Introduction
Lung cancer is the leading cause of cancer-related
mortality and is responsible for approximately 1.59 million deaths
annually worldwide (1). Non-small
cell lung carcinoma (NSCLC) accounts for more than 80% of lung
cancer cases, with adenocarcinoma (ADC) being the most common
histological subtype. Detection of activating somatic mutations in
the epidermal growth factor receptor (EGFR) gene in ADC
(2–5)
has revolutionized diagnosis and treatment and resulted in a shift
towards individual targeted therapy (2). Although several mutations can occur in
exons 18–21 of the tyrosine kinase domain of EGFR, approximately
90% are the 15 bp (E746-750) in-frame deletion in exon 19 or the
L858R point mutation in exon 21 (3,4,6). Mutational status is associated with a
clinical response to EGFR-tyrosine kinase inhibitor (TKI) treatment
(2–7). EGFR-TKIs are more efficacious than
carboplatin-paclitaxel as an initial treatment (8).
Direct DNA sequencing is the classical method used
to detect EGFR mutations; however, this method requires a
sufficient quantity of DNA. In addition, the high costs of
equipment and reagents, and the time-consuming procedure, hamper
its application in clinical practice. To overcome these issues,
various molecular tests for EGFR mutations have been developed,
such as the Scorpion amplified refractory mutation system and
polymerase chain reaction single-strand conformation polymorphism
(9,10). However, many patients in developing
countries do not undergo molecular testing due to the lack of
available equipment and trained personnel. Immunohistochemistry
(IHC) is an inexpensive and accurate method of identifying EGFR
mutations and is available in most pathology laboratories.
Antibodies specific for the exon 19 deletion E746-A750
(delE746-A750) and the L858R mutation have been developed to assess
tumor EGFR status (11–20). A meta-analysis recommended that IHC
but not molecular testing is suitable for detection of EGFR
mutations (21).
As approximately 70% of NSCLC patients are diagnosed
at an advanced stage and are thus ineligible for surgical resection
(22,23), therapeutic decision-making is
dependent on the availability of biopsy samples or cytology
specimens. Compared with biopsy samples, obtaining cytological
specimens is minimally invasive and simple. Several studies have
evaluated cytological specimens using immunocytochemistry (ICC)
with EGFR mutation-specific antibodies (18,24–26);
however, the results were inconsistent (sensitivity 43–100% and
specificity 74–100%) (18,24–26). Two
novel antibodies against EGFR-mutated proteins were developed
recently. SP111 is specific for delE746-A750 and SP125 for the
L858R mutation; both SP111 and SP125 show an efficacy similar to
that of the original clone (27–29). To
date, no study has assessed the utility of SP111 or SP125 for
cytological specimens. Here, we examined the specificity and
sensitivity of IHC and ICC using the SP111 and SP125 antibodies in
surgical and cytological samples from patients with lung
adenocarcinoma.
Patients and methods
Clinical samples and DNA analysis
This study involved 17 patients with pulmonary
adenocarcinoma from whom surgical and cytological samples were
obtained from January 2015 to March 2017 at Osaka University
Medical Hospital. Formalin-fixed paraffin-embedded (FFPE) tissue
sections (5 µm) were prepared. DNA extraction from FFPE samples was
performed according to the standard procedure of the Cobas DNA
Sample Preparation kit (Roche Molecular Systems, Inc., Alameda, CA,
USA). Briefly, the samples were incubated with a protease in
chaotropic lysis/binding buffer to release nucleic acids and
protect genomic DNA from degradation by DNase. The amount of
genomic DNA was spectrophotometrically determined (Nanodrop
ND-1000, Thermo Scientific, Wilmington, DE, USA) and adjusted to 2
ng/µl. DNA (150 ng) was obtained for the Cobas EGFR assay. Target
DNA was amplified and detected using the Cobas 4800 Analyzer (Roche
Molecular Systems, Inc.) according to the manufacturer's
instructions.
The study protocol was approved by the Research
Ethics Board of the Osaka University Research Committee and
conducted according to Institutional Review Board guidelines (cat.
no. 16293).
IHC and ICC of EGFR mutations
FFPE tissue sections (5 µm) were stained
immunohistochemically using the SP111 anti-EGFR delE746-A750 rabbit
monoclonal antibody and SP125 anti-EGFR L858R rabbit monoclonal
antibody (Ventana Medical Systems, Tucson, AZ, USA) and an
automatic staining system (Ventana BenchMark XT; Ventana Medical
Systems). Briefly, the sections were incubated with SP111 or SP125
for 16 min at 37°C, and immunoreactions were detected using the
Ultraview Universal DAB detection kit. The negative control lacked
a primary antibody. The slides were assessed by at least two
pathologists blinded to the EGFR mutation status. The IHC staining
was scored based on the staining intensity and percentage staining
area in the membrane and/or cytoplasm of tumor cells, as follows
(18): 0 no staining; 1+,
light-yellow staining with no obvious particulates or yellow
staining with obvious particulates in <10% of tumor cells; 2+,
yellow staining with obvious particulates in >10% of tumor cells
or brown staining with obvious particulates in <10% of tumor
cells; and 3+, brown staining with obvious particulates in >10%
of tumor cells. Scores of 2+ and 3+ were considered positive
(18).
A cell-transfer technique was used for ICC. A layer
of Malinol medium (Muto Pure Chemicals Co. Ltd., Tokyo, Japan) was
spread uniformly over the top of the cellular material. The slide
was placed into a 70°C oven for 72 h to harden the coating material
and then transferred to a water bath at 50°C for 15 min to soften
the Malinol medium. Using a blade, the liquid coverslip and
attached cells were slowly peeled off the slide. The peeled
membrane was sectioned into 3–5 pieces. Each section was
transferred to another slide, on which ICC was performed using a
similar method to that of IHC. Staining was evaluated as for IHC
(18).
Statistical analysis
Statistical analyses were performed using R version
3.2.2 (https://cran.r-project.org/bin/windows/base/).
Antibody performance was assessed by determining the sensitivity,
specificity, positive predictive value (PPV), and negative
predictive value (NPV). The agreement between IHC or ICC and
molecular testing was calculated using Cohen's kappa score. A κ
value of 0.81–1.0 was defined as nearly perfect agreement, 0.61–0.8
as substantial agreement, 0.41–0.60 as moderate agreement, 0.21 to
0.40 as fair agreement, and 0.00–0.20 as slight agreement.
Results
Patient characteristics
Of the 192 patients with pulmonary adenocarcinoma
who underwent surgical resection from January 2015 to March 2017 at
Osaka University Medical Hospital, cytological specimens were
obtained from 17 patients. The clinicopathological characteristics
of the patients are shown in Table
I. The cytologic specimens comprised exfoliative (bronchial
brush/wash/lavage) (n=9), transbronchial fine-needle aspiration
(FNA; n=7), and transthoracic FNA (n=1) specimens. The age of the
patients was 53–77 years (median, 67.2 years). Twelve and five of
the patients were male and female, respectively. Six cases were
stage IA, seven stage IB, two stage IIA, and two stage IIIA. All
the cases had previously undergone screening for EGFR mutations by
molecular testing. Exon 19 deletion (delE746-A750) was present in
three patients and the L858R mutation in exon 21 in seven patients;
the other patients had neither the L858R mutation nor delE746-A750
(Table I).
 | Table I.Patient characteristics and EGFR
mutations status in primary NSCLC samples and comparative analysis
of IHC and ICC. |
Table I.
Patient characteristics and EGFR
mutations status in primary NSCLC samples and comparative analysis
of IHC and ICC.
|
|
|
|
|
| IHC | ICC |
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Patient no. | Age, years | Sex | Stage | EGFR mutation
status | SP111 | SP125 | Sample type | SP111 | SP125 |
|---|
| 1 | 60 | F | IB | L858R | 0 | 3+ | BB | 0 | 3+ |
| 2 | 77 | M | IB | L858R | 0 | 2+ | FNA | 0 | 1+ |
| 3 | 53 | F | IIA | L858R | 0 | 2+ | FNA | 0 | 3+ |
| 4 | 71 | M | IIA | L858R | 0 | 3+ | BB | 1+ | 3+ |
| 5 | 59 | F | IA | L858R | 0 | 2+ | FNA | 0 | 2+ |
| 6 | 76 | M | IA | L858R | 0 | 2+ | FNA | 0 | 1+ |
| 7 | 61 | M | IB | L858R | 0 | 3+ | FNA | 1+ | 2+ |
| 8 | 72 | M | IA | Exon 19 del | 2+ | 0 | BB | 0 | 0 |
| 9 | 70 | F | IA | Exon 19 del | 3+ | 1+ | FNA | 0 | 0 |
| 10 | 76 | M | IB | Exon 19 del | 3+ | 0 | FNA | 2+ | 0 |
| 11 | 69 | M | IIIA | No mutation | 0 | 0 | BB | 0 | 0 |
| 12 | 72 | M | IIIA | No mutation | 0 | 0 | BB | 0 | 0 |
| 13 | 77 | M | IB | No mutation | 0 | 0 | FNA | 1+ | 1+ |
| 14 | 67 | M | IA | No mutation | 1+ | 0 | BB | 0 | 0 |
| 15 | 53 | M | IB | No mutation | 0 | 0 | BB | 0 | 1+ |
| 16 | 54 | F | IA | No mutation | 0 | 1+ | BB | 0 | 1+ |
| 17 | 75 | M | IB | No mutation | 0 | 1+ | BB | 0 | 1+ |
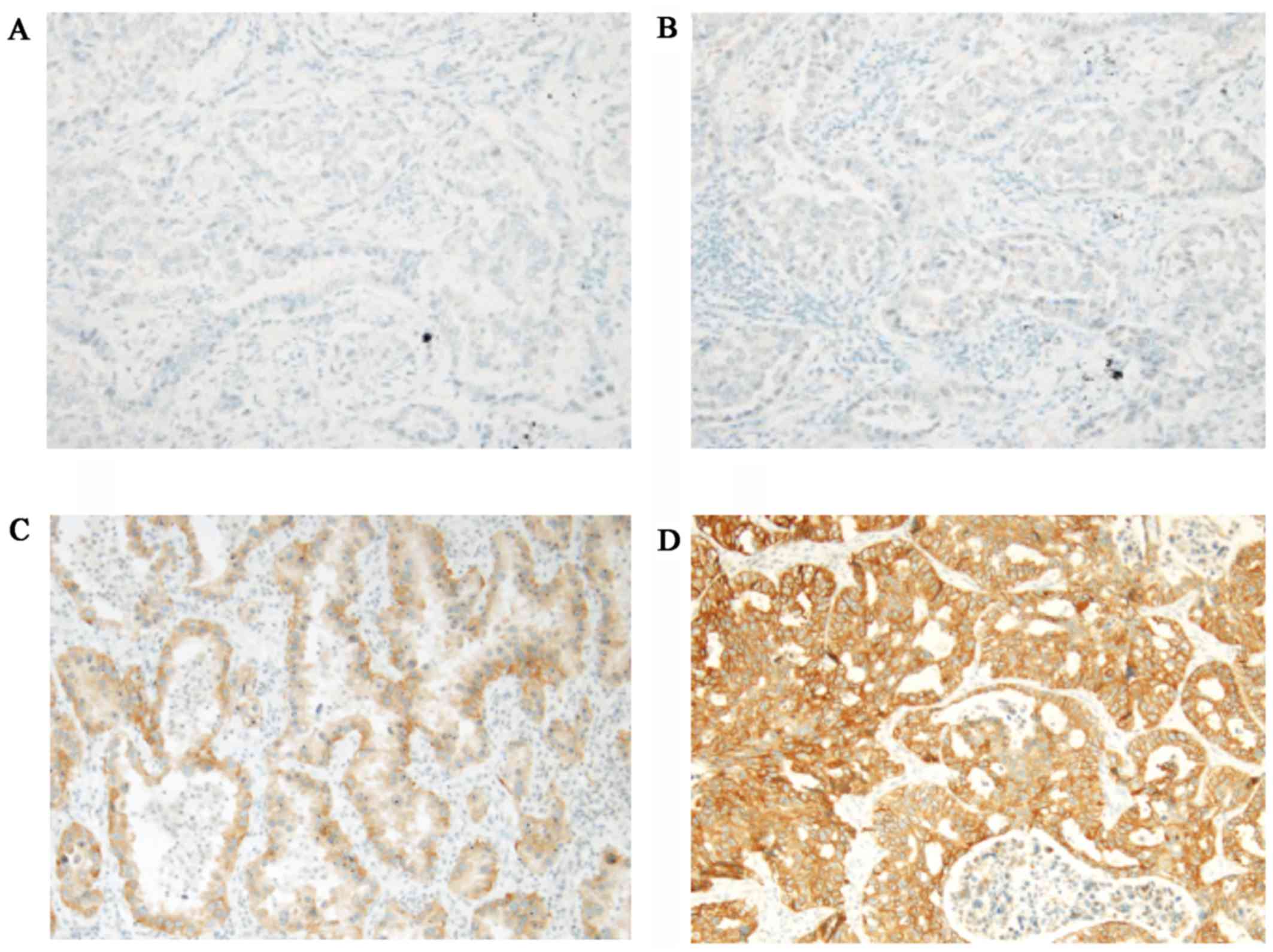
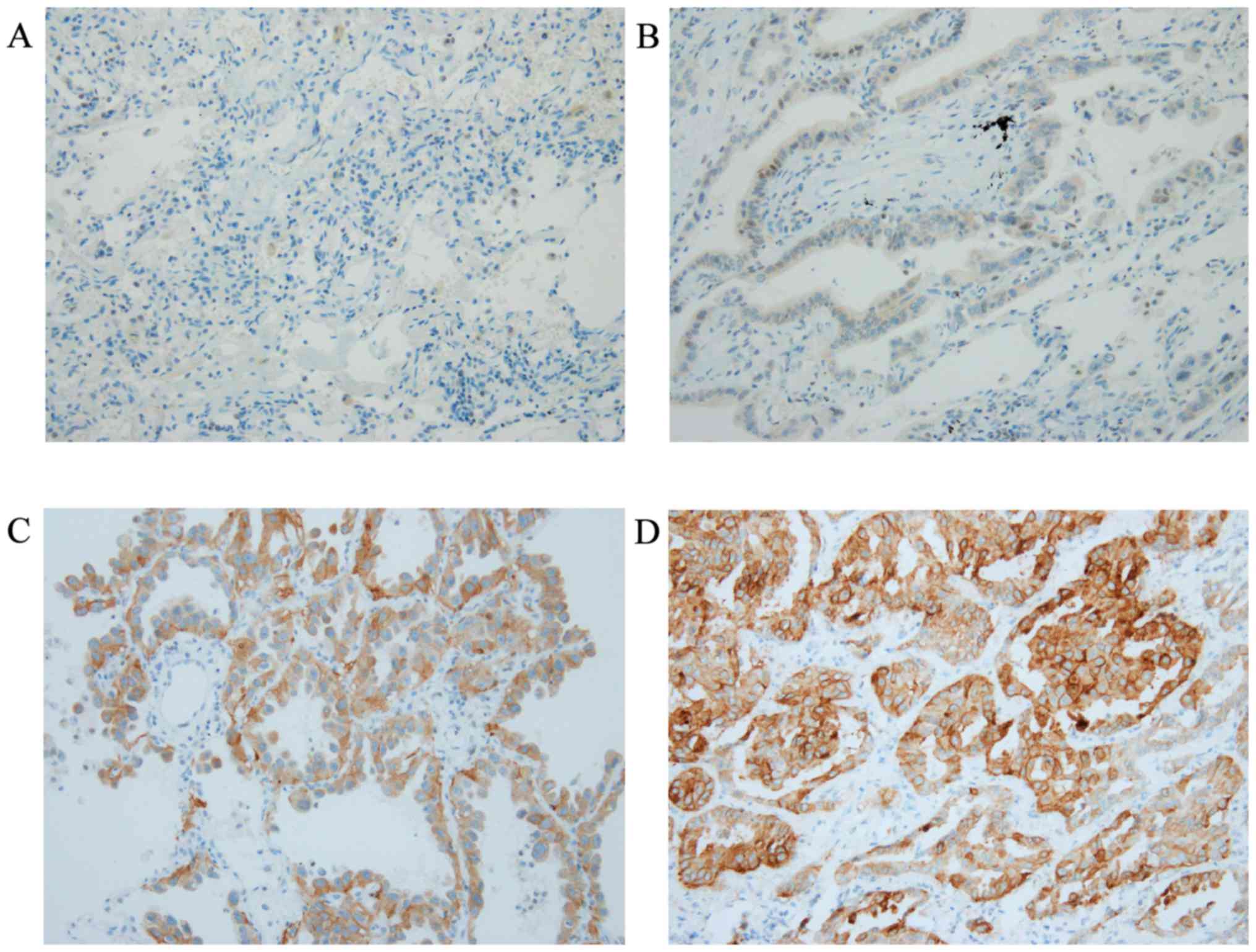
IHC using SP111 and SP125
IHC using SP111 and SP125 yielded scores of 0 to 3+
(Figs. 1 and 2). Using SP111, 13 of 17 cases showed a
score of 0, one a score of 1+, one a score of 2+, and two a score
of 3+ (Table I). Using SP125, 7 of
17 cases showed a score of 0, three a score of 1+, four a score of
2+, and three a score of 3+ (Table
I). The three and seven cases with a score of 2+ or 3+ using
SP111 and SP125, respectively, were evaluated as SP111- and
SP125-positive, respectively, as described previously (18) (Table
II).
 | Table II.Summary of IHC and molecular testing
in resection samples. |
Table II.
Summary of IHC and molecular testing
in resection samples.
| IHC | Exon19del (n=3)
(%) | L858R (n=7)
(%) | No mutation (n=7)
(%) |
|---|
| SP111-positive |
3 (100) | 0 (0) | 0 (0) |
| SP111-negative | 0 (0) |
7 (100) |
7 (100) |
| SP125-positive | 0 (0) |
7 (100) | 0 (0) |
| SP125-negative |
3 (100) | 0 (0) |
7 (100) |
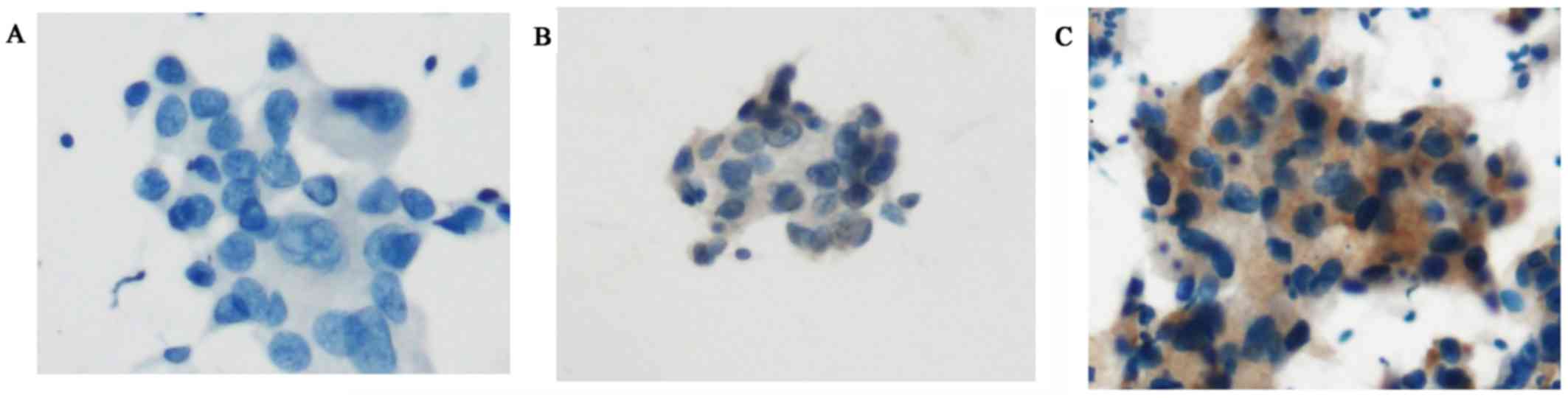
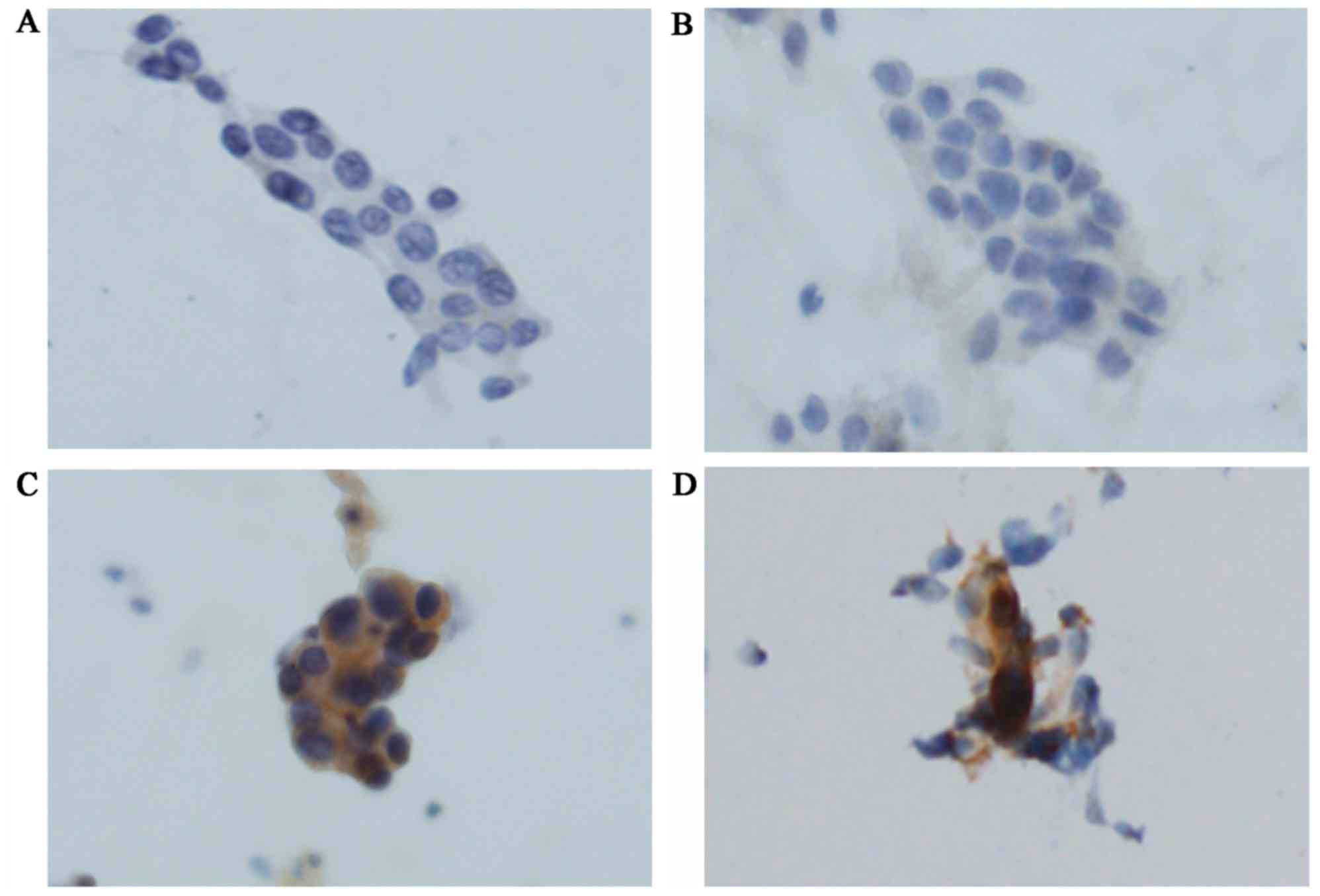
ICC using SP111 and SP125
ICC using SP111 and SP125 yielded four scores
(Figs. 3A-C and 4A-D). Using SP111, 13 of 17 cases showed a
score of 0, three a score of 1+, and one a score of 2+. Using
SP125, 6 of 17 cases showed a score of 0, six a score of 1+, two a
score of 2+, and three a score of 3+. The one case with a score of
2+ using SP111 and the five cases with a score of 2+ or 3+ using
SP125 were evaluated as SP111- and SP125-positive, respectively
(Table III).
 | Table III.Summary of ICC and molecular testing
in resection samples. |
Table III.
Summary of ICC and molecular testing
in resection samples.
| ICC | Exon19del (n=3)
(%) | L858R (n=7)
(%) | No mutation (n=7)
(%) |
|---|
| SP111-positive |
1 (33.3) | 0 (0) | 0 (0) |
| SP111-negative |
2 (66.7) |
7 (100) |
7 (100) |
| SP125-positive | 0 (0) |
5 (71.4) | 0 (0) |
| SP125-negative |
3 (100) |
2 (28.6) |
7 (100) |
EGFR mutations
The seven cases without EGFR mutations were negative
by IHC using SP111 and SP125. The three cases with delE746-A750
were positive using SP111 and negative using SP125. Similarly, the
seven cases with the L858R mutation were positive using SP125 and
negative using SP111. Therefore, the sensitivity and specificity of
both SP111 and SP125 were 100% (Table
II).
The seven cases without EGFR mutations were negative
by ICC using SP111 and SP125. Among the three cases with
delE746-A750, one was positive by ICC using SP111. Thus, the
sensitivity and specificity of SP111 were 33.3 and 100%,
respectively. Five of seven cases with the L858R mutation showed
positive staining for SP125; thus, the sensitivity and specificity
of SP125 were 71.4 and 100%, respectively (Table III).
Comparative analyses between IHC and
ICC
ICC scores of 2+ and 3+ were considered positive.
However, other studies used diverse cut-off values; e.g., Tsai
et al regarded a score of 1+, 2+, or 3+ as positive
(25). We analyzed the concordance
of molecular testing and ICC using various cut-off values by
calculating the PPV, NPV, and Cohen's kappa score. If a score of
≥1+ was considered positive, the sensitivity, specificity, PPV,
NPV, and the κ values were 100, 57.1, 76.9, 100%, and 0.611,
respectively for IHC and 100, 55.6, 66.7, 100%, and 0.239,
respectively, for ICC (Table
IV).
 | Table IV.Comparative analyses between IHC and
ICC in accordance with grading. |
Table IV.
Comparative analyses between IHC and
ICC in accordance with grading.
| Consideration
regarding positive result | Type | Sensitivity, % | Specificity, % | PPV, % | NPV, % | Cohen's Kappa
Score |
|---|
| Score ≥1 | IHC | 100 | 57.1 | 76.9 | 100 | 0.611 |
|
| ICC | 100 | 55.6 | 66.7 | 100 | 0.239 |
| Score ≥2 | IHC | 100 | 100 | 100 | 100 | 1 |
|
| ICC | 60.0 | 100 | 100 | 63.6 | 0.553 |
| Score ≥3 | IHC | 50.0 | 100 | 100 | 58.3 | 0.452 |
|
| ICC | 30.0 | 100 | 100 | 50.0 | 0.261 |
If a score of ≥2+ was considered positive, the
sensitivity, specificity, PPV, NPV, and the κ values were 100, 100,
100, 100%, and 1.0, respectively, for IHC and 60.0, 100, 100,
63.6%, and 0.553, respectively, for ICC (Table IV). If a score of ≥3+ was considered
positive, the abovementioned values were 50.0, 100, 100, 58.3%, and
0.452, respectively, for IHC and 30.0, 100, 100, 50.0%, and 0.261,
respectively, for ICC.
Discussion
Greater understanding of the molecular pathogenesis
of lung cancer has led to the development and application of
targeted therapeutic strategies. Activating somatic mutations in
EGFR are associated with a clinical response to TKIs (3–5). Thus,
molecular testing of EGFR mutations has become routine in clinical
practice (9,10). However, such tests are expensive and
technically difficult to perform in many laboratories. IHC is an
inexpensive and accurate method of identifying EGFR mutations and
is available in most pathology laboratories. Several authors
evaluated the potential of antibodies specific for the delE746-A750
and L858R mutations for screening for EGFR mutations using
surgically resected and biopsy samples (11–21).
Compared with biopsy samples, obtaining cytological specimens is
minimally invasive and simple. Several studies evaluated ICC using
EGFR mutation-specific antibodies (clone 43B2 and clone 6B6) in
cytological specimens (18,24–26). Two
novel antibodies specific for EGFR mutations were developed
recently: SP111, which is specific for delE746-A750, and SP125,
which is specific for the L858R mutation. To date, no report has
evaluated the use of SP111 or SP125 with cytological specimens.
Thus, we evaluated the potential of these novel EGFR
mutation-specific antibodies in ICC.
The present study included 17 cases with surgically
resected and cytological samples available. Seven cases were
positive for the L858R mutation and three cases for delE746-A750;
the remaining seven cases had neither mutation. In ICC, the
sensitivity and specificity of SP111 were 33.3 and 100%,
respectively. In ICC, the sensitivity and specificity of clone 6B6
for delE746-A750 samples were reportedly 66.7 and 83.3%, 100 and
94%, 88 and 96%, and 72.7 and 100%, respectively (18,24–26). The
sensitivity of SP111 was inferior to that of clone 6B6, but its
specificity was comparable or superior. The sensitivity and
specificity of SP125 were 71.4 and 100%, respectively. Previous
studies reported sensitivities and specificities for clone 43B2 of
42.9 and 50%, 100 and 100%, 71 and 86%, and 80 and 93.8%,
respectively (18,24–26).
Thus, the sensitivity of SP125 is comparable with that of clone
43B2, but its specificity is comparable with or superior to that of
clone 43B2.
The sensitivity and specificity of SP111 in ICC were
33.3 and 100%, respectively, and those of SP125 were 71.4 and 100%,
respectively; therefore, the combined sensitivity and specificity
were 60 and 100%, respectively (Table
IV). The combined sensitivity of clones 43B2 and 6B6 in IHC is
reportedly higher than that in ICC (79.7 vs. 50% and 85.2 vs.
66.7%, respectively) (18,26). These findings suggest that ICC using
antibodies to the L858R and delE746-A750 mutations exhibits low
sensitivity. Tumor cells tend to form clusters, which hampers the
evaluation of membrane-positive signals. This issue may be resolved
by disrupting the tumor cell clusters.
The meta-analysis by Chen et al recommended a
four-grade IHC scoring system (in which a score of 2+ or 3+ is
considered positive), not only to reduce differences among readers,
but also to enhance the diagnostic value of mutation-specific
antibodies (21). In this study, the
PPV, NPV, and Cohens' kappa value were highest when a score of ≥2+
was considered positive. Therefore, a cut-off score of 2+ may be
suitable for both IHC and ICC.
Based on the high specificity of ICC using SP111 and
SP125, a positive result may eliminate the need for confirmatory
molecular testing. Patients with positive cytological samples
should immediately start TKI therapy without verification by
molecular testing. Screening for EGFR mutations by ICC may
facilitate therapeutic decision-making, particularly in medical
centers unable to perform molecular testing.
Acknowledgements
The present study was supported by grants from the
Ministry of Education, Culture, Sports, Science and Technology,
Japan (T17K195550).
References
|
1
|
Fact Sheet WHO: 297, . https://www.who.inta. WHO Fact Sheet 297. November
8–2016
|
|
2
|
Travis WD, Rekhtman N, Riley GJ, Geisinger
KR, Asamura H, Brambilla E, Garg K, Hirsch FR, Noguchi M, Powell
CA, et al: Pathologic diagnosis of advanced lung cancer based on
small biopsies and cytology: A paradigm shift. J Thorac Oncol.
5:411–414. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Paez JG, Jänne PA, Lee JC, Tracy S,
Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et
al: EGFR mutations in lung cancer: Correlation with clinical
response to gefitinib therapy. Science. 304:1497–1500. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pao W, Miller V, Zakowski M, Doherty J,
Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, et al:
EGF receptor gene mutations are common in lung cancers from ‘never
smokers’ and are associated with sensitivity of tumors to gefitinib
and erlotinib. Proc Natl Acad Sci USA. 101:pp. 13306–13311. 2004,
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sharma SV, Bell DW, Settleman J and Haber
DA: Epidermal growth factor receptor mutations in lung cancer. Nat
Rev Cancer. 7:169–181. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mitsudomi T and Yatabe Y: Mutations of the
epidermal growth factor receptor gene and related genes as
determinants of epidermal growth factor receptor tyrosine kinase
inhibitors sensitivity in lung cancer. Cancer Sci. 98:1817–1824.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mok TS, Wu YL, Thongprasert S, Yang CH,
Chu DT, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, et
al: Gefitinib or carboplatin-paclitaxel in pulmonary
adenocarcinoma. N Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pao W and Ladanyi M: Epidermal growth
factor receptor mutation testing in lung cancer: Searching for the
ideal method. Clin Cancer Res. 13:4954–4955. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ellison G, Zhu G, Moulis A, Dearden S,
Speake G and McCormack R: EGFR mutation testing in lung cancer: A
review of available methods and their use for analysis of tumour
tissue and cytology samples. J Clin Pathol. 66:79–89. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu J, Kane S, Wu J, Benedettini E, Li D,
Reeves C, Innocenti G, Wetzel R, Crosby K, Becker A, et al:
Mutation-specific antibodies for the detection of EGFR mutations in
non-small-cell lung cancer. Clin Cancer Res. 15:3023–3028. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brevet M, Arcila M and Ladanyi M:
Assessment of EGFR mutation status in lung adenocarcinoma by
immunohistochemistry using antibodies specific to the two major
forms of mutant EGFR. J Mol Diagn. 12:169–176. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kato Y, Peled N, Wynes MW, Yoshida K,
Pardo M, Mascaux C, Ohira T, Tsuboi M, Matsubayashi J, Nagao T, et
al: Novel epidermal growth factor receptor mutation-specific
antibodies for non-small cell lung cancer: Immunohistochemistry as
a possible screening method for epidermal growth factor receptor
mutations. J Thorac Oncol. 5:1551–1558. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kawahara A, Yamamoto C, Nakashima K, Azuma
K, Hattori S, Kashihara M, Aizawa H, Basaki Y, Kuwano M, Kage M, et
al: Molecular diagnosis of activating EGFR mutations in non-small
cell lung cancer using mutation-specific antibodies for
immunohistochemical analysis. Clin Cancer Res. 16:3163–3170. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kitamura A, Hosoda W, Sasaki E, Mitsudomi
T and Yatabe Y: Immunohistochemical detection of EGFR mutation
using mutation-specific antibodies in lung cancer. Clin Cancer Res.
16:3349–3355. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Nakamura H, Mochizuki A, Shinmyo T, Ando
K, Kurimoto N, Yokote K and Takagi M: Immunohistochemical detection
of mutated epidermal growth factor receptors in pulmonary
adenocarcinoma. Anticancer Res. 30:5233–5237. 2010.PubMed/NCBI
|
|
17
|
Kozu Y, Tsuta K, Kohno T, Sekine I,
Yoshida A, Watanabe S, Tamura T, Yokota J, Suzuki K, Asamura H, et
al: The usefulness of mutation-specific antibodies in detecting
epidermal growth factor receptor mutations and in predicting
response to tyrosine kinase inhibitor therapy in lung
adenocarcinoma. Lung Cancer. 73:45–50. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang G, Fan C, Zhang X, Dong Q, Wang L,
Liu Y, Dai S, Yang L, Zhang Y, Yu J and Wang E: Ascertaining an
appropriate diagnostic algorithm using EGFR mutation-specific
antibodies to detect EGFR status in non-small-cell lung cancer.
PLoS One. 8:e591832013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Simonetti S, Molina MA, Queralt C, de
Aguirre I, Mayo C, Bertran-Alamillo J, Sanchez JJ, Gonzalez-Larriba
JL, Jimenez U, Isla D, et al: Detection of EGFR mutations with
mutation-specific antibodies in stage IV non-small-cell lung
cancer. J Transl Med. 8:1352010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xiong Y, Bai Y, Leong N, Laughlin TS,
Rothberg PG, Xu H, Nong L, Zhao J, Dong Y and Li T:
Immunohistochemical detection of mutations in the epidermal growth
factor receptor gene in lung adenocarcinomas using
mutation-specific antibodies. Diagn Pathol. 8:272013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen Z, Liu HB, Yu CH, Wang Y, Wang L and
Song Y: Diagnostic value of mutation-specific antibodies for
immunohistochemical detection of epidermal growth factor receptor
mutations in non-small cell lung cancer: A meta-analysis. PLoS One.
9:e1059402014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang T, Nelson RA, Bogardus A and Grannis
FW Jr: Five-year lung cancer survival: Which advanced stage
nonsmall cell lung cancer patients attain long-term survival?
Cancer. 116:1518–1525. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Molina JR, Adjei AA and Jett JR: Advances
in chemotherapy of non-small cell lung cancer. Chest.
130:1211–1219. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kawahara A, Azuma K, Sumi A, Taira T,
Nakashima K, Aikawa E, Abe H, Yamaguchi T, Takamori S, Akiba J and
Kage M: Identification of non-small-cell lung cancer with
activating EGFR mutations in malignant effusion and cerebrospinal
fluid: Rapid and sensitive detection of exon 19 deletion E746-A750
and exon 21 L858R mutation by immunocytochemistry. Lung Cancer.
74:35–40. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsai TH, Wu SG, Chang YL, Wu CT, Tsai MF,
Wei PF, Yang CH, Yu CJ, Yang PC and Shih JY: Effusion
immunocytochemistry as an alternative approach for the selection of
first-line targeted therapy in advanced lung adenocarcinoma. J
Thorac Oncol. 7:993–1000. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kawahara A, Taira T, Azuma K, Tominaga M,
Hattori S, Kawahara M, Abe H, Yamaguchi T, Akiba J, Takamori S, et
al: A diagnostic algorithm using EGFR mutation-specific antibodies
for rapid response EGFR-TKI treatment in patients with non-small
cell lung cancer. Lung Cancer. 78:39–44. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Allo G, Bandarchi B, Yanagawa N, Wang A,
Shih W, Xu J, Dalby M, Nitta H, To C, Liu N, et al: Epidermal
growth factor receptor mutation-specific immunohistochemical
antibodies in lung adenocarcinoma. Histopathology. 64:826–839.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Seo AN, Park TI, Jin Y, Sun PL, Kim H,
Chang H and Chung JH: Novel EGFR mutation-specific antibodies for
lung adenocarcinoma: Highly specific but not sensitive detection of
an E746_A750 deletion in exon 19 and an L858R mutation in exon 21
by immunohistochemistry. Lung Cancer. 83:316–323. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kim CH, Kim SH, Park SY, Yoo J, Kim SK and
Kim HK: Identification of EGFR mutations by immunohistochemistry
with EGFR mutation-specific antibodies in biopsy and resection
specimens from pulmonary adenocarcinoma. Cancer Res Treat.
47:653–660. 2015. View Article : Google Scholar : PubMed/NCBI
|