Introduction
At present, there is growing evidence suggesting
that microsatellite instability (MSI) may be one possible
predictive marker of immunotherapy response in many cancer types
and in malignant melanoma (MM) (1–7). The
main cause of MSI is a defect in the DNA mismatch repair (MMR)
genes, whose function is to repair the mismatched bases (8). Several repair genes (MSH2, MSH3,
MSH6, MLH1, MLH3, PMS1 and PMS2) are involved in this process
and work by forming heterodimers. The interaction between mismatch
recognition complexes and other proteins such as helicase,
proliferating cell nuclear antigen, replication protein A,
exonuclease 1 is required for the rectification of base-pair
alterations, insertion-deletion loops and hetero-duplexes
instigated during replication and recombination (9). MMR genes aberrations can be
investigated by immunohistochemistry (IHC) for MMR proteins, which
is a quick and simple assay to recognize MMR status (10–12).
There is a clear relationship between the loss of
IHC expression of one or more MMR proteins and the MSI phenomenon
(13,14). It was also known that the MSI status
and the lack of MMR protein expression constitute a positive
prognostic factor for colon cancer patients.
The reason of this evidence has been found in the
enhanced immunogenicity of tumors characterized by MSI and MMR
deficiency (MMR-d), because their defective ability to repair DNA
damages lead to a higher mutational burden and an increased
generation of neoantigens (15).
The involvement of MMR genes was proposed in MM
tumorigenesis (16). MSI was
described in displastic naevi, MMs and MM metastases, suggesting
that the MMR system and consequently MSI could contribute to the
pathogenesis of MM (17).
Differently to colon cancer and other solid malignancies that are
treated with immunotherapy (1,18,19),
potential IHC biomarkers are still limited for MM up to now. This
lack of predictive biomarkers for immunotherapy response in MM
patients is today an important field of investigation, whose aim is
to allow a personalized therapeutic approach. A targeted treatment
in fact maximizes the patient's outcome, avoiding unnecessary risks
for the patient's health and sparing important medical resources,
because of the high costs of the new bioengineered drugs. The aim
of our report was to evaluate the role of MMR IHC proteins
expression as a predictive marker of immunotherapy response.
Patients and methods
The study was conducted in accordance with the World
Medical Association Declaration of Helsinki regarding ethical
conduct of research. The study was regularly approved on October
2014 by the ethics committee of the University of Modena and Reggio
Emilia and written informed consent was provided by patients for
IHC analysis. As a part of a systematic review of the medical
record of patients affected by advanced MM receiving anti PD-1
treatment from 2014 to 2016 at the Dermatology and Oncology
Department of the University of Modena and Reggio Emilia, we
collected the following data: Clinical stage of disease (AJCC 8th
Ed., stage I–IV) (20); primary
tumor location; overall survival (OS), progression-free survival
(PFS) after anti PD-1 therapy; adverse events during anti PD-1
therapy. PFS was determined using the RECIST criteria (21).
IHC analysis of MSH6, MLH1, MSH2 and PMS2 proteins
were carried out on the paraffin-embedded primary tumor samples of
every included patients and on the available metastasis samples.
The sections were cut (3–4 µm) into superfrost plus microscope
slides and allowed to dry at 37°C overnight. The slides were
submitted to antigen retrieval using microwave in 10 mmol/l citrate
buffer, pH 6, at 350 W for 30 min. Immunoperoxidase staining, using
diaminobenzidine as chromogen, was run with the NEX-ES Automatic
Staining System (Ventana). Monoclonal antibodies anti-MSH6 (Clone
44; Transduction Laboratories; BD Biosciences) at 1:2,000,
anti-MLH1 (G168-15) at 1:40, anti-MSH2 (G129-1129; both Pharmingen)
at 1:40 dilution and anti PMS-2 (Biocare Medical) at 1:40 dilution
were used. Nuclei were counterstained with hematoxylin and adjacent
normal tissue in each sample served as positive control. The
complete absence of staining of tumor cells for one of the MMR
proteins was considered indicative of a mismatch repair defect.
Results
We were able to identify 14 patients with advanced
MM that were treated with anti PD-1 during 2014–2016 (Table I). Patients comprised 12 males and 2
females, average age was 71 years, ranging from 47 to 88 years. MM
was located on the head and neck area (n=2, 14%), on the trunk
(n=7, 50%), on the upper limb (n=3, 22%, of which 2 were acral) and
on the lower limb (n=2, 14%, of which 1 was acral); no patient of
our series was affected by mucosal melanoma. The total number of
metastases was 45: 10 were located at the lungs, 8 at the lymph
nodes, 6 at the skin, 3 in the retroperitoneum, 3 at the bone, 3 at
the spleen, 3 at the liver, 3 at the brain, 2 at the muscle, 1 at
the gut, 1 at the stomach, 1 at the pancreas and 1 at the kidney.
Of these, only 23 were available as paraffin embedded samples.
 | Table I.Clinical and pathological features of
the patient cohort. |
Table I.
Clinical and pathological features of
the patient cohort.
| Patient | Sex | Age (years) | Location of the
primary MM | Breslow | BRAF status | Metastasis sites | TNM stage before anti
PD-1 therapy | Line of therapy | Start of anti PD-1
therapy | Last follow-up | Status | Type of response | OS (days) | PFS (days) | MMR status |
|---|
| 1 | M | 82 | Back | 2 | V600E | Skin, lungs,
perirenal | 4b | I | 19/05/16 | 11/07/17 | Alive | CR | 572 | 418 | MMR stable |
| 2 | M | 84 | Head and neck | 11 | WT | Skin, lymph node | 3c | II | 14/04/16 | 16/08/16 | Dead | PD | 154 | 124 | MMR stable |
| 3 | M | 83 | Upper limbs | 4 | WT | Lungs, lymph
node | 4b | I | 12/07/16 | 26/11/16 | Dead | PD | 213 | 81 | MMR stable |
| 4 | M | 73 | Back | 2 | WT | Lung, bone, spleen,
liver, lymph node | 4c | I | 07/07/16 | 27/06/17 | Alive | PR | 406 | 355 | MMR stable |
| 5 | M | 83 | Lower limbs | 3 | WT | Lymph node, muscle,
retroperitoneal | 4a | I | 21/08/16 | 11/07/17 | Alive | CR | 335 | 324 | MMR stable |
| 6 | M | 55 | Back | 15 | V600E | Lymph node, skin | 4a | II | 13/05/16 | 19/11/16 | Dead | PD | 1,040 | 125 | MMR stable |
| 7 | F | 76 | Lower limbs | 7 | WT | Skin | 4a | II | 15/03/16 | 11/07/17 | Alive | PD | 719 | 457 | MMR stable |
| 8 | M | 57 | Back | 4 | V600K | Lymph node, lungs,
skin, liver, spleen, brain | 4c | II | 15/05/16 | 03/09/16 | Dead | PD | 201 | 93 | MMR stable |
| 9 | M | 86 | Acral | 2 | WT | Lungs, pancreatic,
pararenal | 4d | II | 28/01/16 | 12/10/16 | Dead | PD | 368 | 196 | MMR stable |
| 10 | M | 77 | Back | 3 | WT | Lungs, lymph node,
bowel, muscle | 4b | III | 15/09/14 | 19/09/16 | Dead | PD | 1,496 | 517 | MMR stable |
| 11 | F | 48 | Acral | 1,4 | WT | Kidney, lymph node,
lungs, bone, brain, bowel | 4d | V | 21/11/14 | 04/07/17 | Alive | CR | 2,546 | 956 | Loss of MSH6
expression |
| 12 | M | 57 | Chest | 13 | V600E | Skin, liver,
spleen, lungs, bone | 4c | II | 11/10/16 | 11/07/17 | Alive | PD | 94 | 84 | MMR stable |
| 13 | M | 47 | Chest | 3 | V600E | Lungs | 4b | I | 23/08/16 | 11/07/17 | Alive | PR | 361 | 322 | MMR stable |
| 14 | M | 88 | Head and neck | 11 | WT | Lungs, brain | 4d | III | 18/06/15 | 01/07/17 | Dead | PR | 1,088 | 684 | MMR stable |
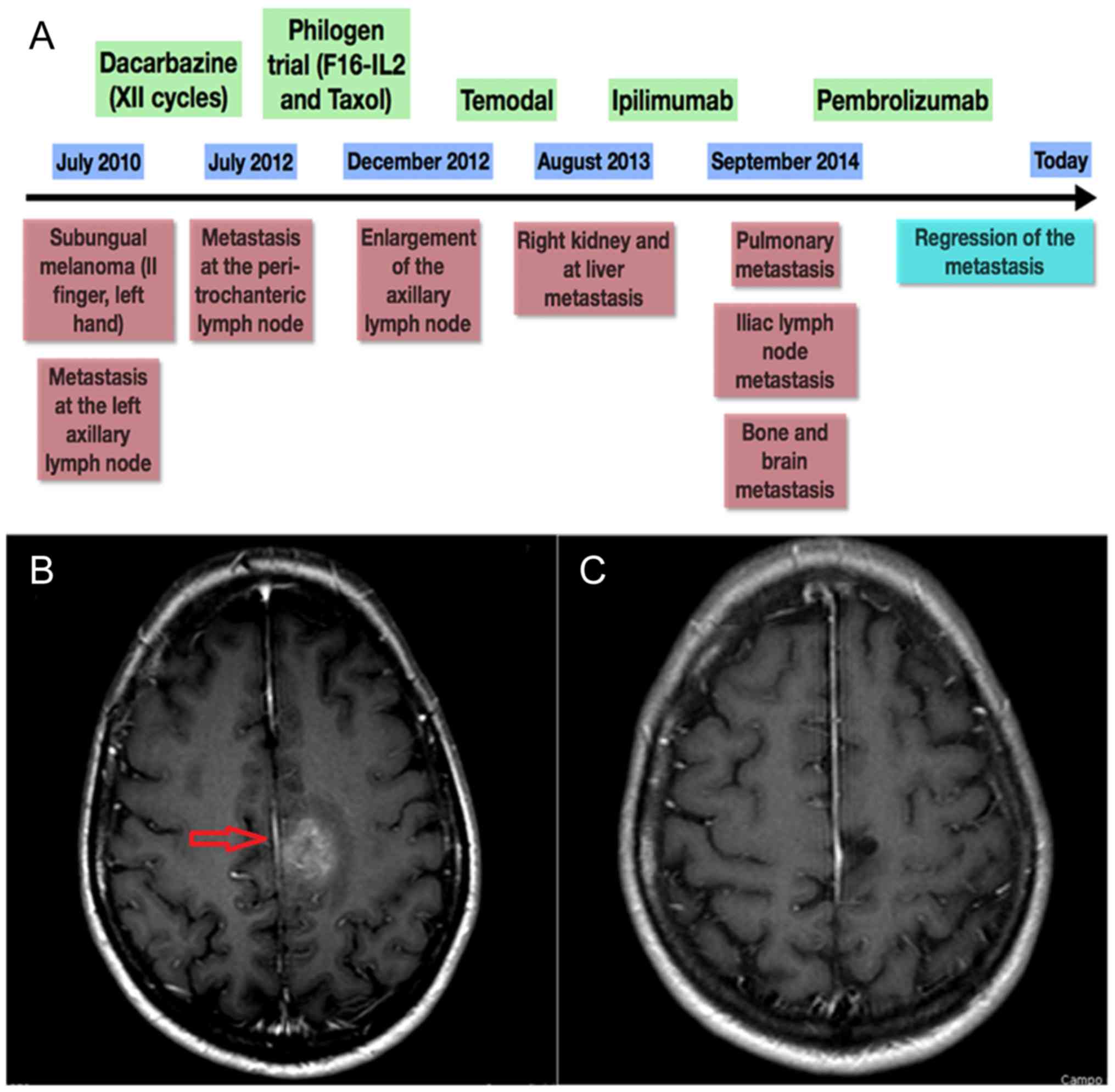
Only three samples showed MMR-d of MMR genes, in
particular of MSH6: they were one primary subungual melanoma and
two metastases (brain and ileus) belonging to the same female
patient, whose complex clinical history is summarized by the time
plot represented in Fig. 1. The IHC
panels of the primary MM and related metastases of our MMR-d and of
an exemplificative MMR normal patient are shown in Figs. 2 and 3, respectively. Remarkably, she showed the
best response to anti PD-1 treatment of our cohort, with PFS and OS
of 956 and 2,546 days, respectively. At present, the patient is
still alive and in complete response. By contrast, MMR-s patients
showed average PFS and OS of 290 and 542 days, respectively.
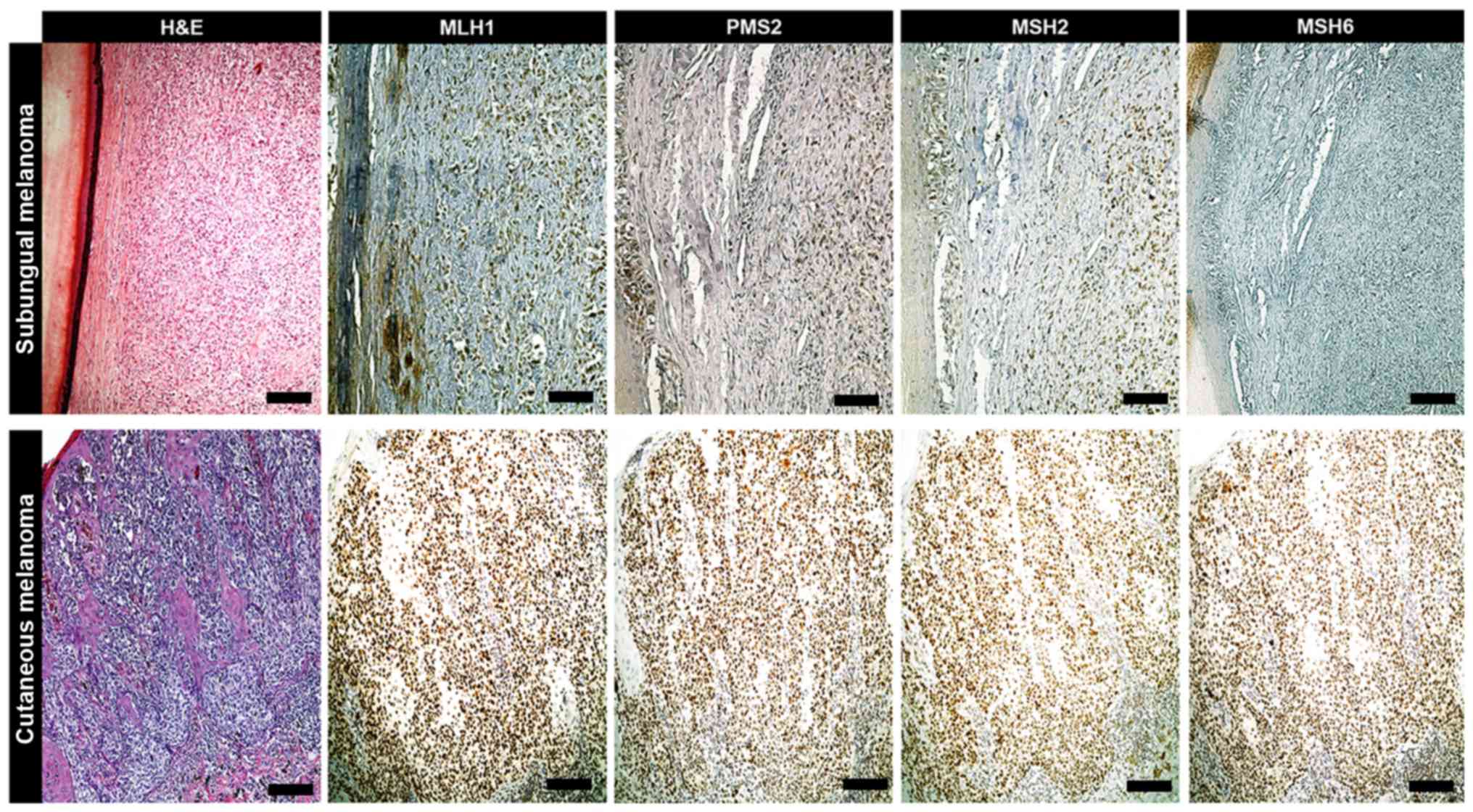 | Figure 2.IHC of primary melanoma. When compared
with MLH1, PMS2 and MSH2 levels, an exclusive loss of MSH6
expression was exhibited in a female case of subungual melanoma
form the patient cohort, as determined via IHC. In the remaining
cases of cutaneous melanoma, the expressions of MLH1, PMS2, MSH2
and MSH6 were within the normal range (scale bars, 100 µm). IHC,
immunohistochemistry; MLH1, clone M1 Ventana; PMS2, clone EPR3947
Ventana; MSH2, clone G219-1129; MSH6, clone 44 Ventana; H&E,
hematoxylin and eosin. |
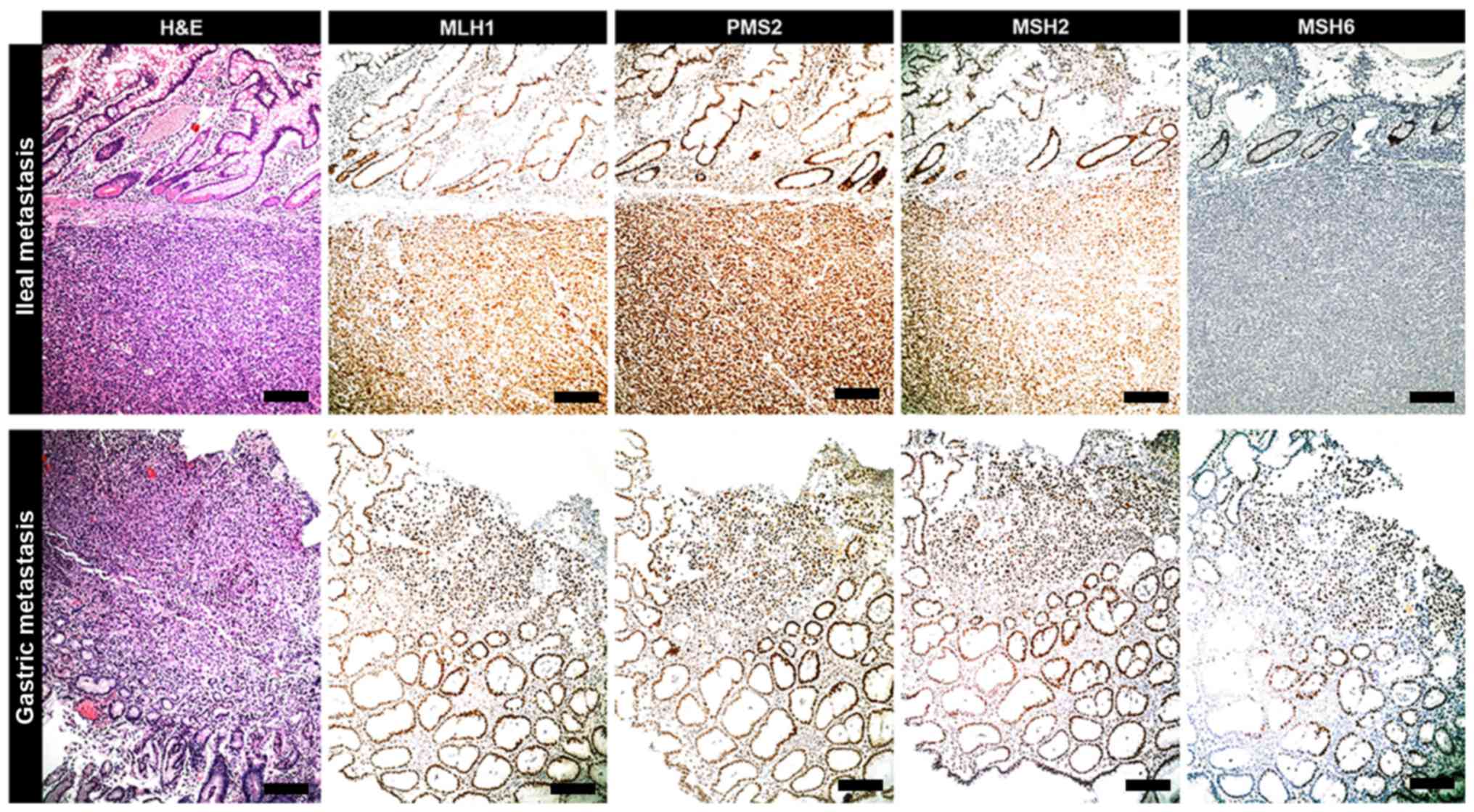 | Figure 3.IHC of metastasis. Ileal metastasis
tumor originating from subungual melanoma exhibits the same IHC
profile of primitive neoplasia. It is characterized by an exclusive
loss of expression of MSH6 protein when compared with MLH1, PMS2
and MSH2 status. In the analyzed metastases of the current cohort
(e.g., gastric metastasis), the expression of MSH6 protein was
preserved as with that of MLH1, PMS2 and MSH2 (scale bars, 100 µm).
IHC, immunohistochemistry; MSH6, clone 44 Ventana; MLH1, clone M1
Ventana; PMS2, clone EPR3947 Ventana; MSH2, clone G219-1129;
H&E, hematoxylin and eosin. |
Discussion
The assessment of MMR protein expression represents
a potential predictive marker which may have crucial importance for
primary and metastatic MM patients. The identification of quick and
simple assays to predict the response to anti PD-1 agents is one of
the goal that the new era of MM immunotherapy should achieve in
order to satisfy the compelling objectives of a personalized
medicine.
Several studies have demonstrated that MMR gene
deficiency is a more widespread phenomenon than first assumed,
being present in many tumor types, such as colon cancer, ovarian
cancer, brain tumors, biliary tract tumors and gastric cancer, but
also in neuroblastoma and endometrial cancer (17). In a recent study by Kim et al
(22) the MMR-d tumor status was
assessed in 430 consecutive solid tumors and was significantly
correlated to the PD-L1 expression. Even though MMR-d is a negative
biomarker for MM chemotherapy sensitivity (23), recent evidences suggest that it may
be a positive biomarker for immunotherapy response. In detail, it
was demonstrated that MSI, which is often a consequence of MMR gene
deficiency, is associated to a better response to immunotherapy in
terms of PFS. As a general consideration, our hypothesis is that
the presence of a mutator tumor phenotype allows an easier escape
of the tumor to the pharmacokinetis of conventional chemotherapy,
but, on the other hand, it produces many neoantigens that increase
the immunogenicity of tumor cells. Thus, it is our hypothesis that
the IHC analysis of MMR gene could be a marker that predict PFS and
OS of MM patients treated by anti-PD-1 agents. In our population,
the best PFS response was reached by the patient that showed MMR-d
tumor status, with lack of expression of MSH6, not associated to
Lynch syndrome or Muir-Torre Syndrome. The clinical history of the
patient was complex and the previous treatment lead to
unsatisfactory results. Noteworthy, anti PD-1 therapy lead to a
dramatic improvement of her management, with the regression of the
brain and other metastasis, that are generally considered the
prognostic factors that most negatively affect patient's survival.
The limitations of our study consisted in the small population size
and the lack of molecular biology investigations. Future studies
are needed to investigate the role of MSI and MMR-d during
immunotherapy in animal model as well as in larger patient
cohorts.
In conclusion, the assessment of MMR protein
expression represents a potential predictive marker, which may have
crucial importance for primary and metastatic MM patients, mainly
as criterion for the adoption of the immunotherapy treatments.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
GPo interpreted patient data and wrote the
manuscript. GPe and AT analyzed and interpreted patient data. RD
and FG enrolled and examined the patients with melanoma. MMac
analyzed the clinical and molecular data. AM performed histological
examinations. GO collected the clinical data and examined the
specimens. SC interpreted the patient data. MMan provided clinical
advice and wrote the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
The present study was approved on October 2014 by
the Ethics Committee of the University of Modena and Reggio Emilia
and written informed consent for participation in the study or use
of their tissue was obtained from all participants.
Patient consent for publication
Written informed consent was obtained for the
publication of data and materials. All data are published with
respect to participants' rights to privacy and to protect their
identity.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MM
|
malignant melanoma
|
|
MMR
|
mismatch repair
|
|
MMR-d
|
mismatch repair deficiency
|
|
MMR-s
|
mismatch repair stable
|
|
MSI
|
microsatellite instability
|
|
IHC
|
immunoistochemistry
|
References
|
1
|
Gargiulo P, Della Pepa C, Berardi S,
Califano D, Scala S, Buonaguro L, Ciliberto G, Brauchli P and
Pignata S: Tumor genotype and immune microenvironment in
POLE-ultramutated and MSI-hypermutated endometrial cancers: New
candidates for checkpoint blockade immunotherapy? Cancer Treat Rev.
48:61–68. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dudley JC, Lin MT, Le DT and Eshleman JR:
Microsatellite instability as a biomarker for PD-1 blockade. Clin
Cancer Res. 22:813–820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bupathi M and Wu C: Biomarkers for immune
therapy in colorectal cancer: Mismatch-repair deficiency and
others. J Gastrointest Oncol. 7:713–720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Czink E, Kloor M, Goeppert B, Fröhling S,
Uhrig S, Weber TF, Meinel J, Sutter C, Weiss KH, Schirmacher P, et
al: Successful immune checkpoint blockade in a patient with
advanced stage microsatellite-unstable biliary tract cancer. Cold
Spring Harb Mol Case Stud. 3:a0019742017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen L, Xiong Y, Li J, Zheng X, Zhou Q,
Turner A, Wu C, Lu B and Jiang J: PD-L1 expression promotes
epithelial to mesenchymal transition in human esophageal cancer.
Cell Physiol Biochem. 42:2267–2280. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Howitt BE, Strickland KC, Sholl LM, Rodig
S, Ritterhouse LL, Chowdhury D, D'Andrea AD, Matulonis UA and
Konstantinopoulos PA: Clear cell ovarian cancers with
microsatellite instability: A unique subset of ovarian cancers with
increased tumor-infiltrating lymphocytes and PD-1/PD-L1 expression.
Oncoimmunology. 6:e12773082017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Benatti P, Gafà R, Barana D, Marino M,
Scarselli A, Pedroni M, Maestri I, Guerzoni L, Roncucci L,
Menigatti M, et al: Microsatellite instability and colorectal
cancer prognosis. Clin Cancer Res. 11:8332–8340. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Richman S: Deficient mismatch repair: Read
all about it (Review). Int J Oncol. 47:1189–1202. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bhattacharya P and Patel TN:
Microsatellite instability and promoter hypermethylation of DNA
repair genes in hematologic malignancies: A forthcoming direction
toward diagnostics. Hematology. 23:77–82. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chapusot C, Martin L, Puig PL, Ponnelle T,
Cheynel N, Bouvier AM, Rageot D, Roignot P, Rat P, Faivre J and
Piard F: What is the best way to assess microsatellite instability
status in colorectal cancer? Study on a population base of 462
colorectal cancers. Am J Surg Pathol. 28:1553–1559. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ponti G, Losi L, Di Gregorio C, Roncucci
L, Pedroni M, Scarselli A, Benatti P, Seidenari S, Pellacani G,
Lembo L, et al: Identification of Muir-Torre syndrome among
patients with sebaceous tumors and keratoacanthomas: Role of
clinical features, microsatellite instability, and
immunohistochemistry. Cancer. 103:1018–1025. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ponti G, Losi L, Pedroni M, Lucci-Cordisco
E, Di Gregorio C, Pellacani G and Seidenari S: Value of MLH1 and
MSH2 mutations in the appearance of Muir-Torre syndrome phenotype
in HNPCC patients presenting sebaceous gland tumors or
keratoacanthomas. J Invest Dermatol. 126:2302–2307. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Garcia JJ, Kramer MJ, O'Donnell RJ and
Horvai AE: Mismatch repair protein expression and microsatellite
instability: A comparison of clear cell sarcoma of soft parts and
metastatic melanoma. Mod Pathol. 19:950–957. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ponti G and Longo C: Microsatellite
instability and mismatch repair protein expression in sebaceous
tumors, keratocanthoma, and basal cell carcinomas with sebaceous
differentiation in Muir-Torre syndrome. J Am Acad Dermatol.
68:509–510. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nebot-Bral L, Coutzac C, Kannouche PL and
Chaput N: Why is immunotherapy effective (or not) in patients with
MSI/MMRD tumors? Bull Cancer. 106:105–113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Alvino E, Marra G, Pagani E, Falcinelli S,
Pepponi R, Perrera C, Haider R, Castiglia D, Ferranti G, Bonmassar
E, et al: High-frequency microsatellite instability is associated
with defective DNA mismatch repair in human melanoma. J Invest
Dermatol. 118:79–86. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ponti G, Losi L, Pellacani G, Wannesson L,
Cesinaro AM, Venesio T, Petti C and Seidenari S: Malignant melanoma
in patients with hereditary nonpolyposis colorectal cancer. Br J
Dermatol. 159:162–168. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Ponti G, Manfredini M, Greco S, Pellacani
G, Depenni R, Tomasi A, Maccaferri M and Cascinu S: BRAF, NRAS and
C-KIT advanced melanoma: Clinico-pathological features,
targeted-therapy strategies and survival. Anticancer Res.
37:7043–7048. 2017.PubMed/NCBI
|
|
19
|
Moscarella E, Pellegrini C, Pampena R,
Argenziano G, Manfredini M, Martorelli C, Ciarrocchi A, Dika E,
Peris K, Antonini A, et al: Dermoscopic similarity is an
independent predictor of BRAF mutational concordance in multiple
melanomas. Exp Dermatol. 28:829–835. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Amin MB, Edge S, Greene F, Byrd DR,
Brookland RK, Washington MK, Gershenwald JE, Compton C, Hess KR,
Sullivan DC, et al: Cancer Staging Manual8th. Springer; New York,
NY: 2017
|
|
21
|
Eisenhauer EA, Therasse P, Bogaerts J,
Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S,
Mooney M, et al: New response evaluation criteria in solid tumours:
Revised RECIST guideline (version 1.1). Eur J Cancer. 45:228–247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim ST, Klempner SJ, Park SH, Park JO,
Park YS, Lim HY, Kang WK, Kim KM and Lee J: Correlating programmed
death ligand 1 (PD-L1) expression, mismatch repair deficiency, and
outcomes across tumor types: Implications for immunotherapy.
Oncotarget. 8:77415–77423. 2017.PubMed/NCBI
|
|
23
|
Lage H, Christmann M, Kern MA, Dietel M,
Pick M, Kaina B and Schadendorf D: Expression of DNA repair
proteins hMSH2, hMSH6, hMLH1, O6-methylguanine-DNA
methyltransferase and N-methylpurine-DNA glycosylase in melanoma
cells with acquired drug resistance. Int J Cancer. 80:744–750.
1999. View Article : Google Scholar : PubMed/NCBI
|