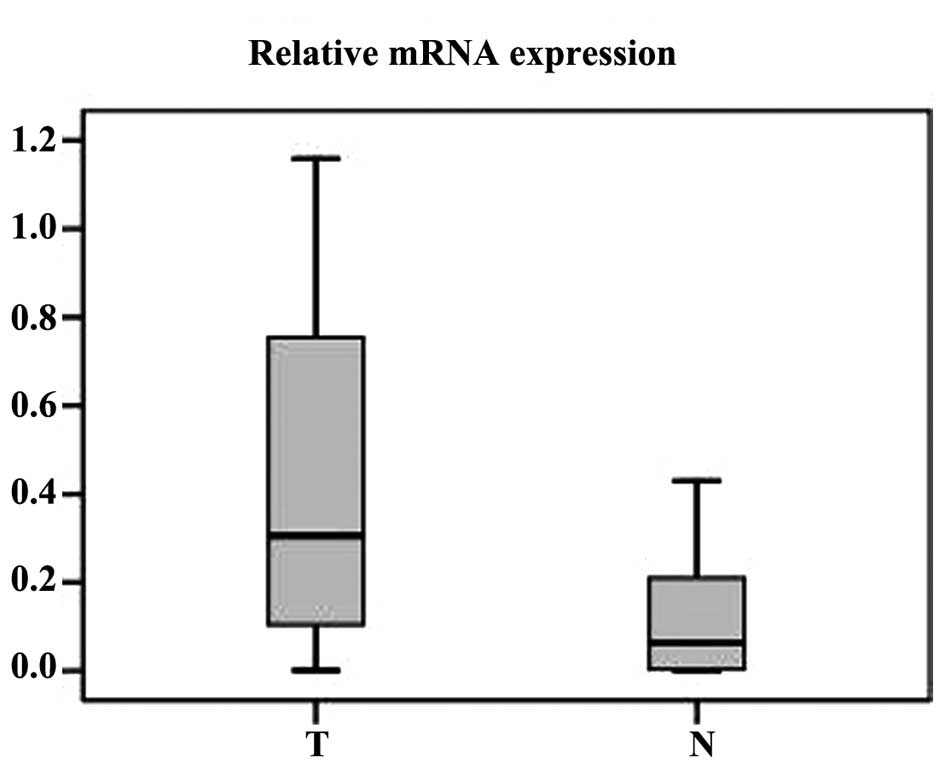
RT-qPCR was performed to measure the expression of
PTTG1 mRNA in 44 ccRCC tumor tissues and normal ADTs. Compared to
normal tissues, 37 ccRCC tumor tissue samples exhibited a higher
expression at the mRNA level (paired-sample t-test, P<0.001;
Fig. 1).
Immunohistochemistry analysis of PTTG1
protein expression in 44 ccRCC and paired ADT samples
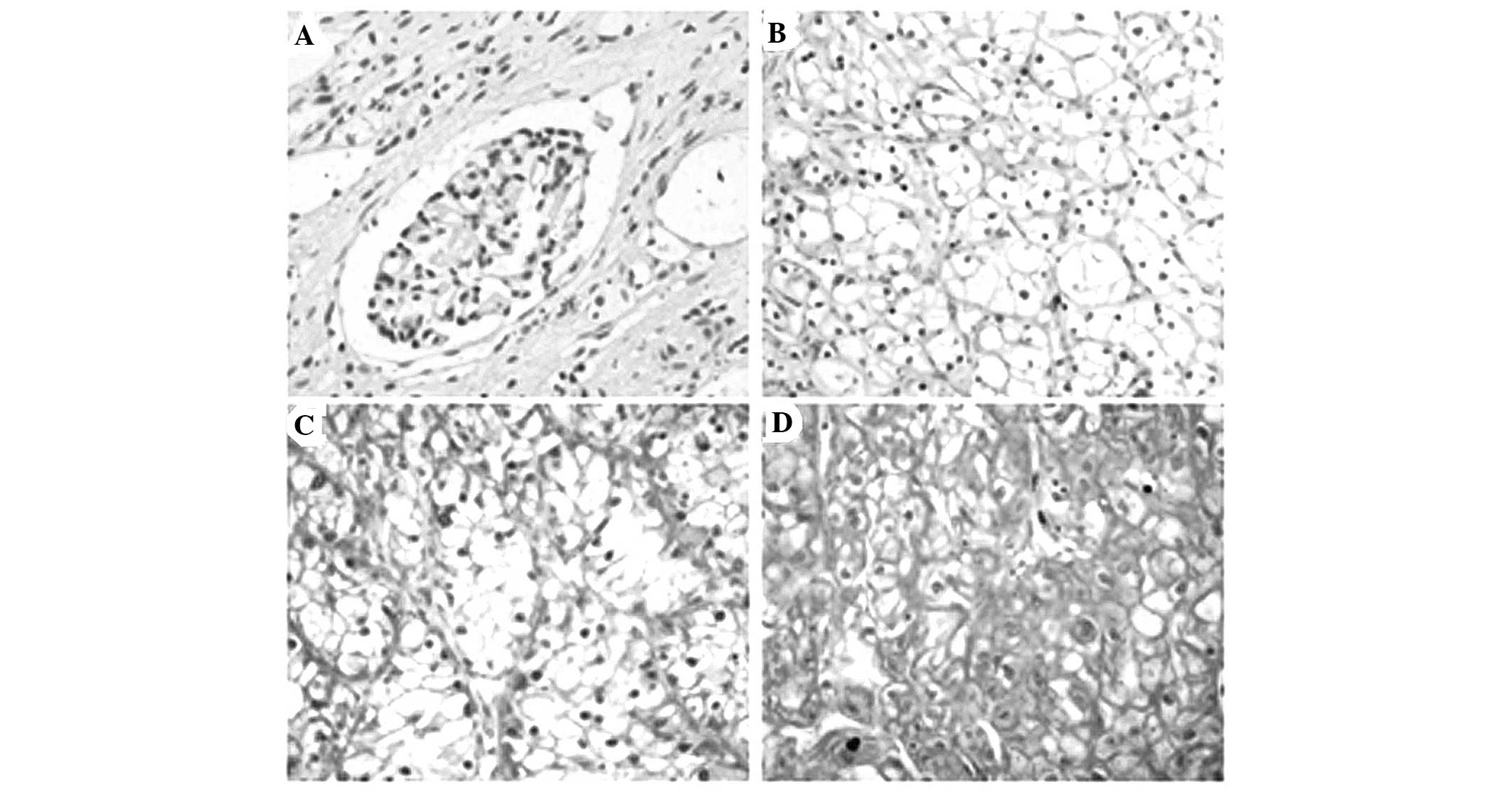
Immunohistochemistry was applied to assess the
expression and subcellular localization of the PTTG1 protein in 44
paraffin-embedded ccRCC and paired normal ADT samples. PTTG1
staining was present mainly in the nuclei and cytoplasm (Fig. 2). In normal renal tissue, PTTG1
protein expression was negative (29/44, score=0) or low (15/44,
score ≤5). The PTTG1 protein expression in the 44 tumor tissue
samples was higher compared to that in the paired ADTs
(paired-sample t-test, P<0.001; Fig. 3).
Immunohistochemical analysis of the
association between PTTG1 protein expression and clinical
characteristics in 192 ccRCC samples
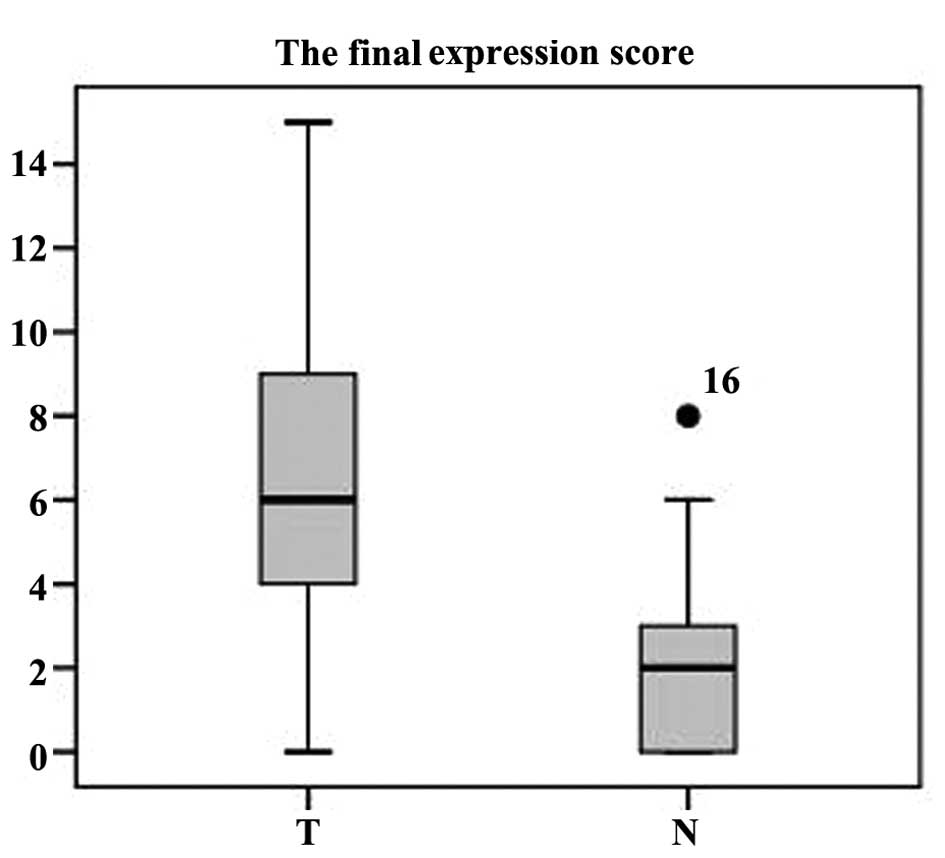
To further assess the correlation between PTTG1
expression and various clinicopathological parameters, a further
immunohistochemical analysis was performed in 192 ccRCC samples. As
shown in Table I, low expression
of PTTG1 (score ≤4) was exhibited by 79 of the 192 tumor samples,
whereas high expression (score ≥5) was exhibited by 113 samples.
Increased expression of PTTG1 in tumor samples was correlated with
T stage (χ2 =10.816, P=0.004), N classification
(χ2 =4.674, P=0.031), metastasis (χ2 =4.169,
P=0.041), recurrence (χ2 =7.903, P=0.005) and Fuhrman
grade (χ2 =14.719, P=0.002), while associations with age
(χ2 =0.560, P=0.454) and gender (χ2 =1.819,
P=0.177) were not identified. High expression of PTTG1 was observed
in 69.5, 62.7 and 77.3% of stage T1, T2 and T3/4 ccRCC cases,
respectively (P=0.004, χ2 test). High expression of
PTTG1 was observed in 69.4 and 53.8% of stage N0 and N1/2 ccRCC
cases, respectively (P=0.041, χ2 test). High expression
of PTTG1 was observed in 74.2 and 54.6% of ccRCC cases with and
without metastasis, respectively (P=0.009, χ2 test).
Finally, high expression of PTTG1 protein was observed in 80.0 and
54.1% of ccRCC cases with and without recurrence, respectively
(P=0.005, χ2 test).
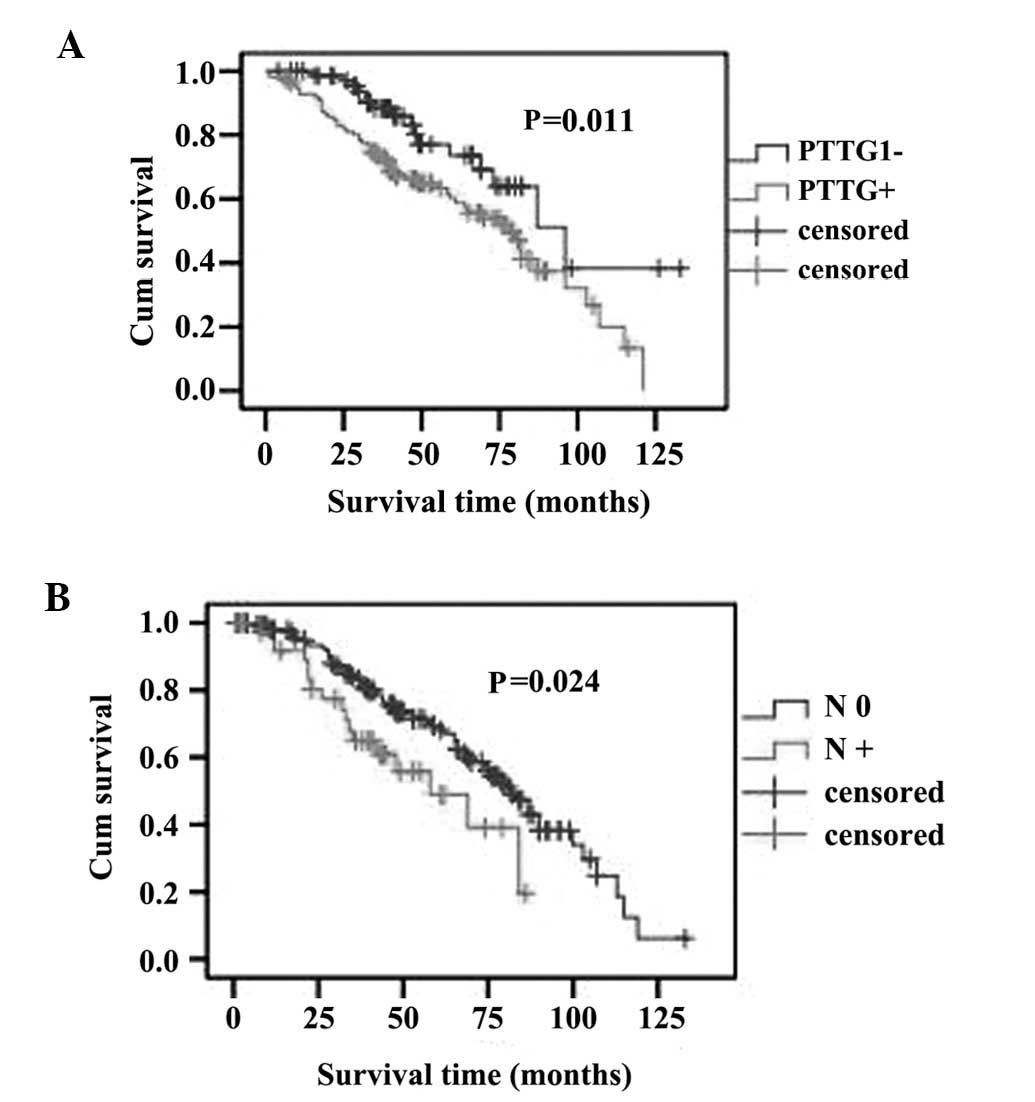
Survival analysis
To further investigate the prognostic value of PTTG1
expression in ccRCC, Kaplan-Meier analysis and the log-rank test
were applied to assess the association between PTTG1 expression
level in ccRCC and prognosis. The level of PTTG1 expression was
found to be correlated with the overall survival of ccRCC patients.
Patients with higher PTTG1 expression (PTTG1+) exhibited poorer
survival rates compared to those with lower expression (PTTG1-). In
the group of PTTG1+ patients, the mean and median survival time was
59.722 and 47 months, respectively; however, the mean and median
survival time in the PTTG1-group was 88.285 and 79 months,
respectively. The log-rank test revealed that the survival rates
were significantly different between the two groups (log-rank;
P=0.011, Fig. 4A). Similarly,
patients without regional lymph node involvement (N0) exhibited a
better prognosis compared to those with regional lymph node
involvement (N+) (log-rank, P=0.024; Fig. 4B).
In addition, the multivariate Cox regression
analysis indicated that PTTG1 expression (P=0.027) and N stage
(P=0.020) were independent prognostic factors for the overall
survival of ccRCC patients (Table
II).
 | Table II.Multivariate Cox regression analysis
for the overall survival rates of clear cell renal cell carcinoma
patients. |
Table II.
Multivariate Cox regression analysis
for the overall survival rates of clear cell renal cell carcinoma
patients.
| Risk factors | Relative risk | 95% confidence
interval | P-value |
|---|
| T stage |
3.001 | 0.366-1.970 |
0.201 |
| M stage |
1.827 | 0.819-3.406 |
0.077 |
| N stage |
7.38 | 2.425-11.307 |
0.02 |
| Age |
0.992 | 0.569-1.451 |
0.285 |
| Gender |
0.622 | 0.489-1.266 |
0.531 |
| Fuhrman grade |
9.579 | 3.110-10.355 |
0.103 |
| PTTG1
expression |
0.838 | 0.358-1.172 |
0.027 |
Discussion
PTTG1 was isolated from rat pituitary tumor cells in
1997 and identified as a pituitary-derived transforming gene
(7). Structural homology led to
the identification of the PTTG1 protein as a vertebrate securin
critical in regulating sister chromatid separation during mitosis
(8). PTTG1 overexpression has been
reported in a variety of endocrine-related tumors, particularly
pituitary, thyroid, breast, ovarian and uterine tumors, as well as
non-endocrine-related cancers involving the central nervous,
pulmonary and gastrointestinal systems. PTTG1 levels were found to
be correlated with tumor invasiveness (14) and PTTG1 has been identified as a
key signature gene associated with tumor metastasis (10).
NIH3T3 cells (15)
and human embryonic kidney 293 cells (16) stably transfected with PTTG1
exhibited increased cell proliferation rates compared to control
vector-transfected cells. Studies in PC3 cells reported
tetracycline-regulated PTTG1 expression and PTTG1-induced cell
growth (17). Inhibition of cell
proliferation by PTTG1 siRNA has been reported in M10 melanoma
(18), HeLa S3 (19) and SH-J1 hepatoma cells (20).
Evidence suggests that an important transforming
mechanism underlying PTTG1 overexpression involves the induction of
chromosomal instability and aneuploidy. p53-deficient MG-63
osteosarcoma cells transiently or stably transfected with
PTTG1-enhanced green fluorescent protein were investigated for
signs of aneuploidy, such as the presence of micronuclei,
macronuclei, or chromosomal bridges (21).
Bernal et al (22) reported that PTTG1 specifically
interacts with p53 in vitro and in vivo and that this
interaction blocks specific binding of p53 to DNA and inhibits its
transcriptional activity. In PTTG1-deficient tumor cells, the
proapoptotic and transactivation functions of p53 were potentiated.
In addition, the overexpression of PTTG1 in hepatocellular
carcinoma cell lines attenuated p53 induction of apoptosis
(20). Thus, these results suggest
a tumorigenic mechanism for PTTG1, as the inhibition of
p53-mediated apoptosis by high securin expression may explain the
survival of tumor cells harboring functional p53 (22). In other studies, siRNA-based
techniques efficiently suppressed endogenous PTTG1 and inhibited
cell proliferation in PC3 prostate cancer cells (17) and hepatocellular carcinoma cell
lines (20, 23).
To the best of our knowledge, this is the first
study to indicate the clinical significance of PTTG1 in ccRCC.
RT-qPCR in 44 ccRCC and paired ADT samples demonstrated a
significant increase of PTTG1 mRNA in the ccRCC samples. Further
immunohistochemical analysis in the 44 paired ccRCC and ADT samples
confirmed the overexpression of PTTG1 protein in the tumor tissues.
These results indicate that PTTG1 may play a significant role in
the initiation and progression of malignancies.
To further investigate the prognostic value of
PTTG1, immunohistochemical analysis was performed to evaluate the
correlation between PTTG1 expression and various
clinicopathological parameters. In this study, we demonstrated that
the increased PTTG1 expression was significantly correlated with
tumor size, Fuhrman grade, stage, N classification, metastasis and
recurrence. According to the Kaplan-Meier analysis, PTTG1 protein
expression in ccRCC was significantly correlated with overall
survival. Patients with high PTTG1 expression levels exhibited a
shorter survival time compared to those with low PTTG1 levels. The
log-rank test revealed that the PTTG1-group exhibited a more
favorable prognosis compared to the PTTG1+ group. In addition, the
TNM stage of ccRCC was found to be closely associated with
prognosis (24). Consistent with
those findings, in the present study, PTTG1 expression and N
classification were independent prognostic factors for the overall
survival of ccRCC patients in the multivariate Cox regression
analysis. Therefore, this study revealed that there were
significant correlations between the PTTG1 expression level and
clinicopathological parameters and, therefore, PTTG1 may represent
a potential prognostic marker and therapeutic target for ccRCC.
To the best of our knowledge, this is the first
study aimed at evaluating the possibility of using PTTG1 as an
indicator of disease progression in the clinical setting, as well
as a prognostic marker for ccRCC patient survival. However, it
should be noted that our study was a single hospital-based,
retrospective study and further multi-centered or community-based
prospective studies are required to verify our findings.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (grant no. 81101524).
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Singer EA, Gupta GN, Marchalik D and
Srinivasan R: Evolving therapeutic targets in renal cell carcinoma.
Curr Opin Oncol. 25:273–280. 2013.PubMed/NCBI
|
|
3
|
Macher-Goeppinger S, Bermejo JL,
Schirmacher P, Pahernik S, Hohenfellner M and Roth W:
Senescence-associated protein p 400 is a prognostic marker in renal
cell carcinoma. Oncol Rep. 30:2245–2253. 2013.PubMed/NCBI
|
|
4
|
Tanaka T, Kuramitsu Y, Wang Y, et al:
Glyoxalase 1 as a candidate for indicating the metastatic potential
of SN12C human renal cell carcinoma cell clones. Oncol Rep.
30:2365–2370. 2013.PubMed/NCBI
|
|
5
|
Wei C, Lai YQ, Li XX and Ye JX:
TGF-β-activated kinase-1: a potential prognostic marker for clear
cell renal cell carcinoma. Asian Pac J Cancer Prev. 14:315–320.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Iwamoto H, Kanda Y, Sejima T, Osaki M,
Okada F and Takenaka A: Serum miR-210 as a potential biomarker of
early clear cell renal cell carcinoma. Int J Oncol. 44:53–58.
2014.PubMed/NCBI
|
|
7
|
Pei L and Melmed S: Isolation and
characterization of a pituitary tumor-transforming gene (PTTG). Mol
Endocrinol. 11:433–441. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zou H, McGarry TJ, Bernal T and Kirschner
MW: Identification of a vertebrate sister-chromatid separation
inhibitor involved in transformation and tumorigenesis. Science.
285:418–422. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Feng ZZ, Chen JW, Yang ZR, Lu GZ and Cai
ZG: Expression of PTTG1 and PTEN in endometrial carcinoma:
correlation with tumorigenesis and progression. Med Oncol.
29:304–310. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoon CH, Kim MJ, Lee H, et al: PTTG1
oncogene promotes tumor malignancy via epithelial to mesenchymal
transition and expansion of cancer stem cell population. J Biol
Chem. 287:19516–19527. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fuhrman SA, Lasky LC and Limas C:
Prognostic significance of morphologic parameters in renal cell
carcinoma. Am J Surg Pathol. 6:655–663. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei C, Wu S, Li X, Wang Y, Ren R, Lai Y
and Ye J: High expression of FER tyrosine kinase predicts poor
prognosis in clear cell renal cell carcinoma. Oncol Lett.
5:473–478. 2013.PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2–ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Demeure MJ, Coan KE, Grant CS, et al:
PTTG1 overexpression in adrenocortical cancer is associated with
poor survival and represents a potential therapeutic target.
Surgery. 154:1405–1416. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kakar SS and Jennes L: Molecular cloning
and characterization of the tumor transforming gene (TUTR1): a
novel gene in human tumorigenesis. Cytogenet Cell Genet.
84:211–216. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamid T, Malik MT and Kakar SS: Ectopic
expression of PTTG1/securin promotes tumorigenesis in human
embryonic kidney cells. Mol Cancer. 4:32005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang SQ, Liao QJ, Wang XW, Xin DQ, Chen
SX, Wu QJ and Ye G: RNAi-mediated knockdown of pituitary
tumor-transforming gene-1 (PTTG1) suppresses the proliferation and
invasive potential of PC3 human prostate cancer cells. Braz J Med
Biol Res. 45:995–1001. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Caporali S, Alvino E, Levati L, et al:
Down-regulation of the PTTG1 proto-oncogene contributes to the
melanoma suppressive effects of the cyclin-dependent kinase
inhibitor PHA-848125. Biochem Pharmacol. 84:598–611. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Solbach C, Roller M, Peters S, Nicoletti
M, Kaufmann M and Knecht R: Pituitary tumor-transforming gene
(PTTG): a novel target for anti-tumor therapy. Anticancer Res.
25:121–125. 2005.PubMed/NCBI
|
|
20
|
Cho-Rok J, Yoo J, Jang YJ, et al:
Adenovirus-mediated transfer of siRNA against PTTG1 inhibits liver
cancer cell growth in vitro and in vivo. Hepatology. 43:1042–1052.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hsu YH, Liao LJ, Yu CH, et al:
Overexpression of the pituitary tumor transforming gene induces p
53-dependent senescence through activating DNA damage response
pathway in normal human fibroblasts. J Biol Chem. 285:22630–22638.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bernal JA, Luna R, Espina A, et al: Human
securin interacts with p 53 and modulates p 53-mediated
transcriptional activity and apoptosis. Nat Genet. 32:306–311.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liang M, Chen X, Liu W, Li S, Li C, Jiang
L and Lv S: Role of the pituitary tumor transforming gene 1 in the
progression of hepatocellular carcinoma. Cancer Biol Ther.
11:337–345. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Levi F, Ferlay J, Galeone C, Lucchini F,
Negri E, Boyle P and La Vecchia C: The changing pattern of kidney
cancer incidence and mortality in Europe. BJU Int. 101:949–958.
2008. View Article : Google Scholar : PubMed/NCBI
|