Introduction
The pancreas consists of the pancreatic duct, acinar
tissue, Langerhans islets, and mesenchymal cells. Tumors derived
from the pancreatic duct epithelium account for the majority of
pancreatic tumors, known as pancreatic duct adenocarcinoma (PDAC).
Pancreatic cancer is extremely lethal with poor prognosis and no
established survival markers. Its 5-year survival rate is only 6%
and remains below 25% even after curative surgery, thereby making
PDAC one of the most lethal tumors (1).
The most frequently detected gene mutation in PDAC
is KRAS, which is found in more than 90% of cases. Other frequently
mutated tumor suppressor genes include p16/CDKN2A, TP53/p53, and
SMAD4/DPC4(2). Inactivation of KRAS,
TP53, p16, and SMAD4 is the most common genetic alteration in human
PDAC (3). Next-generation sequencing
has described these genetic mutations as the ‘big four’ genes
involved in pancreatic cancer (2).
p16 is an important tumor suppressor gene that has been found to
affect the cell cycle (G1 to S) by inactivating cyclin-dependent
kinase inhibitors (4). Inactivation
of p16 is induced by mutation, homozygous deletion, and promoter
methylation (1,5). Mutations in p16 are found in at least
30-50% of pancreatic cancer cases (3,6). Several
reports have suggested that inactivation of p16 is significantly
associated with its protein expression by immunohistochemistry
(IHC) (7,8). Recent studies have also reported an
association between p16 inactivation and poor prognosis (5,8).
However, the correlation between p16 expression on IHC and
prognosis remains controversial.
Therefore, we performed immunohistochemical staining
of samples from 103 PDAC patients to assess the relationship
between p16 expression and clinicopathological features, including
prognosis. In some cases, we quantified p16 mRNA expression through
RNA sequencing to investigate the correlation between p16
inactivation and its expression on IHC.
Materials and methods
Patients and tissue samples
From January 2013 to December 2017, 103 patients
underwent elective pancreatic resection at the Division of
Hepato-Biliary-Pancreatic Surgery, Chiba Cancer Center (Chiba,
Japan), with a final histopathologic diagnosis of PDAC and no
neoadjuvant therapy. Patients with intraductal papillary mucinous
neoplasms (IPMNs) with minimal invasive component were excluded.
TNM and grading were in accordance with the World Health
Organization (WHO) recommendations Union Internationale Contre le
Cancer (UICC) 8th edition. Freshly removed pancreatic tissue
samples were immediately fixed in formalin for at least 12 h and
embedded in paraffin. Each resected specimen was stained with
hematoxylin and eosin (H&E) and subsequently, microscopically
diagnosed by at least two pathologists. The present study was
approved by the ethics committee of the Chiba Cancer Center, Japan.
All methods were performed in accordance with the relevant
guidelines and regulations.
p16 immunohistochemistry
We measured p16 levels by IHC using anti-human
p16INK4a mouse monoclonal antibody (E6H4; Roche).
Five-µm-thick sections were obtained from formalin-fixed,
paraffin-embedded tissues and set aside for CINtec p16 Histology
[E6H4] with a VENTANA Optiview DAB universal kit and a VENTANA
BenchMark ULTRA automated slide stainer (Roche). Heat-induced
antigen retrieval was carried out using Cell Conditioning 1 (CC1;
Ventana Medical Systems) for 24 min at 95̊C, and the primary
antibody was applied to the sample for 4 min.
The IHC results were scored based on the percentage
positivity of staining. p16 protein expression was evaluated by two
pathologists at a percentage of every 5% of the staining area of
all tumor cells. For statistical comparisons, cases in which
p16-positive cells exceeded 10% of the total tumor cells were
considered positive. In normal pancreas, p16 positivity was
observed in the islets of Langerhans with scattered non-specific
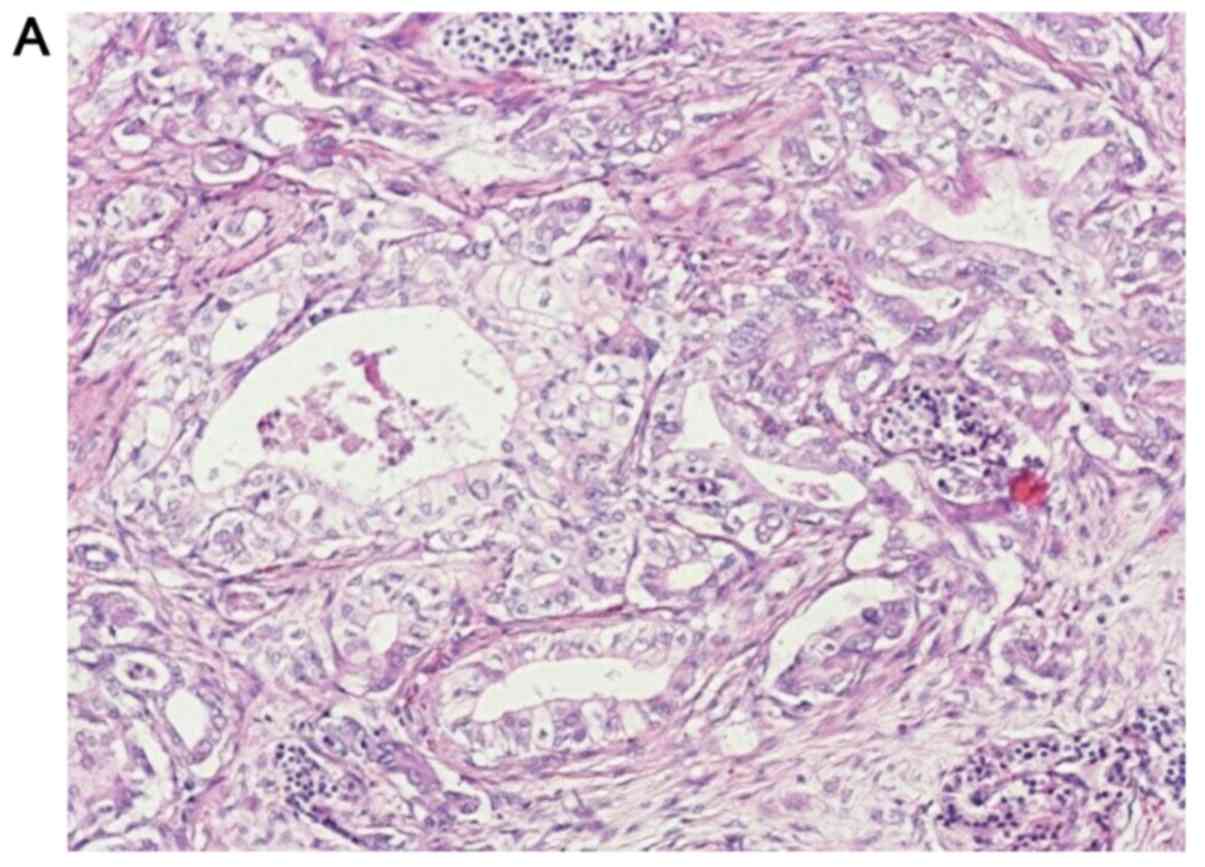
cytoplasmic positivity in the ductal and acinar cells (Fig. 1B), and this was determined to be the
positive control. The findings obtained for the normal pancreas
were compared with those obtained for tumor cells. An 80% agreement
between pathologists in the immunostaining evaluation was set as
the criterion. When the pathologists disagreed with regard to the
evaluations, a decision was reached based on consultation.
RNA sequencing (RNA-seq)
Total RNA was isolated from frozen tissue blocks
containing approximately 50-100 mg PDAC tissues following the
manufacturer's instructions. The frozen tissues from our hospital's
biobank were ground using liquid nitrogen and homogenized. RNA was
extracted using the miRNeasy Mini kit (QIAGEN), and the quality,
quantity, and integrity of the total RNA were evaluated using a
NanoDrop One/Onec UV-Vis Spectrophotometer (Thermo
Fisher Scientific) and Bioanalyzer 2100 (Agilent Technologies).
Samples with an RNA quality score (RIN value) of >7.0 were used
for RNA-seq. rRNA was excluded from the total RNA using
RiboMinus™ Eukaryote System v2. mRNA was barcoded with
Ion Xpress™ RNA-Seq Barcode 1-16 kit (Thermo Fisher Scientific),
and the library was generated using Ion Total RNA-Seq kit v2
(Thermo Fisher Scientific). The libraries were constructed for
next-generation sequencing (NGS) using an Ion Proton™ instrument
(Thermo Fisher Scientific) with 2x75-base pair (bp) paired-end
protocol. In total, 8 libraries were sequenced, generating 34-60
million pairs of reads per sample. The quantity of the sequencing
data was analyzed by a bioinformatician using BAM files from NGS.
The number of reads mapping to the annotated genomic features was
quantified from BAM files using feature count from the Subread
package (http://subread.sourceforge.net/).
Statistical variables and
analyses
Age was divided into two groups with 70 as the
median: ≤70 and 70<. Lymph nodes, margin status, cytology,
lymphatic invasion, neural invasion, vascular invasion,
differentiation, and TNM staging (UICC 8th edition) were defined
based on the pathological search results. Lymph nodes were positive
for lymph node metastasis or negative for lymph node metastasis.
The margin status was R2, R1, and R0 for gross stump positive,
histopathological stump positive, and histopathological stump
negative, respectively. Cytology was defined as CY1 when cancer
cells were found by peritoneal washing cytology; otherwise, it was
defined as CY0. Lymphatic invasion, neural invasion, and vascular
invasion were each divided into four stages: ly0, ly1, ly2, ly3;
ne0, ne1, ne2, ne3; and v0, v1, v2, v3, respectively. Ly0, ne0, and
v0 were defined as without lymphatic invasion, neural invasion, and
vascular invasion, respectively. Vascular invasion was divided into
two groups: V0, v1 and v2, v3, because there was only one patient
of v0. Pancreatic cancer tissue was classified according to the
degree of differentiation: Well, moderate, and poor. Here,
differentiation was divided into two groups for convenience:
Well/moderate and poor. Overall survival was defined as the period
between the surgery and final observation (in months). For samples
extracted from an infinite population, we assumed a sample ratio of
0.5 for activated p16 and 1 for inactivated p16 with 95% confidence
and 5% error. The required sample size was 385, but the actual
sample size might be small. For the statistical analyses,
Mann-Whitney U test and chi-square test were performed. A survival
curve was prepared using the Kaplan-Meier method, and log-rank test
assessed significant differences. P<0.05 was considered to
indicate a statistically significant difference.
Results
Clinical pathological background of
patients
PDAC tissues were obtained from 103 patients (59
males, 44 females), who had surgeries for pancreatic cancer, and
diagnosed with PDAC by pathologists. Age ranged from 50-87 with 70
as the median. We divided PDAC patients into two groups in terms of
median age. There were 50 patients older than 70 years and 53
patients younger than 70 years. There were 45 patients with
well-differentiated tumors that were mainly of the tissue type, 50
with moderately differentiated tumors, and 8 with poorly
differentiated tumors. Lymphatic invasion, neural invasion, and
vascular invasion were each scored in 4 grades with 0 as negative.
Twenty-seven PDAC patients were negative for lymphatic invasion,
whereas only six patients were negative for neural invasion, and
only one was negative for vascular invasion. The number of patients
with weak vascular invasion (v1) was 10. There were 86 patients
with negative pathological margins and 91 with negative cytology.
According to the TNM classification of UICC 8th edition, there were
12 patients with stage IA disease, 14 with stage IB, 5 with stage
IIA, 36 with stage IIB, and 36 with stage III (Table I).
 | Table IClinicopathological background,
outcome and comparison between positive and negative p16
groups. |
Table I
Clinicopathological background,
outcome and comparison between positive and negative p16
groups.
| Variable | Number (%) | Positive p16
expression group | Negative p16
expression group | P-value |
|---|
| Number (%) | 103 | 48 (46.6) | 55 (53.4) | |
| Sex | | | | 0.11a |
|
Male | 59 (57.3) | 23 (23.3) | 36 (35.0) | |
|
Female | 44 (42.7) | 25 (24.3) | 19 (18.4) | |
| Median age | 70 | 71.5 | 69 | 0.33b |
| Age range | 50-87 | 50-83 | 51-87 | |
| Lymph nodes | | | | 1.00a |
|
Negative | 31 (30.1) | 14 (13.6) | 17 (16.5) | |
|
Positive | 72 (69.9) | 34 (33.0) | 38 (36.9) | |
| Margin status | | | | 0.06a |
|
R0 | 86 (83.5) | 44 (42.7) | 42 (40.8) | |
|
R1/R2 | 17 (16.5) | 4 (3.9) | 13 (12.6) | |
| Cytology | | | | 0.18a |
|
CY0 | 91 (88.3) | 40 (38.8) | 51 (49.5) | |
|
CY1 | 12 (11.7) | 8 (7.8) | 4 (3.9) | |
| Lymphatic
invasion | | | | 0.37a |
|
Negative | 17 (16.5) | 15 (14.6) | 12 (11.7) | |
|
Positive | 76 (73.8) | 33 (32.0) | 43 (41.7) | |
| Neural
invasion | | | | 0.41a |
|
Negative | 6 (5.8) | 4 (3.9) | 2 (1.9) | |
|
Positive | 97 (94.2) | 44 (42.7) | 53 (51.5) | |
| Vascular
invasion | | | | 1.00a |
|
Negative (v
0/1) | 21 (20.4) | 10 (9.7) | 11 (10.7) | |
|
Positive (v
2/3) | 82 (79.6) | 38 (36.9) | 44 (42.7) | |
|
Differentiation | | | | 0.72a |
|
Well/Moderate | 95 (92.2) | 45 (43.7) | 50 (48.5) | |
|
Poor | 8 (7.8) | 3 (2.9) | 5 (4.9) | |
| T factor (UICC
8th) | | | | 1.00a |
|
T1/2 | 79 (76.7) | 37 (35.9) | 42 (40.8) | |
|
T3 | 24 (21.4) | 11 (10.7) | 13 (12.6) | |
| Stage (UICC
8th) | | | | 0.83c |
|
IA | 12 (11.7) | 4 (3.9) | 8 (7.8) | |
|
IB | 14 (13.6) | 7 (6.8) | 7 (6.8) | |
|
IIA | 5 (4.9) | 3 (2.9) | 2 (1.9) | |
|
IIB | 36 (35.0) | 18 (17.5) | 18 (17.5) | |
|
III | 36 (35.0) | 16 (15.5) | 20 (19.4) | |
Expression of p16 protein in
PDACs
The loss of p16 protein expression was noted in 55
out of 103 (53.4%) tumors as determined by IHC (Fig. 1). We observed 7 out of 55 ductal
adenocarcinomas with weak staining (≤10%) for p16 by IHC. The 55
weakly to negatively stained tumors were grouped together in the
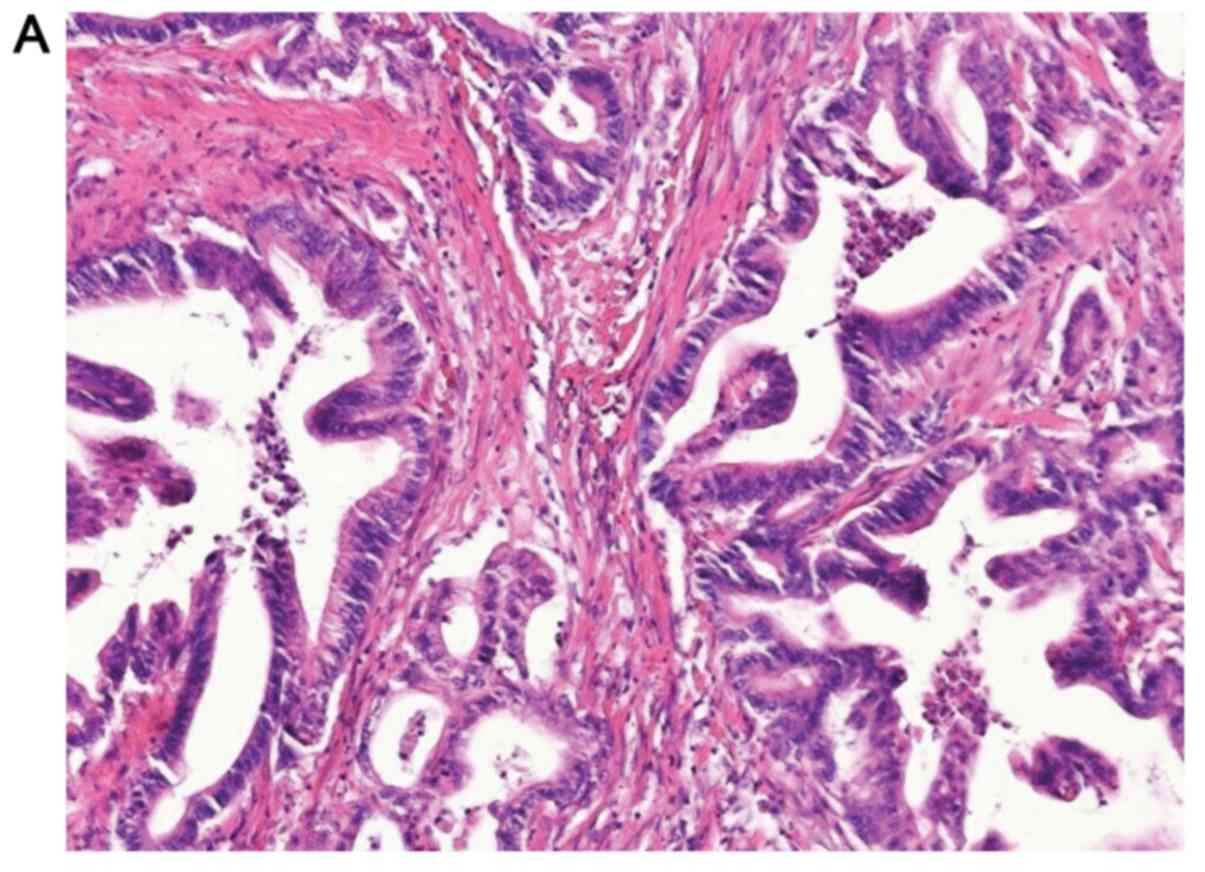
negative p16 expression group. Meanwhile, 48 out of 103 (46.6%)
tumors were stained positively with strong to moderate staining
(>10%) for p16 by IHC (Fig. 2)
and were included in the positive p16 expression group (Table I). Overall, 46.6% of the positive
patients were considered positive as a result of exceeding 10% of
the total tumor cells as described in Table I.
Clinicopathological outcomes
No correlation was found between p16 status and sex,
age, TNM stage, or histological differentiation (Mann-Whitney U
test, chi-square test, Fisher's exact test; P>0.05), as shown in
Table I. The survival curves for
sex, age, histological differentiation, pathological margin status,
cytology, lymphatic invasion, neural invasion, vascular invasion,
and TNM grade were plotted using the Kaplan-Meier method and
analyzed using the log-rank test. Four factors were found to be
significantly associated with prognosis (Table II): Lymph node metastasis
(P<0.001), cytology (P=0.006), neural invasion (P=0.009), and T
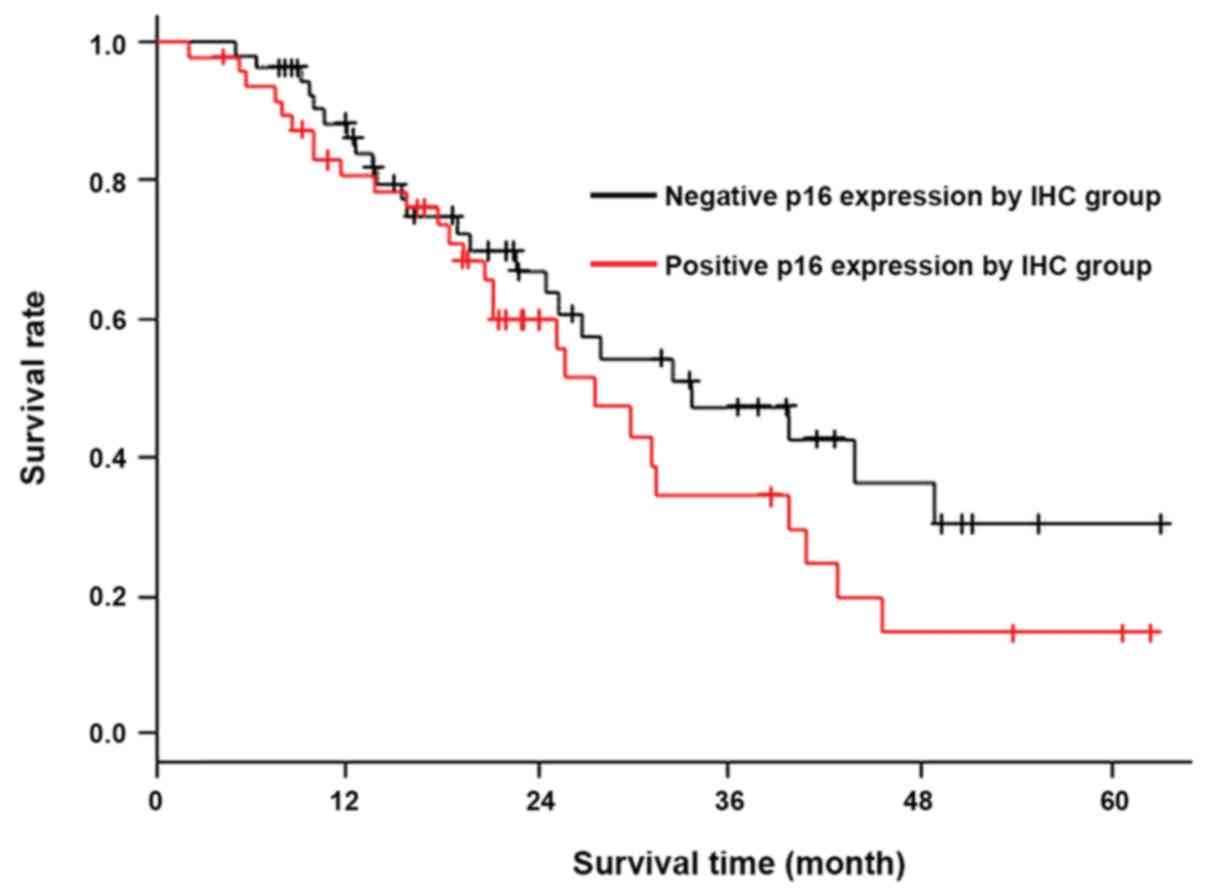
factor (UICC 8th; P=0.005). Therefore, p16-negative status on IHC
was not significantly associated with poor prognosis according to
the Kaplan-Meier method (P=0.181), as shown in Fig. 3. The multivariate Cox proportional
regression analysis was not performed because p16 was not
significantly different in the univariate analysis.
 | Table IIUnivariate analysis of prognostic
factors for overall survival. |
Table II
Univariate analysis of prognostic
factors for overall survival.
| Variable | No. of patients
(%) | Median (95%
confidence interval) | P-value |
|---|
| Sex | | | 0.270 |
|
Male | 59 (57.3) | 27.1
(20.2-32-2) | |
|
Female | 44 (42.7) | 42.5
(23.4-N/A) | |
| Age | | | 0.518 |
|
≤70 | 53 (51.5) | 30.2
(21.6-40.8) | |
|
>70 | 50 (48.5) | 24.9
(19.7-43.5) | |
|
Range | 53-87 | | |
| Follow-up (M) | | | |
|
Median | 19.1 | | |
|
Range | 2-61.2 | | |
| Lymph nodes | | | <0.001 |
|
Negative | 31 (30.0) | 47.5
(29.9-N/A) | |
|
Positive | 72 (70.0) | 24.5
(18.4-29.7) | |
| Margin status | | | 0.812 |
|
R0 | 86 (83.5) | 28.8
(24.0-40.8) | |
|
R1-R2 | 17 (16.5) | 24.5
(14.9-N/A) | |
| Cytology | | | 0.006 |
|
CY0 | 91 (88.3) | 38.5
(24.1-40.8) | |
|
CY1 | 12 (11.7) | 15.1
(8.15-N/A) | |
| Lymphatic
invasion | | | 0.151 |
|
Negative | 27 (26.2) | 41.4
(24.5-N/A) | |
|
Positive | 76 (73.8) | 27.1
(20.2-32.2) | |
| Neural
invasion | | | 0.009 |
|
Negative | 6 (5.8) | N/A | |
|
Positive | 97 (94.2) | 26.7
(21.6-32.2) | |
| Vascular
invasion | | | 0.081 |
|
Negative
(v0/1) | 21 (20.4) | N/A | |
|
Positive
(v2/3) | 82 (79.6) | 27.1
(23.4-32.2) | |
|
Differentiation | | | 0.975 |
|
Well/Moderate | 95 (92.2) | 28.8
(24.0-37.0) | |
|
Poor | 8 (7.8) | 11.6
(1.91-N/A) | |
| T factor (UICC
8th) | | | 0.005 |
|
T1/2 | 79 (76.7) | 31.4
(24.5-43.5) | |
|
T3 | 24 (23.3) | 19.1
(11.9-28.4) | |
| Stage (UICC
8th) | | | 0.13 |
|
IA | 12 (11.7) | 39.3 | |
|
IB | 14 (13.6) | 22.1 | |
|
IIA | 5 (4.9) | 22.3 | |
|
IIB | 36 (35.0) | 18.2 (20.2-39.0
(Ⅰ,Ⅱ)) | |
|
III | 36 (35.0) | 17.5 (13.2-28.4
(Ⅲ)) | |
| p16 IHC (Median:
10%) | | | 0.18 |
|
Positive
(>10%) | 48 (46.6) | 26.7
(23.7-47.5) | |
|
Negative
(≤10%) | 55 (53.4) | 32.6
(20.0-38.5) | |
| p16 IHC (Average:
32%) | | | 0.08 |
|
Positive
(>32%) | 37 (35.9) | 24.5
(18.4-29.9) | |
|
Negative
(≤32%) | 66 (64.1) | 32.2
(24.1-46.8) | |
Correlation between protein expression
on IHC and mRNA expression using RNA-seq of p16
Of the 103 patients, 8 were registered in our
biobank, and we analyzed mRNA expression by RNA-seq of samples from
these 8 patients. The relationship between protein expression level
of p16 on IHC and mRNA expression level of p16 (CDKN2A) using
RNA-seq was confirmed. The protein expression level of p16 on IHC
was observed in percentage by every 5%. Five of the eight cases did
not express the p16 protein on IHC, whereas three had mRNA
expression levels below 0.5. Three of the eight cases showed p16
protein expression level over 10%, and these same cases had mRNA
expression levels of over 3.5. Here, Spearman's rank correlation
coefficient test showed a correlation between protein expression on
IHC and mRNA expression using RNA-seq (P=0.021) as shown in
Table III.
 | Table IIIComparison of p16 expression by IHC
and RNA-seq. |
Table III
Comparison of p16 expression by IHC
and RNA-seq.
| Case no. | P16 expression by
IHCa | P16 expression by
RNA-seq | P-value | rb |
|---|
| | | | 0.021 | 0.784 |
|
1 | 0 | 2.728 | | |
|
2 | 0 | 0.269 | | |
|
3 | 0 | 3.349 | | |
|
4 | 90 | 3.787 | | |
|
5 | 0 | 0.265 | | |
|
6 | 0 | 0.233 | | |
|
7 | 40 | 4.399 | | |
|
8 | 40 | 4.521 | | |
Discussion
Analyses using genetically modified mice revealed
that the initiation of pancreatic tumorigenesis required KRAS gene
mutation, and tumorigenesis is accelerated by the presence of p16
or TP53 mutation (9). The p16 gene
product belongs to an important group of proteins that negatively
regulates the G1 phase of the cell cycle. It binds to
cyclin-dependent kinases, (CDK)4 and CDK6, and inhibits their
interaction with cyclin D1. The inhibition of the cyclin CDK4/6
complex prevents the phosphorylation of retinoblastoma (Rb) protein
and the release of E2F, subsequently leading to the inhibition of
the transition from G1 to S phase in the cell cycle (4). Therefore, dysfunction in p16 induces Rb
protein phosphorylation, and the cell cycle shifts from the G1 to
the S phase, resulting in the synthesis of deoxyribonucleic acid
(DNA). Consequently, genetic abnormalities induce the inactivation
of the p16 gene and provide a growth advantage to cells involved in
tumorigenesis (10).
The inactivation of the p16 gene occurs through
intragenic mutations with loss of heterozygosity (40%), homozygous
deletion (40%), and methylation-associated transcriptional
silencing (15%) (5) and has been
reported in approximately 95% of PDAC cases (11-13).
An examination of 25 PDAC cases showed that p16 was inactivated or
mutated in 80% of tumors (6).
Another examination with the same sample size showed that the
inactivation of p16 was significantly associated with a negative
p16 expression on IHC (7). Ohtsubo
et al found that p16 inactivation tended to be more detected
in patients with immunohistochemically negative p16 expression than
in those with positive expression, after the examination of 60
pancreatic carcinoma cases (8).
Although the details remain unclear, p16
inactivation may not be necessary to achieve p16 negative
expression on IHC. In this study, we divided the cases into two
groups with the median value (10%) of p16 expression range on IHC
and evaluated the relationship between the two groups. There were
no significant differences in the clinicopathological factors
between the groups (Tables II and
IV). The results did not change
when average values were used. p16 inactivation has been
significantly associated with poor prognosis, lymphatic metastasis,
and lymphatic invasion (5,8). However, the correlation between p16
expression on IHC and prognosis remains controversial because of
inconsistent results. Some studies have reported an association
between negative p16 expression on IHC and poor prognosis (14), whereas others have found no
significant relationship (15,16). In
the present study, a negative p16 expression was not significantly
associated with a poor prognosis; in fact, rather than a poor
prognosis, negative p16 expression was associated with better
prognosis. In other words, positive p16 expression tended to be
associated with poor prognosis (Fig.
3).
 | Table IVClinicopathological background,
outcome and comparison between positive and negative p16 groups
(average). |
Table IV
Clinicopathological background,
outcome and comparison between positive and negative p16 groups
(average).
| Variable | Number (%) | Positive p16
expression group | Negative p16
expression group | P-value |
|---|
| Number (%) | 103 | 37 (35.9) | 66 (64.1) | |
| Sex | | | | 0.41a |
|
Male | 59 (57.3) | 19 (18.4) | 40 (38.8) | |
|
Female | 44 (42.7) | 18 (17.5) | 26 (25.2) | |
| Median age | 70 | 69 | 71 | 0.46b |
| Age range | 50-87 | 50-80 | 51-87 | |
| Lymph nodes | | | | 0.38a |
|
Negative | 31 (30.1) | 9 (8.7) | 22 (21.4) | |
|
Positive | 72 (69.9) | 28 (27.2) | 44 (42.7) | |
| Margin status | | | | 0.28a |
|
R0 | 86 (83.5) | 33 (32.0) | 53 (51.5) | |
|
R1/R2 | 17 (16.5) | 4 (3.9) | 13 (12.6) | |
| Cytology | | | | 0.11a |
|
CY0 | 91 (88.3) | 30 (29.1) | 61 (59.2) | |
|
CY1 | 12 (11.7) | 7 (6.8) | 5 (4.9) | |
| Lymphatic
invasion | | | | 0.11a |
|
Negative | 17 (16.5) | 11 (10.7) | 6 (5.8) | |
|
Positive | 76 (73.8) | 26 (25.2) | 60 (58.2) | |
| Neural
invasion | | | | 0.24a |
|
Negative | 6 (5.8) | 4 (3.9) | 2 (1.9) | |
|
Positive | 97 (94.2) | 33 (32.0) | 64 (62.1) | |
| Vascular
invasion | | | | 0.80a |
|
Negative (v
0/1) | 21 (20.4) | 8 (7.8) | 13 (12.6) | |
|
Positive (v
2/3) | 82 (79.6) | 29 (28.1) | 53 (51.5) | |
|
Differentiation | | | | 0.71a |
|
Well/moderate | 95 (92.2) | 35 (44.0) | 60 (58.2) | |
|
Poor | 8 (7.8) | 2 (1.9) | 6 (5.8) | |
| T factor (UICC
8th) | | | | 0.63a |
|
T1/2 | 79 (76.7) | 27 (26.2) | 52 (50.5) | |
|
T3 | 24 (21.4) | 10 (9.7) | 14 (13.6) | |
| Stage (UICC
8th) | | | | 0.48c |
|
IA | 12 (11.7) | 3 (2.9) | 9 (8.7) | |
|
IB | 14 (13.6) | 3 (2.9) | 11 (10.7) | |
|
IIA | 5 (4.9) | 3 (2.9) | 2 (1.9) | |
|
IIB | 36 (35.0) | 15 (14.6) | 21 (20.4) | |
|
III | 36 (35.0) | 13 (12.6) | 23 (22.3) | |
In other tumors, such as laryngeal squamous
carcinoma, positive p16 expression has been significantly
associated with poor prognosis (17). Zhao et al (18) suggested that positive p16 expression
in non-small-cell lung carcinoma was associated with poor outcome.
In colon adenocarcinomas, p16 overexpression has been shown to
correlate with the clinical features of poorer prognosis, such as
sex, distal location, tumor grade, and stage (19). Meanwhile, in breast cancer, p16
overexpression was detected in approximately 20% of tumors and was
significantly associated with unfavorable prognostic factors
(20).
Among the four gene mutations frequently found in
pancreatic cancer, KRAS, TP 53, and SMAD 4 have also been related
to prognosis (5,8,14,15).
Positive lymph node metastasis and the presence of KRAS mutation
have been identified as independent prognostic markers according to
a multivariate analysis (5,14). Another multivariate analysis found
that the number of driver gene alterations among these four genes
remained independently associated with overall survival (21). Consistent with other reports, our
findings revealed the significant association between lymph node
metastasis and poor prognosis. However, negative p16 expression was
not necessarily associated with poor prognosis; instead, it was
associated with better prognosis. The examination of p16 in
combination with other genes such as KRAS, p53, and SMAD4 may find
correlations with prognosis and other clinicopathological factors
(14). In addition, we evaluated the
inactivation state of p16 through the expression level of p16 mRNA
using RNA-seq and found a correlation with protein expression level
on IHC in a small number of cases (Table III). However, we could not evaluate
the inactivation state of p16 using other factors. If the
relationship among the factors causing inactivation of p16, i.e.,
mutation, homozygous deletion, and promoter methylation, expression
level using RNA-seq, and protein expression on IHC can be examined
in more samples, more insights into the inactivation of p16 and
expression on IHC can be obtained.
Here are some limitations of our methods. We found
no significant difference in p16 expression status, and it was not
associated with poor prognosis. This can be due to our small sample
size, i.e., 103, as the statistically required size was 385.
Moreover, confounding factors, such as mutated KRAS, might worsen
the prognosis, but we have not investigated the relationship
between KRAS and prognosis. Unfortunately, we have not evaluated
the KRAS protein by immunostaining, which is one of the limitations
of our study.
Therefore, to confirm the relationship between the
inactivation of p16 and p16 expression on IHC, we evaluated p16
expression using RNA-seq. From a small sample of 8 cases, we
presented a correlation between p16 expression on IHC and mRNA
expression using RNA-seq (Table
III). In pancreatic cancer, inactivation of p16 has been
assessed by exon sequencing and has been reported to occur by
mutation, homozygous deletion, and promoter methylation of the p16
gene (1,5,22). In
this study, the number of samples might again be too small, and
therefore, the correlation between the factors causing inactivation
of p16 gene, namely mutation, homozygous deletion, and promoter
methylation, and mRNA expression could not be confirmed. In this
study, we considered the inactivation of p16 as a decrease in mRNA
expression level, extracted it from the information of RNA-seq, and
obtained a correlation with the range of staining on IHC. However,
p16 inactivation did not correlate with clinicopathological data.
As a limitation of RNA-seq, we do not use controls because we did
not analyze expression fluctuations and did not detect
differentially expressed genes, only expression level analysis and
normalization of p16 (CDKN2A).
As far as we searched, there were no reports
examining p16 inactivation and p16 immunostaining at the same time
in pancreatic cancer, and it was considered a novel report.
The inactivation status of p16 was evaluated using
mRNA expression level, and it was related to protein expression
level on IHC. If the p16 protein expression on IHC was low, p16
would have been inactivated. We defined low p16 protein expression
level on IHC (<10%) as negative p16 expression (Table III). The p16 expression status,
i.e., positive or negative, on IHC was not significantly associated
with clinicopathological factors including Overall survival
(Tables I and II).
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YI and IH analyzed and interpreted the patient data
regarding the pancreatic cancer was a major contributor in writing
the manuscript. FI, SC, HA, HYa, HN, HYo, WT collected tissue
samples and extracted mRNA from pancreatic tissue. MI performed the
pathological examination of the pancreatic cancer and interpreted
the tissue on IHC. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
This study was approved by the Chiba Cancer Center
Review Board (H29-006). All procedures followed were in accordance
with the ethical standards of the responsible committee on human
experimentation and with the Helsinki Declaration of 1964 and its
later amendments. Informed consent was obtained from all patients
in this study.
Patient consent for publication
Written informed consent was obtained from the
patients for publication of this study and accompanying
clinicopathological data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Spath C, Nitsche U, Muller T, Michalski C,
Erkan M, Kong B and Kleeff J: Strategies to improve the outcome in
locally advanced pancreatic cancer. Minerva Chir. 70:97–106.
2015.PubMed/NCBI
|
|
2
|
Waddell N, Pajic M, Patch AM, Chang DK,
Kassahn KS, Bailey P, Johns AL, Miller D, Nones K, Quek K, et al:
Whole genomes redefine the mutational landscape of pancreatic
cancer. Nature. 518:495–501. 2015.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Balachandran VP, Luksza M, Zhao JN,
Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U,
Senbabaoglu Y, et al: Identification of unique neoantigen qualities
in long-term survivors of pancreatic cancer. Nature. 551:512–516.
2017.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Russo AA, Tong L, Lee JO, Jeffrey PD and
Pavletich NP: Structural basis for inhibition of the
cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature. 395:237–243. 1998.PubMed/NCBI View
Article : Google Scholar
|
|
5
|
Schlitter AM, Segler A, Steiger K,
Michalski CW, Jäger C, Konukiewitz B, Pfarr N, Endris V,
Bettstetter M, Kong B, et al: Molecular, morphological and survival
analysis of 177 resected pancreatic ductal adenocarcinomas (PDACs):
Identification of prognostic subtypes. Sci Rep.
7(41064)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Ryan DP, Hong TS and Bardeesy N:
Pancreatic adenocarcinoma. N Engl J Med. 371:1039–1049.
2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Attri J, Srinivasan R, Majumdar S, Radotra
BD and Wig J: Alterations of tumor suppressor gene p16INK4a in
pancreatic ductal carcinoma. BMC Gastroenterol.
5(22)2005.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ohtsubo K, Watanabe H, Yamaguchi Y, Hu YX,
Motoo Y, Okai T and Sawabu N: Abnormalities of tumor suppressor
gene p16 in pancreatic carcinoma: Immunohistochemical and genetic
findings compared with clinicopathological parameters. J
Gastroenterol. 38:663–671. 2003.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Moskaluk CA, Hruban RH and Kern SE: p16
and K-ras gene mutations in the intraductal precursors of human
pancreatic adenocarcinoma. Cancer Res. 57:2140–2143.
1997.PubMed/NCBI
|
|
10
|
Sellers WR, Rodgers JW and Kaelin WG Jr: A
potent transrepression domain in the retinoblastoma protein induces
a cell cycle arrest when bound to E2F sites. Proc Natl Acad Sci
USA. 92:11544–11548. 1995.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Caldas C, Hahn SA, da Costa LT, Redston
MS, Schutte M, Seymour AB, Weinstein CL, Hruban RH, Yeo CJ and Kern
SE: Frequent somatic mutations and homozygous deletions of the p16
(MTS1) gene in pancreatic adenocarcinoma. Nat Genet. 8:27–32.
1994.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Schutte M, Hruban RH, Geradts J, Maynard
R, Hilgers W, Rabindran SK, Moskaluk CA, Hahn SA, Schwarte-Waldhoff
I, Schmiegel W, et al: Abrogation of the Rb/p16 tumor-suppressive
pathway in virtually all pancreatic carcinomas. Cancer Res.
57:3126–3130. 1997.PubMed/NCBI
|
|
13
|
Ueki T, Toyota M, Sohn T, Yeo CJ, Issa JP,
Hruban RH and Goggins M: Hypermethylation of multiple genes in
pancreatic adenocarcinoma. Cancer Res. 60:1835–1839.
2000.PubMed/NCBI
|
|
14
|
Oshima M, Okano K, Muraki S, Haba R, Maeba
T, Suzuki Y and Yachida S: Immunohistochemically detected
expression of 3 major genes (CDKN2A/p16, TP53, and SMAD4/DPC4)
strongly predicts survival in patients with resectable pancreatic
cancer. Ann Surg. 258:336–346. 2013.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Masetti M, Acquaviva G, Visani M, Tallini
G, Fornelli A, Ragazzi M, Vasuri F, Grifoni D, Di Giacomo S,
Fiorino S, et al: Long-term survivors of pancreatic adenocarcinoma
show low rates of genetic alterations in KRAS, TP53 and SMAD4.
Cancer Biomark. 21:323–334. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Jeong J, Park YN, Park JS, Yoon DS, Chi HS
and Kim BR: Clinical significance of p16 protein expression loss
and aberrant p53 protein expression in pancreatic cancer. Yonsei
Med J. 46:519–525. 2005.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Larque AB, Conde L, Hakim S, Alos L, Jares
P, Vilaseca I, Cardesa A and Nadal A: P16(INK4a)
overexpression is associated with CDKN2A mutation and worse
prognosis in HPV-negative laryngeal squamous cell carcinomas.
Virchows Arch. 466:375–382. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Zhao W, Huang CC, Otterson GA, Leon ME,
Tang Y, Shilo K and Villalona MA: Altered p16(INK4) and RB1
expressions are associated with poor prognosis in patients with
nonsmall cell lung cancer. J Oncol. 2012(957437)2012.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Lam AK, Ong K, Giv MJ and Ho YH: p16
expression in colorectal adenocarcinoma: Marker of aggressiveness
and morphological types. Pathology. 40:580–585. 2008.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Milde-Langosch K, Bamberger AM, Rieck G,
Kelp B and Loning T: Overexpression of the p16 cell cycle inhibitor
in breast cancer is associated with a more malignant phenotype.
Breast Cancer Res Treat. 67:61–70. 2001.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yachida S, White CM, Naito Y, Zhong Y,
Brosnan JA, Macgregor-Das AM, Morgan RA, Saunders T, Laheru DA,
Herman JM, et al: Clinical significance of the genetic landscape of
pancreatic cancer and implications for identification of potential
long-term survivors. Clin Cancer Res. 18:6339–6347. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cancer Genome Atlas Research Network.
Electronic address: uriandrew_aguirre@dfci.harvard.edusimpleandrew_aguirre@dfci.harvard.edu;
Cancer Genome Atlas Research Network: Integrated genomic
characterization of pancreatic ductal adenocarcinoma. Cancer Cell
32: 185-203.e13, 2017.
|