Reciprocal chromosomal translocations are recurrent
features of several hematological malignancies (http://cgap.nci.nih.gov/Chromosomes/Mitelman;
http://atlasgeneticsoncology.org/). The
cloning of the genes located at the breakpoints of chromosomal
translocations in leukemia and lymphoma has led to the
identification of new genes involved in carcinogenesis. These
rearrangements generate new genes, called fusion genes, or lead to
the activation of a proto-oncogene by relocation near active
regulatory sequences. The second mechanism is the hallmark of
lymphoma and B-cell chronic lymphocytic leukemia (CLL).
CLL represents the most common hematological
malignancy in Western countries, with a highly heterogeneous
clinical course and prognosis, and a time-to-progression varying
from months to several years. Some patients live for prolonged
periods without therapy, while others rapidly develop progressive
disease and require treatment (1,2).
Transformation into a fast-growing diffuse large
B-cell lymphoma is encountered in ~5% of CLL patients and it is
referred to as Richter's syndrome (7,8). Under
rare circumstances, plasmablastic lymphoma or plasmablastic
transformation may be observed, representing an unusual example of
Richter's syndrome.
Numerous studies have searched for reliable
prognostic markers capable of predicting the progression and
outcome of this disease. They include two different clinical
staging systems, one described by Rai et al (9) and the other by Binet et al
(10). Other prognostic markers, such
as CD38 and ZAP-70 expression and IG heavy chain variable region
(IGHV) mutational status, are being evaluated or used
(2,11). More recently, next-generation
sequencing has identified mutations in a few genes that may have a
prognostic impact (12,13).
Between 2000 and 2014, the Cytogenetics Laboratory
of the Brest University Hospital collected blood or bone marrow
samples from CLL patients diagnosed and/or followed up in 10
hospitals in Brittany. The clinical diagnosis of CLL in each
patient was based on a persistent lymphocytosis of >5×109
cells/l and a typical immunophenotypic picture (CD5+,
CD19+, CD20+, CD23+ and weak
expression of sIg) (15–17). A total of 396 patients were referred
at diagnosis and 275 during follow-up.
Peripheral blood and bone marrow were cultured for
72 h. Cell stimulation was performed with tetradecanoylphorbol
acetate from 2000 to 2010, and with the immunostimulatory
CpG-oligonucleotide DSP30 and interleukin-2 from 2010 onwards. The
chromosomes were R-banded and the karyotype was described according
to the International System for Human Cytogenetics Nomenclature
(18).
iFISH was performed on fixed cells from the cultures
using the Vysis CLL FISH Probe kit (Abbott Molecular, Rungis,
France). The CLL panel includes two set of probes: A first set
consisting of LSI TP53 SpectrumOrange and ATM SpectrumGreen probes,
and a second set consisting of LSI D13S319 SpectrumOrange/13q34
SpectrumAqua and CEP 12 SpectrumGreen probes. A total of 300
interphase nuclei were studied for each set of probes. Based on the
recommendations of the CLL Research Consortium FISH standardization
project, FISH cut-off was set at 10% for each hybridization
(19).
Metaphase and iFISH using the IGH Breakapart Probe
(Cytocell, Compiègne, France or Abbott Molecular) was not
systematically performed on all samples, but only on those in which
an abnormality of chromosome 14 was suspected. Translocations
involving the IG κ locus (IGK) or IG λ locus (IGL)
genes were assessed with the IGK Breakapart Probe or the IGL
Breakapart Probe (Cytocell).
iFISH with the CLL FISH Probe kit was performed on
25 patients carrying an IG translocation (Table II). Mono-allelic deletion of 13q14
was identified in 9 patients (36%), including a bi-allelic in 2
patients. Trisomy 12 was found in 12 patients (48%). Deletions of
11q22 and 17p13 were observed in 1 (4%) and 3 (12%) patients,
respectively (Table II). Recently,
we reported our iFISH results on 638 patients. Del(13q14) was found
in 65% and a trisomy 12 in 22.1% of the patients. Deletions of
11q22 and 17p13 were observed in 13.3 and 8.6% of the patients,
respectively (20).
For years, banding karyotyping has been hampered by
the low mitotic index of CLL cells, and the majority of the studies
currently rely on iFISH with an IGH probe to determine the
frequency of IGH translocations. A total of 18 studies were
retrieved from the literature (Table
III). The overall frequency was 8.3% (327/3,922) but the range
varied considerably, from 1.9% in a study from the USA, to 26.1% in
a Canadian study. Although a geographic/ethnic uneven distribution
cannot be ruled out, it is likely that differences in diagnosis
(PLL included or not), referral, access to care and healthcare
systems may have affected the frequency. Even for those
investigated at diagnosis, we cannot exclude that socioeconomic
status or access to healthcare may have delayed the time at which
diagnosis was made, with the consequence that patients in some
series were at a more advanced stage (20,21).
A limited number of studies attempted to identify
the partner genes. Even in those cases, they used commercially
available Dual Color, Dual Fusion Translocation Probes to detect
IGH/B-cell lymphoma 2 (BCL2), IGH/cyclin D1
(CCND1), sometimes completed by v-myc avian myelocytomatosis
viral oncogene homolog (MYC) and BCL3 Dual Color
Breakapart rearrangement probes. Furthermore, none of these studies
used probes targeting the IGK and IGL genes and,
therefore, underestimated the true frequency of IG
translocations.
We conducted a thorough search in the literature
looking for IG translocations, using the Mitelman Database of
Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman) as the
starting point.
Some translocations have been found to be frequent,
while others have only been reported in a few or single cases. At
present, 31 partner chromosome bands have been described, but the
partner gene has only been identified in 10 translocations. We
identified 4 chromosomal bands that had never been shown to be
involved in IG translocations among the 35 patients in the Brest
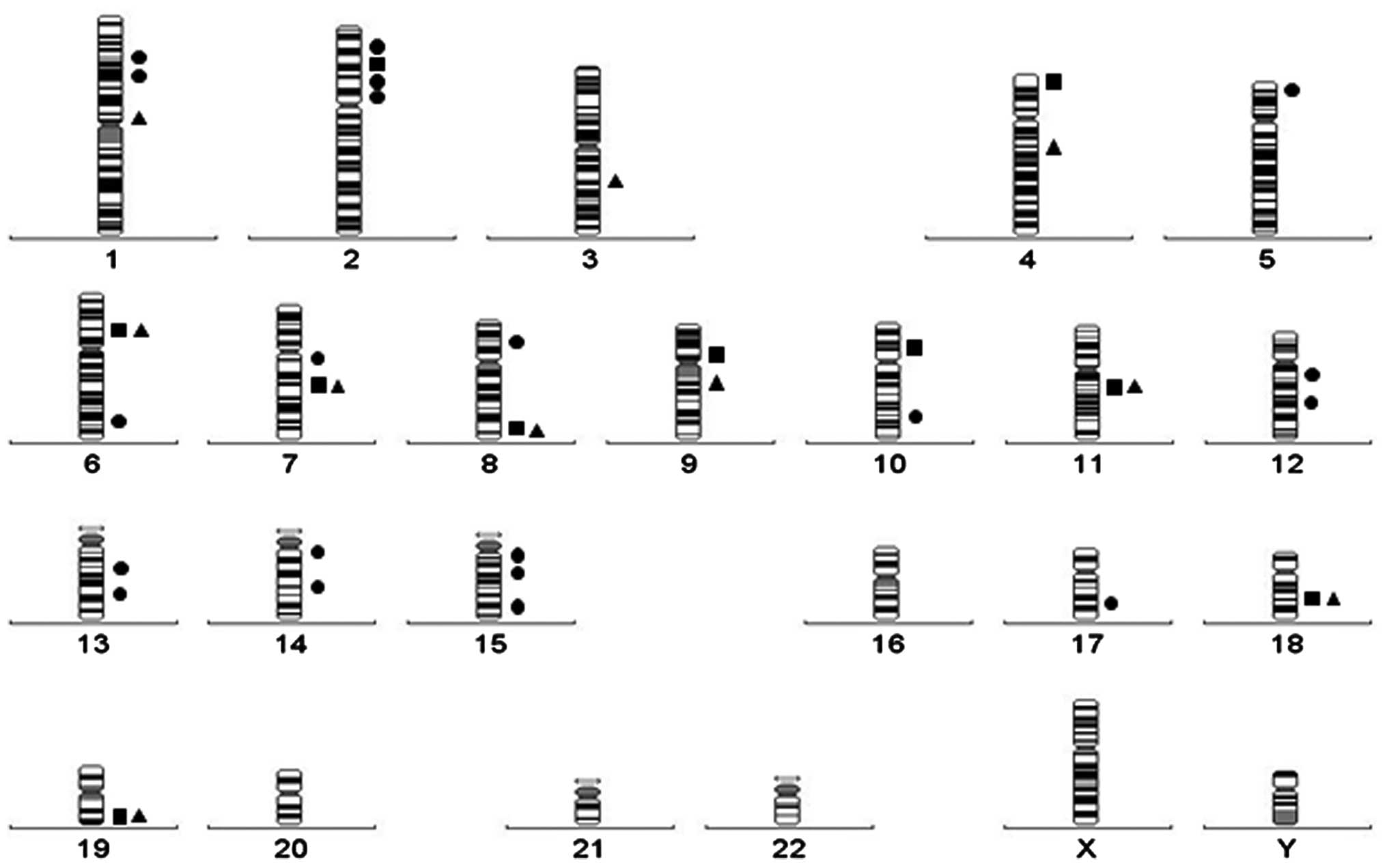
cohort, bringing the total to 35 (Fig.
1). The number of bands involved in IG translocations is
significantly lower compared with that involved in Ets variant 6 or
Runt-related transcription factor 1 gene translocations (48 and 55
bands, respectively) (22,23). The difference is likely the result of
the B-cell lineage specificity of the partner genes deregulated by
the IG translocations.
As previously mentioned, IG translocations relocate
genes near active regulatory sequences, leading to their
overexpression. However, two exceptions are known. In 2004, Schmidt
et al (24) reported a patient
with B-cell CLL carrying a t(12;14)(q23;q32), in whom the
carbohydrate (chondroitin 4) sulfotransferase 11 gene
(CHST11) was fused to IGH to create a chimeric gene.
Fusion RNAs may be translated into truncated proteins and lead to
deregulation of the CHST11 protein trafficking across intracellular
membranes, resulting in loss of function, as observed with other
fusions (23).
All 3 translocations, t(14;18) (q32;q21),
t(2;18)(p11;q21.3) and t(18;22)(q21.3;q11), and their molecular
consequences, IGH/BCL2, IGK/BCL2 and IGL/BCL2,
have been reported (28,29). These translocations are present at
diagnosis or arise during evolution and are usually associated with
additional karyotypic changes, more particularly with trisomy 12
(30–32).
A total of 144 cases reported in the literature were
reviewed. A t(14;18) was identified in 111 cases (77.1%), a t(2;18)
in 8 cases (5.5%) and a t(18;22) in 25 cases (17.4%). Trisomy 12
was found in 71 cases (49.3%). The IG translocation was the sole
abnormality in 39 cases (27.1%) and part of a complex karyotype
(defined as composed of ≥3 abnormalities) in 37 cases (25.7%).
These are found in typical CLL (sometimes during
Richter's transformation) and in CLL/PL (29,30,32).
Atypical cytological characteristics (increased number of lymphoid
cells with irregular nuclear contour, plasmacytoid features or PLs)
and/or immunophenotypic profile (lack of CD23 and
intermediate/strong CD20 expression) is reported and hypothesized
to be linked to trisomy 12 (29,33,34).
IG/BCL2 fusion is significantly associated with mutated
IGHV status (30–32), which led Baseggio et al
(30) to conclude at a post-germinal
center cellular origin.
This translocation appears at diagnosis in the
primary clone in the majority of the cases, and as a secondary
change during karyotypic progression in a limited number of cases
(39). This is rarely the sole
cytogenetic aberration, with trisomy 12 being the most frequent
associated abnormality (40–45).
A total of 123 cases reported in the literature were
reviewed. No variant translocation was found. Trisomy 12 was
identified in 72 cases (58.5%). The translocation was the sole
abnormality in 13 cases (10.6%) and part of a complex karyotype in
52 cases (42.3%).
CLL with t(14;19) is associated with atypical
morphological (small cells, often with nuclear indentations) and
immunophenotypical characteristics (40,41,46). The
translocation is found in all three subtypes and during Richter's
transformation (42,43,45,47). The
vast majority of t(14;19) express unmutated IGHV genes
(39–41,45,46), which
is significantly higher compared with the 46% reported in the
literature for typical CLL (48).
Patients have an aggressive clinical course and the overall
prognosis is poor (42,43,45,46).
Although the (11;14)(q13;q32) translocation was
previously considered to be the hallmark of mantle cell lymphoma
(MCL), it is currently identified in 10–20% of PLL and 2–5% of CLL
cases (56). CLL cases in which this
translocation has been found are usually atypical in terms of
morphology (majority of small lymphocytes with PLs and/or large
lymphocytes) and immunophenotype (CD5+,
CD19+, sIg+, FMC7+ and
CD10−) (57–60). Recognition of this cytogenetic subset
of atypical CLL is crucial, as, given its poor prognosis, it may
require early treatment (61).
We identified 106 CLL or PLL cases associated with
t(11;14) in the literature. Only one variant translocation,
t(11;22)(q13;q11), was found (62).
Trisomy 12 was identified in 5 cases (4.7%). The translocation was
the sole abnormality in 21 cases (19.6%) and part of a complex
karyotype in 64 cases (59.8%).
Distinction of t(11;14) translocation-associated CLL
and MCL in the leukemic phase is not unequivocal (60,64). It is
hypothesized that MCL and atypical CLL with the t(11;14) represent
the extremes of a spectrum of disorders of follicle mantle lineage
(60).
This abnormality is rare in typical CLL and is
associated with increased PLs (CLL with occasional PLs, CLL/PL and
PLL) (68–71). Male and elderly patients are
predominantly affected, as in CLL/PL patients (69–71).
This translocation may be acquired in the chronic
phase, but is associated with an advanced clinical stage at
presentation (68,71). It is usually included in a complex
karyotype. It may represent a secondary abnormality contributing to
disease progression and carries a poor prognosis (68–70).
We reviewed 38 cases from the literature. A t(8;14)
was identified in 22 cases (57.9%), a t(2;8) in 5 cases (13.2%) and
a t(8;22) in 11 cases (28.9%). Trisomy 12 was rarely associated,
being identified in only 3 cases (7.9%). The IG translocation was
the sole abnormality in 8 cases (21.1%) and part of a complex
karyotype in 18 cases (47.4%).
The (2;14) translocation appears to be an early
event, as it has been found to be the sole karyotypic abnormality
at diagnosis. It is also present in the main clone, with subclones
containing additional abnormalities in other patients (77,78). Only
6 cases were retrieved from the literature, 2 of which were
associated with trisomy 12, and 3 of which were present in
subclones with a complex karyotype.
All patients thus far analyzed express ZAP-70 and
all but one also carried unmutated IGVH genes (78,79).
Therefore, it is expected that t(2;14) is associated with an
aggressive disease and a poor prognosis, which appears to be the
case, although data is sparse (77,78).
The t(10;14)(p11-p13;q32) and t(10;22)(p12;q11) were
identified in 5 and 1 CLL cases, respectively (82). The translocation was identified during
clinical progression or Richter's transformation and appears to
carry a poor prognosis (82).
These translocations are not associated with a
particular subtype of CLL, or with IGVH mutation status;
they do not appear to be driver abnormalities in CLL genesis, but
are rather a marker of disease progression (82).
Although the t(4;14) has long been known to be a
recurrent abnormality in multiple myeloma, it was first described
by Bacher et al (90) in 2
patients exhibiting the immunophenotype of CLL and CLL/PL. Cerny
et al (91) reported a CLL
patient with the t(4;22)(p16;q11) and a typical immunophenotype, in
whom the lymphoid proliferation was composed of small lymphocytes
(with round nuclei, condensed chromatin, indistinct nucleoli and
scant cytoplasm) and PLs (larger nucleolated lymphocytes). A fourth
case was reported by Geller et al (92) in a patient with CLL/PL and a typical
immunophenotype. A total of 2 cases had a trisomy 12.
These 4 patients exhibited no consistent
characteristics, other than the presence of PLs. The translocation
was identified at diagnosis or during evolution. The cells were
negative or positive for CD38 and ZAP-70, and had a mutated or
non-mutated IGVH status. Although the number of cases
reported is small, this translocation should be considered as
indicative of adverse prognosis.
The t(4;14)(p16;q32) is a unique example of IG
translocation, as it simultaneously deregulates 2 genes with
oncogenic potential. Indeed, MMSET domain (also referred to
as Wolf-Hirschhorn syndrome candidate 1) and FGFR3 reside on
either side of the 4p16 breakpoint. After the translocation,
MMSET remains on the derivative chromosome 4, while
FGFR3 moves to the derivative chromosome 14 (93–95). The
MMSET protein has histone methyltransferase activity and may act as
a transcriptional regulator controlling cell cycle and apoptosis
(96,97). The FGFR3 protein is a tyrosine protein
kinase that acts as a cell surface receptor for fibroblast growth
factors and triggers downstream mitogen-activated protein kinase
and phosphatidylinositol 3-kinase signaling. It plays an essential
role in the regulation of cell growth and differentiation (98,99).
Although these translocations, particularly
t(2;7)(p11;q21), have been reported in several cases of splenic
marginal zone lymphoma (SMZL), they were unequivocally identified
in 12 CLL patients (100–104). Unfortunately, no data on cell
morphology, immunophenotype, or prognosis, are available.
Of note, in SMZL and CLL, t(2;7) is more frequent
compared with the two other translocations, t(7;14) and t(7;22). No
explanation has been provided as to why IGK is more likely
than IGH to juxtapose to CDK6, contrary to the other
translocations in which IGH is much more frequently
rearranged (104). Of the 11 cases
retrieved from the literature, 7 had a (2;7)(p11;q21) translocation
(63.6%), 3 had a t(7;14) (27.3%) and 1 had a t(7;22)(q21;q11)
(9.1%). Our series included 1 case with t(2;7) and 1 case with
t(7;22). The translocation was the sole abnormality or included in
a complex karyotype in 4 cases each.
Although the t(9;14)(p13;q32) and its variants have
been mostly identified in diffuse large B-cell lymphoma (109), the t(9;14) has also been found in 7
CLL cases (111–114). Few data are available on the cell
morphology, but plasmacytoid differentiation is a common
characteristic.
Although no trisomy 12 was found, a duplication of
part of its long arm was identified in 2 cases. The t(9;14) was
included in a complex karyotype in 4 patients, but present as the
sole abnormality in the remaining 3 patients.
The main characteristics of the translocations for
which the partner gene was identified are summarized in Table IV. IG translocations result in the
deregulated expression of genes involved in several pathways. All
the partner genes thus far identified are involved in the control
of cell proliferation and/or apoptosis (127). In the majority of the cases, the
translocated partner gene becomes transcriptionally deregulated as
a consequence of its transposition into the IG locus (128).
Although the number of cases with a given
translocation is sometimes low, it appears that CLL associated with
IG translocations is characterized by atypical cell morphology,
including plasmacytoid characteristics, and the propensity of being
enriched in PLs. The IGHV mutational status varies between
translocations, those with unmutated IGHV presumably
involving cells at an earlier stage of the B-cell lineage.
With the exception of t(14;18), prognosis appears
to be poor for the translocations for which sufficient data is
available (123,129,130).
Davids et al (131)
demonstrated that the time to first treatment was significantly
shorter among CLL patients harboring 14q32 translocations without
t(14;18), compared with those with t(14;18). Furthermore, the
presence of an IGH translocation associated with a del(13q)
was shown to confer a poorer prognosis compared with del(13q) alone
(132).
Although most centers currently use FISH to
identify trisomy 12 and deletions of 13q14, 11q22 and 17p13, which
are known to have prognostic significance, it is evident that
searching for translocations involving not only IGH, but
also IGL and IGK, by banding and molecular
cytogenetics, will add new information. Furthermore, as the
prognosis depends on the partner gene involved in the
translocation, it is important to identify this partner gene, at
least in recurrent translocations, to ensure the patients receive
optimal treatment.
We would like to thank all the physicians from the
different hospitals who sent us samples for cytogenetic analyses.
We are grateful to the technicians of the cytogenetic laboratory
for their skilled work.
|
1
|
Dores GM, Anderson WF, Curtis RE, Landgren
O, Ostroumova E, Bluhm EC, Rabkin CS, Devesa SS and Linet MS:
Chronic lymphocytic leukaemia and small lymphocytic lymphoma:
Overview of the descriptive epidemiology. Br J Haematol.
139:809–819. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nabhan C and Rosen ST: Chronic lymphocytic
leukemia: A clinical review. JAMA. 312:2265–2276. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Swerdlow SH, Campo E, Harris NL, Jaffe ES,
Pileri SA, Stein H, Thiele J and Vardiman JW: WHO Classification of
Tumours of Hematopoietic and Lymphoid Tissues (4th). IARC press.
Lyon: 2008.
|
|
4
|
Galton DA, Goldman JM, Wiltshaw E,
Catovsky D, Henry K and Goldenberg GJ: Prolymphocytic leukaemia. Br
J Haematol. 27:7–23. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kjeldsberg CR and Marty J: Prolymphocytic
transformation of chronic lymphocytic leukemia. Cancer.
48:2447–2457. 1981. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
DiGiuseppe JA and Borowitz MJ: Clinical
utility of flow cytometry in the chronic lymphoid leukemias. Semin
Oncol. 25:6–10. 1998.PubMed/NCBI
|
|
7
|
Armitage JO, Dick FR and Corder MP:
Diffuse histiocytic lymphoma complicating chronic lymphocytic
leukemia. Cancer. 41:422–427. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Parikh SA, Rabe KG, Call TG, Zent CS,
Habermann TM, Ding W, Leis JF, Schwager SM, Hanson CA, Macon WR, et
al: Diffuse large B-cell lymphoma (Richter syndrome) in patients
with chronic lymphocytic leukaemia (CLL): A cohort study of newly
diagnosed patients. Br J Haematol. 162:774–782. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rai KR, Sawitsky A, Cronkite EP, Chanana
AD, Levy RN and Pasternack BS: Clinical staging of chronic
lymphocytic leukemia. Blood. 46:219–234. 1975.PubMed/NCBI
|
|
10
|
Binet JL, Lepoprier M, Dighiero G, Charron
D, D'Athis P, Vaugier G, Beral HM, Natali JC, Raphael M, Nizet B,
et al: A clinical staging system for chronic lymphocytic leukemia:
Prognostic significance. Cancer. 40:855–864. 1977. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Van Bockstaele F, Verhasselt B and
Philippé J: Prognostic markers in chronic lymphocytic leukemia: A
comprehensive review. Blood Rev. 23:25–47. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Villamor N, Conde L, Martínez-Trillos A,
Cazorla M, Navarro A, Beà S, López C, Colomer D, Pinyol M, Aymerich
M, et al: NOTCH1 mutations identify a genetic subgroup of chronic
lymphocytic leukemia patients with high risk of transformation and
poor outcome. Leukemia. 27:1100–1106. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jeromin S, Weissmann S, Haferlach C,
Dicker F, Bayer K, Grossmann V, Alpermann T, Roller A, Kohlmann A,
Haferlach T, et al: SF3B1 mutations correlated to cytogenetics and
mutations in NOTCH1, FBXW7, MYD88, XPO1 and TP53 in 1160 untreated
CLL patients. Leukemia. 28:108–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Puiggros A, Puigdecanet E, Salido M,
Ferrer A, Abella E, Gimeno E, Nonell L, Herranz MJ, Galván AB,
Rodríguez-Rivera M, et al: Genomic arrays in chronic lymphocytic
leukemia routine clinical practice: Are we ready to substitute
conventional cytogenetics and fluorescence in situ hybridization
techniques? Leuk Lymphoma. 54:986–995. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matutes E, Owusu-Ankomah K, Morilla R,
Marco Garcia J, Houlihan A, Que TH and Catovsky D: The
immunological profile of B-cell disorders and proposal of a scoring
system for the diagnosis of CLL. Leukemia. 8:1640–1645.
1994.PubMed/NCBI
|
|
16
|
Cheson BD, Bennett JM, Grever M, Kay N,
Keating MJ, O'Brien S and Rai KR: National Cancer
Institute-sponsored Working Group guidelines for chronic
lymphocytic leukemia: Revised guidelines for diagnosis and
treatment. Blood. 87:4990–4997. 1996.PubMed/NCBI
|
|
17
|
Hallek M, Cheson BD, Catovsky D,
Caligaris-Cappio F, Dighiero G, Döhner H, Hillmen P, Keating MJ,
Montserrat E, Rai KR, et al: International Workshop on Chronic
Lymphocytic Leukemia: Guidelines for the diagnosis and treatment of
chronic lymphocytic leukemia: A report from the International
Workshop on Chronic Lymphocytic Leukemia updating the National
Cancer Institute-Working Group 1996 guidelines. Blood.
111:5446–5456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shaffer LG, McGowan-Jordan J and Schmid M:
An International System for Human Cytogenetic Nomenclature. Karger
AG, Basel: 2013.
|
|
19
|
Smoley SA, Van Dyke DL, Kay NE, Heerema
NA, Dell' Aquila ML, Dal Cin P, Koduru P, Aviram A, Rassenti L,
Byrd JC, et al: Standardization of fluorescence in situ
hybridization studies on chronic lymphocytic leukemia (CLL) blood
and marrow cells by the CLL Research Consortium. Cancer Genet
Cytogenet. 203:141–148. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
De Braekeleer M, Le Bris MJ, Basinko A,
Morel F and Douet-Guilbert N: Incidence of chromosomal anomalies
detected by interphase fluorescent in situ hybridization in chronic
lymphoid leukemia. Int J Hematol Oncol. 4:133–141. 2015. View Article : Google Scholar
|
|
21
|
De Braekeleer M, De Braekeleer E and
Douet-Guilbert N: Geographic/ethnic variability of chromosomal and
molecular abnormalities in leukemia. Expert Rev Anticancer Ther.
15:1093–1102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
De Braekeleer E, Douet-Guilbert N, Morel
F, Le Bris MJ, Férec C and De Braekeleer M: RUNX1 translocations
and fusion genes in malignant hemopathies. Future Oncol. 7:77–91.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
De Braekeleer E, Douet-Guilbert N, Morel
F, Le Bris MJ, Basinko A and De Braekeleer M: ETV6 fusion genes in
hematological malignancies: A review. Leuk Res. 36:945–961. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schmidt HH, Dyomin VG, Palanisamy N,
Itoyama T, Nanjangud G, Pirc-Danoewinata H, Haas OA and Chaganti
RS: Deregulation of the carbohydrate (chondroitin 4)
sulfotransferase 11 (CHST11) gene in a B-cell chronic lymphocytic
leukemia with a t(12;14)(q23;q32). Oncogene. 23:6991–6996. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Aamot HV, Bjørnslett M, Delabie J and Heim
S: t(14;22)(q32;q11) in non-Hodgkin lymphoma and myeloid leukaemia:
Molecular cytogenetic investigations. Br J Haematol. 130:845–851.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kominami R: Role of the transcription
factor Bcl11b in development and lymphomagenesis. Proc Jpn Acad Ser
B Phys Biol Sci. 88:72–87. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Obata M, Kominami R and Mishima Y: BCL11B
tumor suppressor inhibits HDM2 expression in a p53-dependent
manner. Cell Signal. 24:1047–1052. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dyer MJ, Zani VJ, Lu WZ, O'Byrne A, Mould
S, Chapman R, Heward JM, Kayano H, Jadayel D, Matutes E, et al:
BCL2 translocations in leukemias of mature B cells. Blood.
83:3682–3688. 1994.PubMed/NCBI
|
|
29
|
Lin P, Jetly R, Lennon PA, Abruzzo LV,
Prajapati S and Medeiros LJ: Translocation (18;22)(q21;q11) in
B-cell lymphomas: A report of 4 cases and review of the literature.
Hum Pathol. 39:1664–1672. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Baseggio L, Geay MO, Gazzo S, Berger F,
Traverse-Glehen A, Ffrench M, Hayette S, Callet-Bauchu E, Verney A,
Morel D, et al: In non-follicular lymphoproliferative disorders,
IGH/BCL2-fusion is not restricted to chronic lymphocytic leukaemia.
Br J Haematol. 158:489–498. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Tang G, Banks HE, Sargent RL, Medeiros LJ
and Abruzzo LV: Chronic lymphocytic leukemia with
t(14;18)(q32;q21). Hum Pathol. 44:598–605. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Put N, Meeus P, Chatelain B, Rack K,
Boeckx N, Nollet F, Graux C, Van Den Neste E, Janssens A, Madoe V,
et al: Translocation t(14;18) is not associated with inferior
outcome in chronic lymphocytic leukemia. Leukemia. 23:1201–1204.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sen F, Lai R and Albitar M: Chronic
lymphocytic leukemia with t(14;18) and trisomy 12. Arch Pathol Lab
Med. 126:1543–1546. 2002.PubMed/NCBI
|
|
34
|
Matutes E, Oscier D, Garcia-Marco J, Ellis
J, Copplestone A, Gillingham R, Hamblin T, Lens D, Swansbury GJ and
Catovsky D: Trisomy 12 defines a group of CLL with atypical
morphology: Correlation between cytogenetic, clinical and
laboratory features in 544 patients. Br J Haematol. 92:382–388.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cleary ML, Smith SD and Sklar J: Cloning
and structural analysis of cDNAs for bcl-2 and a hybrid
bcl-2/immunoglobulin transcript resulting from the t(14;18)
translocation. Cell. 47:19–28. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hua C, Zorn S, Jensen JP, Coupland RW, Ko
HS, Wright JJ and Bakhshi A: Consequences of the t(14;18)
chromosomal translocation in follicular lymphoma: Deregulated
expression of a chimeric and mutated BCL-2 gene. Oncogene Res.
2:263–275. 1988.PubMed/NCBI
|
|
37
|
Tsujimoto Y and Croce CM: Analysis of the
structure, transcripts, and protein products of bcl-2, the gene
involved in human follicular lymphoma. Proc Natl Acad Sci USA.
83:5214–5218. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Oltvai ZN, Milliman CL and Korsmeyer SJ:
Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that
accelerates programmed cell death. Cell. 74:609–619. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Martín-Subero JI, Ibbotson R, Klapper W,
Michaux L, Callet-Bauchu E, Berger F, Calasanz MJ, De Wolf-Peeters
C, Dyer MJ, Felman P, et al: A comprehensive genetic and
histopathologic analysis identifies two subgroups of B-cell
malignancies carrying a t(14;19)(q32;q13) or variant
BCL3-translocation. Leukemia. 21:1532–1544. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Huh YO, Abruzzo LV, Rassidakis GZ,
Parry-Jones N, Schlette E, Brito-Bapabulle V, Matutes E,
Wotherspoon A, Keating MJ, Medeiros LJ, et al: The
t(14;19)(q32;q13)-positive small B-cell leukaemia: A
clinicopathologic and cytogenetic study of seven cases. Br J
Haematol. 136:220–228. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Huh YO, Schweighofer CD, Ketterling RP,
Knudson RA, Vega F, Kim JE, Luthra R, Keating MJ, Medeiros LJ and
Abruzzo LV: Chronic lymphocytic leukemia with t(14;19)(q32;q13) is
characterized by atypical morphologic and immunophenotypic features
and distinctive genetic features. Am J Clin Pathol. 135:686–696.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Michaux L, Mecucci C, Stul M, Wlodarska I,
Hernandez JM, Meeus P, Michaux JL, Scheiff JM, Noël H, Louwagie A,
et al: BCL3 rearrangement and t(14;19)(q32;q13) in
lymphoproliferative disorders. Genes Chromosomes Cancer. 15:38–47.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Michaux L, Dierlamm J, Wlodarska I, Bours
V, Van den Berghe H and Hagemeijer A: t(14;19)/BCL3 rearrangements
in lymphoproliferative disorders: A review of 23 cases. Cancer
Genet Cytogenet. 94:36–43. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
McKeithan TW, Takimoto GS, Ohno H,
Bjorling VS, Morgan R, Hecht BK, Dubé I, Sandberg AA and Rowley JD:
BCL3 rearrangements and t(14;19) in chronic lymphocytic leukemia
and other B-cell malignancies: A molecular and cytogenetic study.
Genes Chromosomes Cancer. 20:64–72. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Chapiro E, Radford-Weiss I, Bastard C,
Luquet I, Lefebvre C, Callet-Bauchu E, Leroux D, Talmant P,
Mozziconacci MJ, Mugneret F, et al: The most frequent
t(14;19)(q32;q13)-positive B-cell malignancy corresponds to an
aggressive subgroup of atypical chronic lymphocytic leukemia.
Leukemia. 22:2123–2127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Schweighofer CD, Huh YO, Luthra R, Sargent
RL, Ketterling RP, Knudson RA, Barron LL, Medeiros LJ, Keating MJ
and Abruzzo LV: The B cell antigen receptor in atypical chronic
lymphocytic leukemia with t(14;19)(q32;q13) demonstrates remarkable
stereotypy. Int J Cancer. 128:2759–2764. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shin SY, Park CJ, Lee KH, Huh J, Chi HS
and Seo EJ: An illustrative case of t(14;19)/BCL3 rearrangement as
a karyotypic evolution of chronic lymphocytic leukemia. Ann
Hematol. 92:1717–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Murray F, Darzentas N, Hadzidimitriou A,
Tobin G, Boudjogra M, Scielzo C, Laoutaris N, Karlsson K,
Baran-Marzsak F, Tsaftaris A, et al: Stereotyped patterns of
somatic hypermutation in subsets of patients with chronic
lymphocytic leukemia: Implications for the role of antigen
selection in leukemogenesis. Blood. 111:1524–1533. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
McKeithan TW, Ohno H and Diaz MO:
Identification of a transcriptional unit adjacent to the breakpoint
in the 14;19 translocation of chronic lymphocytic leukemia. Genes
Chromosomes Cancer. 1:247–255. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ohno H, Takimoto G and McKeithan TW: The
candidate proto-oncogene bcl-3 is related to genes implicated in
cell lineage determination and cell cycle control. Cell.
60:991–997. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Bours V, Franzoso G, Azarenko V, Park S,
Kanno T, Brown K and Siebenlist U: The oncoprotein Bcl-3 directly
transactivates through kappa B motifs via association with
DNA-binding p50B homodimers. Cell. 72:729–739. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Dechend R, Hirano F, Lehmann K, Heissmeyer
V, Ansieau S, Wulczyn FG, Scheidereit C and Leutz A: The Bcl-3
oncoprotein acts as a bridging factor between NF-kappaB/Rel and
nuclear co-regulators. Oncogene. 18:3316–3323. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Orlowski RZ and Baldwin AS Jr: NF-kappaB
as a therapeutic target in cancer. Trends Mol Med. 8:385–389. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Baldwin AS: Control of oncogenesis and
cancer therapy resistance by the transcription factor NF-kappaB. J
Clin Invest. 107:241–246. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kashatus D, Cogswell P and Baldwin AS:
Expression of the Bcl-3 proto-oncogene suppresses p53 activation.
Genes Dev. 20:225–235. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Huret JL: t(11;14)(q13;q32). Atlas Genet
Cytogenet Oncol Haematol. 2:129–131. 1998.
|
|
57
|
Cuneo A, de Angeli C, Roberti MG, Piva N,
Bigoni R, Gandini D, Rigolin GM, Moretti S, Cavazzini P, del Senno
L, et al: Richter's syndrome in a case of atypical chronic
lymphocytic leukaemia with the t(11;14)(q13;q32): Role for a p53
exon 7 gene mutation. Br J Haematol. 92:375–381. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Späth-Schwalbe E, Flath B, Kaufmann O,
Thiel G, Brinckmann R, Dietel M and Possinger K: An unusual case of
leukemic non-Hodgkin's lymphoma with blastic transformation. Ann
Hematol. 79:217–221. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Cuneo A, Balboni M, Piva N, Rigolin GM,
Roberti MG, Mejak C, Moretti S, Bigoni R, Balsamo R, Cavazzini P,
et al: Atypical chronic lymphocytic leukaemia with
t(11;14)(q13;q32): Karyotype evolution and prolymphocytic
transformation. Br J Haematol. 90:409–416. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
De Angeli C, Gandini D, Cuneo A, Moretti
S, Bigoni R, Roberti MG, Bardi A, Castoldi GL and del Senno L:
BCL-1 rearrangements and p53 mutations in atypical chronic
lymphocytic leukemia with t(11;14)(q13;q32). Haematologica.
85:913–921. 2000.PubMed/NCBI
|
|
61
|
Cuneo A, Bigoni R, Negrini M, Bullrich F,
Veronese ML, Roberti MG, Bardi A, Rigolin GM, Cavazzini P, Croce
CM, et al: Cytogenetic and interphase cytogenetic characterization
of atypical chronic lymphocytic leukemia carrying BCL1
translocation. Cancer Res. 57:1144–1150. 1997.PubMed/NCBI
|
|
62
|
Komatsu H, Yoshida K, Seto M, Iida S,
Aikawa T, Ueda R and Mikuni C: Overexpression of PRAD1 in a mantle
zone lymphoma patient with a t(11;22)(q13;q11) translocation. Br J
Haematol. 85:427–429. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Nishida Y, Takeuchi K, Tsuda K, Ugai T,
Sugihara H, Yamakura M, Takeuchi M and Matsue K: Acquisition of
t(11;14) in a patient with chronic lymphocytic leukemia carrying
both t(14;19)(q32;q13.1) and +12. Eur J Haematol. 91:179–182. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Matutes E, Carrara P, Coignet L,
Brito-Babapulle V, Villamor N, Wotherspoon A and Catovsky D: FISH
analysis for BCL-1 rearrangements and trisomy 12 helps the
diagnosis of atypical B cell leukaemias. Leukemia. 13:1721–1726.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Rimokh R, Berger F, Bastard C, Klein B,
French M, Archimbaud E, Rouault JP, Lucia Santa B, Duret L,
Vuillaume M, et al: Rearrangement of CCND1 (BCL1/PRAD1) 3′
untranslated region in mantle-cell lymphomas and
t(11q13)-associated leukemias. Blood. 83:3689–3696. 1994.PubMed/NCBI
|
|
66
|
Sherr CJ: The Pezcoller lecture: Cancer
cell cycles revisited. Cancer Res. 60:3689–3695. 2000.PubMed/NCBI
|
|
67
|
Bates S, Bonetta L, MacAllan D, Parry D,
Holder A, Dickson C and Peters G: CDK6 (PLSTIRE) and CDK4 (PSK-J3)
are a distinct subset of the cyclin-dependent kinases that
associate with cyclin D1. Oncogene. 9:71–79. 1994.PubMed/NCBI
|
|
68
|
Reddy K, Satyadev R, Bouman D, Hibbard MK,
Lu G and Paolo R: Burkitt t(8;14)(q24;q32) and cryptic deletion in
a CLL patient: Report of a case and review of literature. Cancer
Genet Cytogenet. 166:12–21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Huh YO, Lin KI, Vega F, Schlette E, Yin
CC, Keating MJ, Luthra R, Medeiros LJ and Abruzzo LV: MYC
translocation in chronic lymphocytic leukaemia is associated with
increased prolymphocytes and a poor prognosis. Br J Haematol.
142:36–44. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Put N, Van Roosbroeck K, Konings P, Meeus
P, Brusselmans C, Rack K, Gervais C, Nguyen-Khac F, Chapiro E,
Radford-Weiss I, et al: BCGHo and the GFCH: Chronic lymphocytic
leukemia and prolymphocytic leukemia with MYC translocations: A
subgroup with an aggressive disease course. Ann Hematol.
91:863–873. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Asirvatham JR, Brody J, Vora R, Kolitz JE,
Fields SZ, Sreekantaiah C and Zhang X: Prognostic significance of
isolated t(8:14) in chronic lymphocytic leukemia. Leuk Lymphoma.
55:685–688. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Taub R, Kirsch I, Morton C, Lenoir G, Swan
D, Tronick S, Aaronson S and Leder P: Translocation of the c-myc
gene into the immunoglobulin heavy chain locus in human Burkitt
lymphoma and murine plasmacytoma cells. Proc Natl Acad Sci USA.
79:7837–7841. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Kelly K and Siebenlist U: The role of
c-myc in the proliferation of normal and neoplastic cells. J Clin
Immunol. 5:65–77. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Li Z, Van Calcar S, Qu C, Cavenee WK,
Zhang MQ and Ren B: A global transcriptional regulatory role for
c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA.
100:8164–8169. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Dang CV, Resar LM, Emison E, Kim S, Li Q,
Prescott JE, Wonsey D and Zeller K: Function of the c-Myc oncogenic
transcription factor. Exp Cell Res. 253:63–77. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Hoffman B and Liebermann DA: Apoptotic
signaling by c-MYC. Oncogene. 27:6462–6472. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Satterwhite E, Sonoki T, Willis TG, Harder
L, Nowak R, Arriola EL, Liu H, Price HP, Gesk S, Steinemann D, et
al: The BCL11 gene family: Involvement of BCL11A in lymphoid
malignancies. Blood. 98:3413–3420. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Yin CC, Lin KI, Ketterling RP, Knudson RA,
Medeiros LJ, Barron LL, Huh YO, Luthra R, Keating MJ and Abruzzo
LV: Chronic lymphocytic leukemia with t(2;14)(p16;q32) involves the
BCL11A and IgH genes and is associated with atypical morphologic
features and unmutated IgVH genes. Am J Clin Pathol. 131:663–670.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Küppers R, Sonoki T, Satterwhite E, Gesk
S, Harder L, Oscier DG, Tucker PW, Dyer MJS and Siebert R: Lack of
somatic hypermutation of IG V(H) genes in lymphoid malignancies
with t(2;14)(p13;q32) translocation involving the BCL11A gene.
Leukemia. 16:937–939. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Liu P, Keller JR, Ortiz M, Tessarollo L,
Rachel RA, Nakamura T, Jenkins NA and Copeland NG: Bcl11a is
essential for normal lymphoid development. Nat Immunol. 4:525–532.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Pulford K, Banham AH, Lyne L, Jones M,
Ippolito GC, Liu H, Tucker PW, Roncador G, Lucas E, Ashe S, et al:
The BCL11AXL transcription factor: Its distribution in normal and
malignant tissues and use as a marker for plasmacytoid dendritic
cells. Leukemia. 20:1439–1441. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Rouhigharabaei L, Ferreiro JF, Put N,
Michaux L, Tousseyn T, Lefebvre C, Gardiner A, De Kelver W,
Demuynck H, Verschuere J, et al: BMI1, the polycomb-group gene, is
recurrently targeted by genomic rearrangements in progressive
B-cell leukemia/lymphoma. Genes Chromosomes Cancer. 52:928–944.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Li Z, Cao R, Wang M, Myers MP, Zhang Y and
Xu RM: Structure of a Bmi-1-Ring1B polycomb group ubiquitin ligase
complex. J Biol Chem. 281:20643–20649. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Raaphorst FM, Otte AP and Meijer CJ:
Polycomb-group genes as regulators of mammalian lymphopoiesis.
Trends Immunol. 22:682–690. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Ringrose L and Paro R: Polycomb/Trithorax
response elements and epigenetic memory of cell identity.
Development. 134:223–232. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Sauvageau M and Sauvageau G: Polycomb
group proteins: Multi-faceted regulators of somatic stem cells and
cancer. Cell Stem Cell. 7:299–313. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Siddique HR and Saleem M: Role of BMI1, a
stem cell factor, in cancer recurrence and chemoresistance:
Preclinical and clinical evidences. Stem Cells. 30:372–378. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Silva J, García JM, Peña C, García V,
Domínguez G, Suárez D, Camacho FI, Espinosa R, Provencio M, España
P, et al: Implication of polycomb members Bmi-1, Mel-18, and Hpc-2
in the regulation of p16INK4a, p14ARF, h-TERT, and c-Myc expression
in primary breast carcinomas. Clin Cancer Res. 12:6929–6936. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Jacobs JJ, Scheijen B, Voncken JW, Kieboom
K, Berns A and van Lohuizen M: Bmi-1 collaborates with c-Myc in
tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF.
Genes Dev. 13:2678–2690. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Bacher U, Haferlach T, Schnittger S, Weiss
T, Burkhard O, Bechtel B, Kern W and Haferlach C: Detection of a
t(4;14)(p16;q32) in two cases of lymphoma showing both the
immunophenotype of chronic lymphocytic leukemia. Cancer Genet
Cytogenet. 200:170–174. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Cerny J, Yu H and Miron PM: Novel FGFR3
rearrangement t(4;22)(p16;q11.2) in a patient with chronic
lymphocytic leukemia/small lymphocytic lymphoma. Ann Hematol.
92:1433–1435. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Geller MD, Pei Y, Spurgeon SE, Durum C and
Leeborg NJ: Chronic lymphocytic leukemia with a FGFR3
translocation: case report and literature review of an uncommon
cytogenetic event. Cancer Genet. 207:340–343. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Kalff A and Spencer A: The t(4;14)
translocation and FGFR3 overexpression in multiple myeloma:
Prognostic implications and current clinical strategies. Blood
Cancer J. 2:e892012. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Chesi M, Nardini E, Lim RSC, Smith KD,
Kuehl WM and Bergsagel PL: The t(4;14) translocation in myeloma
dysregulates both FGFR3 and a novel gene, MMSET, resulting in
IgH/MMSET hybrid transcripts. Blood. 92:3025–3034. 1998.PubMed/NCBI
|
|
95
|
Chesi M, Nardini E, Brents LA, Schröck E,
Ried T, Kuehl WM and Bergsagel PL: Frequent translocation
t(4;14)(p16.3;q32.3) in multiple myeloma is associated with
increased expression and activating mutations of fibroblast growth
factor receptor 3. Nat Genet. 16:260–264. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lauring J, Abukhdeir AM, Konishi H, Garay
JP, Gustin JP, Wang Q, Arceci RJ, Matsui W and Park BH: The
multiple myeloma associated MMSET gene contributes to cellular
adhesion, clonogenic growth, and tumorigenicity. Blood.
111:856–864. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Martinez-Garcia E, Popovic R, Min DJ,
Sweet SM, Thomas PM, Zamdborg L, Heffner A, Will C, Lamy L, Staudt
LM, et al: The MMSET histone methyl transferase switches global
histone methylation and alters gene expression in t(4;14) multiple
myeloma cells. Blood. 117:211–220. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hart KC, Robertson SC and Donoghue DJ:
Identification of tyrosine residues in constitutively activated
fibroblast growth factor receptor 3 involved in mitogenesis, Stat
activation, and phosphatidylinositol 3-kinase activation. Mol Biol
Cell. 12:931–942. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
L'Hôte CG and Knowles MA: Cell responses
to FGFR3 signalling: Growth, differentiation and apoptosis. Exp
Cell Res. 304:417–431. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Vahdati M, Graafland H and Emberger JM:
Karyotype analysis of B-lymphocytes transformed by Epstein-Barr
virus in 21 patients with B cell chronic lymphocytic leukemia. Hum
Genet. 63:327–331. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Oscier DG, Gardiner A and Mould S:
Structural abnormalities of chromosome 7q in chronic
lymphoproliferative disorders. Cancer Genet Cytogenet. 92:24–27.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Fink SR, Smoley SA, Stockero KJ,
Paternoster SF, Thorland EC, Van Dyke DL, Shanafelt TD, Zent CS,
Call TG, Kay NE, et al: Loss of TP53 is due to rearrangements
involving chromosome region 17p10 approximately p12 in chronic
lymphocytic leukemia. Cancer Genet Cytogenet. 167:177–181. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Hayette S, Tigaud I, Callet-Bauchu E,
Ffrench M, Gazzo S, Wahbi K, Callanan M, Felman P, Dumontet C,
Magaud JP, et al: In B-cell chronic lymphocytic leukemias, 7q21
translocations lead to overexpression of the CDK6 gene. Blood.
102:1549–1550. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Douet-Guilbert N, Tous C, Le Flahec G,
Bovo C, Le Bris MJ, Basinko A, Morel F and De Braekeleer M:
Translocation t(2;7)(p11;q21) associated with the CDK6/IGK
rearrangement is a rare but recurrent abnormality in B-cell
lymphoproliferative malignancies. Cancer Genet. 207:83–86. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Corcoran MM, Mould SJ, Orchard JA,
Ibbotson RE, Chapman RM, Boright AP, Platt C, Tsui LC, Scherer SW
and Oscier DG: Dysregulation of cyclin dependent kinase 6
expression in splenic marginal zone lymphoma through chromosome 7q
translocations. Oncogene. 18:6271–6277. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Brito-Babapulle V, Gruszka-Westwood AM,
Platt G, Andersen CL, Elnenaei MO, Matutes E, Wotherspoon AC,
Weston-Smith SG and Catovsky D: Translocation t(2;7)(p12;q21-22)
with dysregulation of the CDK6 gene mapping to 7q21-22 in a
non-Hodgkin's lymphoma with leukemia. Haematologica. 87:357–362.
2002.PubMed/NCBI
|
|
107
|
Ruas M, Gregory F, Jones R, Poolman R,
Starborg M, Rowe J, Brookes S and Peters G: CDK4 and CDK6 delay
senescence by kinase-dependent and p16INK4a-independent mechanisms.
Mol Cell Biol. 27:4273–4282. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Handschick K, Beuerlein K, Jurida L,
Bartkuhn M, Müller H, Soelch J, Weber A, Dittrich-Breiholz O,
Schneider H, Scharfe M, et al: Cyclin-dependent kinase 6 is a
chromatin-bound cofactor for NF-κB-dependent gene expression. Mol
Cell. 53:193–208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Grossel MJ and Hinds PW: From cell cycle
to differentiation: An expanding role for cdk6. Cell Cycle.
5:266–270. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Matushansky I, Radparvar F and Skoultchi
AI: CDK6 blocks differentiation: Coupling cell proliferation to the
block to differentiation in leukemic cells. Oncogene. 22:4143–4149.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Offit K, Parsa NZ, Filippa D, Jhanwar SC
and Chaganti RS: t(9;14)(p13;q32) denotes a subset of low-grade
non-Hodgkin's lymphoma with plasmacytoid differentiation. Blood.
80:2594–2599. 1992.PubMed/NCBI
|
|
112
|
Finn WG, Kay NE, Kroft SH, Church S and
Peterson LC: Secondary abnormalities of chromosome 6q in B-cell
chronic lymphocytic leukemia: A sequential study of karyotypic
instability in 51 patients. Am J Hematol. 59:223–229. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Dicker F, Schnittger S, Haferlach T, Kern
W and Schoch C: Immunostimulatory oligonucleotide-induced metaphase
cytogenetics detect chromosomal aberrations in 80% of CLL patients:
A study of 132 CLL cases with correlation to FISH, IgVH status, and
CD38 expression. Blood. 108:3152–3160. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Dascalescu CM, Péoc'h M, Callanan M, Jacob
MC, Sotto MF, Gressin R, Sotto JJ and Leroux D: Deletion 7q in
B-cell low-grade lymphoid malignancies: A cytogenetic/fluorescence
in situ hybridization and immunopathologic study. Cancer Genet
Cytogenet. 109:21–28. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Busslinger M, Klix N, Pfeffer P, Graninger
PG and Kozmik Z: Deregulation of PAX-5 by translocation of the Emu
enhancer of the IgH locus adjacent to two alternative PAX-5
promoters in a diffuse large-cell lymphoma. Proc Natl Acad Sci USA.
93:6129–6134. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Iida S, Rao PH, Nallasivam P, Hibshoosh H,
Butler M, Louie DC, Dyomin V, Ohno H, Chaganti RSK and Dalla-Favera
R: The t(9;14)(p13;q32) chromosomal translocation associated with
lymphoplasmacytoid lymphoma involves the PAX-5 gene. Blood.
88:4110–4117. 1996.PubMed/NCBI
|
|
117
|
Ohno H, Ueda C and Akasaka T: The
t(9;14)(p13;q32) translocation in B-cell non-Hodgkin's lymphoma.
Leuk Lymphoma. 36:435–445. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Barberis A, Widenhorn K, Vitelli L and
Busslinger M: A novel B-cell lineage-specific transcription factor
present at early but not late stages of differentiation. Genes Dev.
4:849–859. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Eberhard D, Jiménez G, Heavey B and
Busslinger M: Transcriptional repression by Pax5 (BSAP) through
interaction with corepressors of the Groucho family. EMBO J.
19:2292–2303. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Sonoki T, Harder L, Horsman DE, Karran L,
Taniguchi I, Willis TG, Gesk S, Steinemann D, Zucca E,
Schlegelberger B, et al: Cyclin D3 is a target gene of
t(6;14)(p21.1;q32.3) of mature B-cell malignancies. Blood.
98:2837–2844. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Wlodarska I, Dierickx D, Vanhentenrijk V,
Van Roosbroeck K, Pospísilová H, Minnei F, Verhoef G, Thomas J,
Vandenberghe P and De Wolf-Peeters C: Translocations targeting
CCND2, CCND3, and MYCN do occur in t(11;14)-negative mantle cell
lymphomas. Blood. 111:5683–5690. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Cavazzini F, Hernandez JA, Gozzetti A,
Rossi Russo A, De Angeli C, Tiseo R, Bardi A, Tammiso E, Crupi R,
Lenoci MP, et al: Chromosome 14q32 translocations involving the
immunoglobulin heavy chain locus in chronic lymphocytic leukaemia
identify a disease subset with poor prognosis. Br J Haematol.
142:529–537. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Shaughnessy J Jr, Gabrea A, Qi Y, Brents
L, Zhan F, Tian E, Sawyer J, Barlogie B, Bergsagel PL and Kuehl M:
Cyclin D3 at 6p21 is dysregulated by recurrent chromosomal
translocations to immunoglobulin loci in multiple myeloma. Blood.
98:217–223. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Motokura T, Keyomarsi K, Kronenberg HM and
Arnold A: Cloning and characterization of human cyclin D3, a cDNA
closely related in sequence to the PRAD1/cyclin D1 proto-oncogene.
J Biol Chem. 267:20412–20415. 1992.PubMed/NCBI
|
|
125
|
Gumina MR, Xu C and Chiles TC: Cyclin D3
is dispensable for human diffuse large B-cell lymphoma survival and
growth: Evidence for redundancy with cyclin E. Cell Cycle.
9:820–828. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Decker T, Schneller F, Hipp S, Miething C,
Jahn T, Duyster J and Peschel C: Cell cycle progression of chronic
lymphocytic leukemia cells is controlled by cyclin D2, cyclin D3,
cyclin-dependent kinase (cdk) 4 and the cdk inhibitor p27.
Leukemia. 16:327–334. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Willis TG and Dyer MJS: The role of
immunoglobulin translocations in the pathogenesis of B-cell
malignancies. Blood. 96:808–822. 2000.PubMed/NCBI
|
|
128
|
Küppers R and Dalla-Favera R: Mechanisms
of chromosomal translocations in B cell lymphomas. Oncogene.
20:5580–5594. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Nowakowski GS, Dewald GW, Hoyer JD,
Paternoster SF, Stockero KJ, Fink SR, Smoley SA, Remstein ED,
Phyliky RL, Call TG, et al: Interphase fluorescence in situ
hybridization with an IGH probe is important in the evaluation of
patients with a clinical diagnosis of chronic lymphocytic
leukaemia. Br J Haematol. 130:36–42. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Cavazzini F, Rizzotto L, Sofritti O,
Daghia G, Cibien F, Martinelli S, Ciccone M, Saccenti E, Dabusti M,
Elkareem AA, et al: Clonal evolution including 14q32/IGH
translocations in chronic lymphocytic leukemia: Analysis of
clinicobiologic correlations in 105 patients. Leuk Lymphoma.
53:83–88. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Davids MS, Vartanov A, Werner L, Neuberg
D, Dal Cin P and Brown JR: Controversial fluorescence in situ
hybridization cytogenetic abnormalities in chronic lymphocytic
leukaemia: New insights from a large cohort. Br J Haematol.
170:694–703. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Gerrie AS, Bruyere H, Chan MJ, Dalal CB,
Ramadan KM, Huang SJ, Toze CL and Gillan TL: Immunoglobulin heavy
chain (IGH@) translocations negatively impact treatment-free
survival for chronic lymphocytic leukemia patients who have an
isolated deletion 13q abnormality. Cancer Genet. 205:523–527. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Gerrie AS, Huang SJ, Bruyere H, Dalal C,
Hrynchak M, Karsan A, Ramadan KM, Smith AC, Tyson C, Toze CL, et
al: Population-based characterization of the genetic landscape of
chronic lymphocytic leukemia patients referred for cytogenetic
testing in British Columbia, Canada: The role of provincial
laboratory standardization. Cancer Genet. 207:316–325. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Lu G, Kong Y and Yue C: Genetic and
immunophenotypic profile of IGH@ rearrangement detected by
fluorescence in situ hybridization in 149 cases of B-cell chronic
lymphocytic leukemia. Cancer Genet Cytogenet. 196:56–63. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Shanafelt TD, Witzig TE, Fink SR, Jenkins
RB, Paternoster SF, Smoley SA, Stockero KJ, Nast DM, Flynn HC,
Tschumper RC, et al: Prospective evaluation of clonal evolution
during long-term follow-up of patients with untreated early-stage
chronic lymphocytic leukemia. J Clin Oncol. 24:4634–4641. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Aoun P, Blair HE, Smith LM, Dave BJ, Lynch
J, Weisenburger DD, Pavletic SZ and Sanger WG: Fluorescence in situ
hybridization detection of cytogenetic abnormalities in B-cell
chronic lymphocytic leukemia/small lymphocytic lymphoma. Leuk
Lymphoma. 45:1595–1603. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Nelson BP, Gupta R, Dewald GW, Paternoster
SF, Rosen ST and Peterson LC: Chronic lymphocytic leukemia FISH
panel: Impact on diagnosis. Am J Clin Pathol. 128:323–332. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Flanagan MB, Sathanoori M, Surti U, Soma L
and Swerdlow SH: Cytogenetic abnormalities detected by fluorescence
in situ hybridization on paraffin-embedded chronic lymphocytic
leukemia/small lymphocytic lymphoma lymphoid tissue biopsy
specimens. Am J Clin Pathol. 130:620–627. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Jenderny J, Goldmann C, Thede R, Ebrecht M
and Korioth F: Detection of clonal aberrations by cytogenetic
analysis after different culture methods and by FISH in 129
patients with chronic lymphocytic leukemia. Cytogenet Genome Res.
144:163–168. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Haferlach C, Dicker F, Schnittger S, Kern
W and Haferlach T: Comprehensive genetic characterization of CLL: A
study on 506 cases analysed with chromosome banding analysis,
interphase FISH, IgV(H) status and immunophenotyping. Leukemia.
21:2442–2451. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Alhourani E, Rincic M, Othman MA, Pohle B,
Schlie C, Glaser A and Liehr T: Comprehensive chronic lymphocytic
leukemia diagnostics by combined multiplex ligation dependent probe
amplification (MLPA) and interphase fluorescence in situ
hybridization (iFISH). Mol Cytogenet. 7:792014. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Döhner H, Stilgenbauer S, Benner A,
Leupolt E, Kröber A, Bullinger L, Döhner K, Bentz M and Lichter P:
Genomic aberrations and survival in chronic lymphocytic leukemia. N
Engl J Med. 343:1910–1916. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Berkova A, Pavlistova L, Babicka L,
Houskova L, Tajtlova J, Balazi P, Cmunt E, Schwarz J, Karban J,
Trneny M, et al: Combined molecular biological and molecular
cytogenetic analysis of genomic changes in 146 patients with B-cell
chronic lymphocytic leukemia. Neoplasma. 55:400–408.
2008.PubMed/NCBI
|
|
144
|
Amare PS, Gadage V, Jain H, Nikalje S,
Manju S, Mittal N, Gujral S and Nair R: Clinico-pathological impact
of cytogenetic subgroups in B-cell chronic lymphocytic leukemia:
Experience from India. Indian J Cancer. 50:261–267. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Yoon JH, Kim Y, Yahng SA, Shin SH, Lee SE,
Cho BS, Eom KS, Kim YJ, Lee S, Kim HJ, et al: Validation of Western
common recurrent chromosomal aberrations in Korean chronic
lymphocytic leukaemia patients with very low incidence. Hematol
Oncol. 32:169–177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Xu W, Li JY, Pan JL, Qiu HR, Shen YF, Li
L, Wu YF and Xue YQ: Interphase fluorescence in situ hybridization
detection of cytogenetic abnormalities in B-cell chronic
lymphocytic leukemia. Int J Hematol. 85:430–436. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Qiu HX, Xu W, Cao XS, Zhou M, Shen YF, Xu
YL, Sun XM, Liu Q, Wang R, Qiu HR, et al: Cytogenetic
characterisation in Chinese patients with chronic lymphocytic
leukemia: A prospective, multicenter study on 143 cases analysed
with interphase fluorescence in situ hybridisation. Leuk Lymphoma.
49:1887–1892. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Wang DM, Miao KR, Fan L, Qiu HR, Fang C,
Zhu DX, Qiu HX, Xu W and Li JY: Intermediate prognosis of 6q
deletion in chronic lymphocytic leukemia. Leuk Lymphoma.
52:230–237. 2011. View Article : Google Scholar : PubMed/NCBI
|