Introduction
With 1.8 million new cases and almost 861,000 deaths
in 2018 globally, colorectal cancer (CRC) is the third most
commonly diagnosed cancer in males and the second in females
according to the World Health Organization (1). Improvements in earlier cancer
detection and management, in combination with an increased
understanding of the molecular and genetic basis of the disease
will aid in better treatment decision and may also guide future
therapeutic approaches (2).
The microsatellite instability (MSI) pathway is
amongst the most important molecular pathways identified to cause
CRC, along with other pathways, such as the chromosomal
instability, CpG island methylator phenotype and serrated pathways
(3). MSI is the accumulation of
repeat length mutations in short DNA sequences and arises from
defects in the DNA mismatch repair (MMR) system, which corrects any
errors made by DNA polymerases during DNA replication (4). Tumours with MSI have a better
prognosis than microsatellite stable CRC (5). Detection of MSI status performed using
PCR-based methods or deficiency status of MMR proteins, including
DNA mismatch repair protein Mlh1 (MLH1), DNA mismatch repair
protein Msh2 (MSH2), DNA mismatch repair protein Msh6 (MSH6) and
mismatch repair endonuclease PMS2 (PMS2), detected using
immunohistochemistry (IHC) is routinely advocated to detect
hereditary cancer, such as in Lynch syndrome, and also to predict
the prognosis/chemotherapy response including the response to most
recent immunotherapies (6).
CRC is known to be a heterogenous disease
characterized by different molecular subtypes. Consensus molecular
subtyping is most beneficial in the identification of a specific
targeted therapy for both initial treatments and in metastatic
settings (7-9).
Similar attempts to classify CRC have been made by evaluating
multiple markers, such as caudal-type homeobox protein 2 (CDX2),
BRCA1, p53, adenomatous polyposis coli, β-catenin and other DNA
repair proteins such as MMR proteins, O6-methylguanine
DNA methyltransferase and excision repair cross-complementing 1,
and correlate them with survival and response to therapy (10). CDX2 has been well-established as a
diagnostic marker for CRC and its downregulation is associated with
poor differentiation and MMR deficiency (11). However, less is known about the
utility of BRCA1 in CRC, except in a few studies where the
expression of BRCA1 predicting a better survival rate has been
reported (12,13). The role of BRCA1 as a tumour
suppressor gene was confirmed by its action in DNA damage repair
(DDR), mediated mainly via homologous recombination-based pathways
(14). Apart from mutations,
epigenetic mechanisms, such as those mediated by miRNAs, are also
known to mediate loss of BRCA1 function (15). miRNAs are small noncoding RNAs of
19-22 base pairs that are generated by a series of enzymatic
processes in the nucleus and cytoplasm. Apart from affecting
multiple cellular processes, several miRs, such as miR-16, miR-24,
miR-188 and the miR-183-96-182 cluster have been implicated in DNA
damage response and DNA repair (16,17).
Defects in DDR drive cancer development by fostering DNA mutations,
but also provide cancer-specific vulnerabilities that can be
therapeutically exploited. The recent approval and use of multiple
newer modalities of cancer therapy, such as using poly ADP ribose
polymerase inhibitors through the approach of synthetic lethality,
or checkpoint inhibitors and immunomodulatory drugs, have made them
promising candidates for the treatment of multiple types of cancers
(18-20).
Although the incidence of CRC in India is much lower
than in the West, a higher proportion of right sided and grade III
tumours has been reported and is associated with significant
mortality and morbidity (21). With
an interest to evaluate and correlate clinical features with known
prognostic markers, the present study was performed on a
retrospective series from the pathology archives. The present study
analysed the association between MMR proteins, CDX2 and BRCA1 to
determine the utility of their inter-relationship in the
identification of subclasses amenable to specific therapies.
Materials and methods
Selection of primary colorectal cancer
samples
Seventy-six colorectal cancer tumour blocks were
identified and selected for the study. These cases were examined
and reported by the Department of Pathology, St. John's Medical
College and Hospital, Bangalore, India between 2013 and 2017. The
present retrospective study was approved by the Institutional
Ethical Committee, St John's Medical College and Hospital.
Representative formalin-fixed and paraffin-embedded tumour blocks
from each of the selected cases were obtained for the study. All
tumours with a >50% tumour content, as estimated by a
pathologist from the consecutive series, were chosen for analysis.
Clinico-pathological characteristics, such as age, sex, grade,
pathological and lymph node stage, histological type, lymphocytic
response, lymphovascular invasion and tumour site were obtained
from the clinical and histopathological records from the
hospital.
Tissue microarray (TMA) construction
and IHC of CDX2, BRCA1 and mismatch repair proteins MSH2, MSH6,
MLH1 and PMS2
TMA was constructed using the Quick Ray manual
tissue microarrayer (Unitma Co., Ltd.). A master block grid plan
was made with adequate precautions to ensure an accurate
orientation and unambiguous specimen identification. Block
construction was performed according to the manufacturer's
instructions. Two cores of 1.5 mm each were taken from each tumour
block. Sections were cut and stained with hematoxylin and eosin
(H&E) to confirm the adequate representation of each tumour.
Cores with <100 interpretable tumour cells were excluded from
the analysis.
IHC was performed for CDX2, BRCA1 and MMR proteins
(MLH1, PMS2, MSH2 and MSH6), according to standard procedures.
Briefly, 5-µm thick sections were placed on poly-L-lysine-coated
slides and subjected to deparaffinization in xylene and rehydrated
in graded alcohol. Following blocking endogenous peroxidase with a
3% hydrogen peroxide solution, antigen retrieval was performed in
0.01 mol/l EDTA buffer at pH 8 using a heat triggered multi-epitope
retrieval system (PathnSitu Biotechnologies Pvt. Ltd.) for 15 min
at 90˚C. Primary blocking was done with 1% BSA (Sigma-Aldrich;
Merck KGaA) for 30 min at room temperature. Details of the primary
antibody clone, source and the dilutions are shown in Table SI. Primary antibodies were applied
for 1 h at room temperature. Sections were further incubated with
secondary antibody (cat. no. K5007; EnVision Detection System;
Dako; Agilent Technologies, Inc.) for 20 min as per the kit
instructions, followed by colour development using 3,
3-diaminobenzidine for 10 min. Sections were counterstained with
haematoxylin for 5 min at room temperature and mounted after
dehydration in graded alcohol and xylene. Appropriate positive and
negative controls were run for each batch.
Evaluation of CDX2, BRCA1 and MMR
proteins
For CDX2 and BRCA1 scoring, each tumour core was
scored for intensity as follows: 0=no staining; 1=weak staining;
2=moderate staining; 3=strong staining. The percentage of cells
stained was estimated from 0-100%. The histochemical score (H score
range, 0-300) was calculated by multiplying the intensity and the
percentage of staining. Although BRCA1 staining was observed in
both the nucleus and the cytoplasm, only its nuclear presence was
evaluated, indicating BRCA1 functional ability. A nuclear H score
of ≥10 was considered as positive BRCA1 and CDX2 expression. The H
score was used for the analysis of both proteins to obtain a
quantitative estimate with a broad dynamic range from
0-300(22). In the absence of any
standard diagnostic criteria for proteins such as BRCA1 and CDX2,
the H score method was followed for quantitation.
MMR proteins were scored only on the percentage of
stained tumour cells, irrespective of staining intensity. Standard
guidelines were followed where any presence of MMR proteins in the
nucleus is considered adequate and acceptable to record as intact
expression (23). Intact normal
staining of non-tumour cells was considered as an internal positive
control. Cases with ≥10% tumour cells showing nuclear staining were
considered positive (intact expression). Weak focal nuclear stain
in <10% of tumour cells was considered focal expression and a
complete absence of nuclear stain in the presence of the positive
internal control (lymphocytes and stromal cells) was considered
negative (loss of expression). The dual loss of either MSH2 and
MSH6 or MLH1 and PMS2, or the isolated loss of PMS2 was considered
as an MMR-deficient group (dMMR). Intact/focal protein presence of
either MSH2/MSH6 or MLH1/PMS2 was considered as an MMR-proficient
group (pMMR).
H&E-stained microscopic sections of tumors were
simultaneously evaluated by two trained pathologists to confirm the
presence of above proteins in the tumor cells in comparison to
normal cells (24). Each tissue
microarray core was examined microscopically by the same
pathologists independently to count ~1,500 cells in three different
areas on the tissue section. The mean expression levels of both the
observers were taken as final scores. In case of disagreement, the
final score was determined by consensus after re-examination.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR) of miRNAs
The methods used for nucleic acid extraction and
RT-qPCR have been described in detail in our previous publication
(25). In brief, two 20-µm sections
taken from each patient's tumour block were deparaffinized using
heat, and then subjected to overnight digestion using proteinase K
(cat. no. 19133; Qiagen GmbH). Total RNA was then extracted using
TRI reagent according to manufacturer's instructions (cat. no.
T9424; Sigma-Aldrich; Merck KGaA). RNA quantification was performed
using the Qubit RNA BR Assay kit (cat. no. Q10210; Invitrogen;
Thermo Fisher Scientific, Inc.) on a Qubit 2.0 Fluorometer (cat.
no. Q32866; Invitrogen; Thermo Fisher Scientific, Inc). Samples
with a yield of ≥500 ng RNA and adequate transcript preservation to
show amplification by RT-qPCR were used for subsequent
experiments.
miRNA present in total RNA extracted as described
above was converted to cDNA using stem-loop primers specific for
the chosen miR, according to published protocols. Detailed
methodology for quantification of miR using qPCR is provided in our
previous publication (26).
Briefly, 50 ng of total RNA was used for cDNA conversion using the
TaqMan microRNA Reverse Transcription kit (cat. no. 4366596;
Applied Biosystems; Thermo Fisher Scientific, Inc.). The expression
levels of a selected set of DDR miRs (miR183, miR96 and miR182)
were determined, along with the control miR (RNU48). The assay IDs
(cat. no. 4427975; Applied Biosystems; Thermo Fisher Scientific,
Inc,) and sequences of the miRs are shown in Tables SII and SIII. Cycle threshold (Ct) values for the
test miRs were normalized relative to the mean Ct value of the
control miR for each sample as ΔCt. The relative normalized units
of expression of the test miRs were calculated as 15-ΔCt,
representing the dynamic range of the assay as being 15 Cts.
Statistical analysis
Descriptive analysis was used to tabulate the
clinical variables amongst the various groups segregated based on
the presence or absence of the chosen protein markers. All markers
were expressed as the proportion of cases noted as positive or
negative. Parametric or non-parametric tests of significance, such
as paired Student's t-test or the Mann Whitney U test, based on the
normality of distribution, were applied to determine the levels of
significance in the distribution of chosen variables between the
groups. The correlation between protein expression levels was
calculated using Pearson's correlation coefficient. In the absence
of prior available data for combined loss of BRCA1 and MMR
proteins, sample size estimates were not attempted. Analysis was
performed on XLStat software (version 2019.4.2; Addinsoft).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Clinical characteristics of the
cohort
The mean and median age of the CRC patients was 54.9
and 55 years, respectively. Of the 76 tumours studied, two were
uninterpretable in CDX2 and BRCA1 IHC, one tumour yielded
insufficient RNA and one tumour had loss of all MMR proteins, and
therefore, only 72 (95%) tumours could be satisfactorily
interpreted. A slight male preponderance was noted with 58% (42/72)
in the group of tumours selected. The right and left sided tumours
were equal in ratio. Most tumours belonged to stage II (38%) and
stage III (43%). Details on the tumour grade were available for 70%
(51/72) of tumours, and most (84%) of them were grade II tumours.
Approximately 40% (29/72) of the tumours had a high lymphocytic
response and lympho-vascular invasion was present in 43% (31/72) of
tumours.
Of the 36 right sided tumors, 12 (33%) were mucinous
tumors. The presence of the CDX2, BRCA1 proteins and the MMR status
were further examined among mucinous and non-mucinous tumours. No
statistically significant difference was observed between the two
groups (data not shown).
Staining pattern and distribution of
all proteins
CDX2 is known to be widely expressed in the large
intestine (11) and showed nuclear
staining with bright intensity. BRCA1 staining intensity was lower
compared with CDX2 expression and varied across tumours. The
intensity of MMR protein staining also varied widely.
Expression of MMR proteins
Among the four MMR proteins studied, MSH2 and PMS2
had intact/focal expression in 89% of the tumours. Table I details the expression levels of
all the MMR proteins. MLH1 protein was detected in a small
proportion of tumours (42%). MSH6 was always intact (60%) in the
presence of MSH2, while the presence of PMS2 was observed in 34/42
tumours when MLH1 was absent. Tumours with the dual loss of MSH2
and MSH6 (11%) had intact protein status for either MLH1 and/or
PMS2, and vice versa (10%). The dual presence of MLH1 and PMS2 was
observed in 29% of the tumours, while only one tumour had isolated
loss of PMS2. A total of 22% (16/72) of the tumours were
categorized as dMMR, as they had a dual loss of MSH2/MSH6 or MLH1/
PMS2, or the isolated loss of PMS2. Representative microscopic IHC
images of all the MMR proteins showing positive nuclear stain are
shown in Fig. 1A-D. As shown in
Fig. 1, MLH1 presented with the
lowest staining intensity compared to other MMR proteins. Table II shows the association of MMR
status with clinical variables. None of the clinical variables were
significantly different between the MMR proficient and deficient
subtypes, aside from dMMR tumours, which had a higher lymph node
spread (P-0.02). Among the dMMR tumours, 75% of them belonged to
males and almost 70% of them belonged to stage III, although this
was not statistically significant.
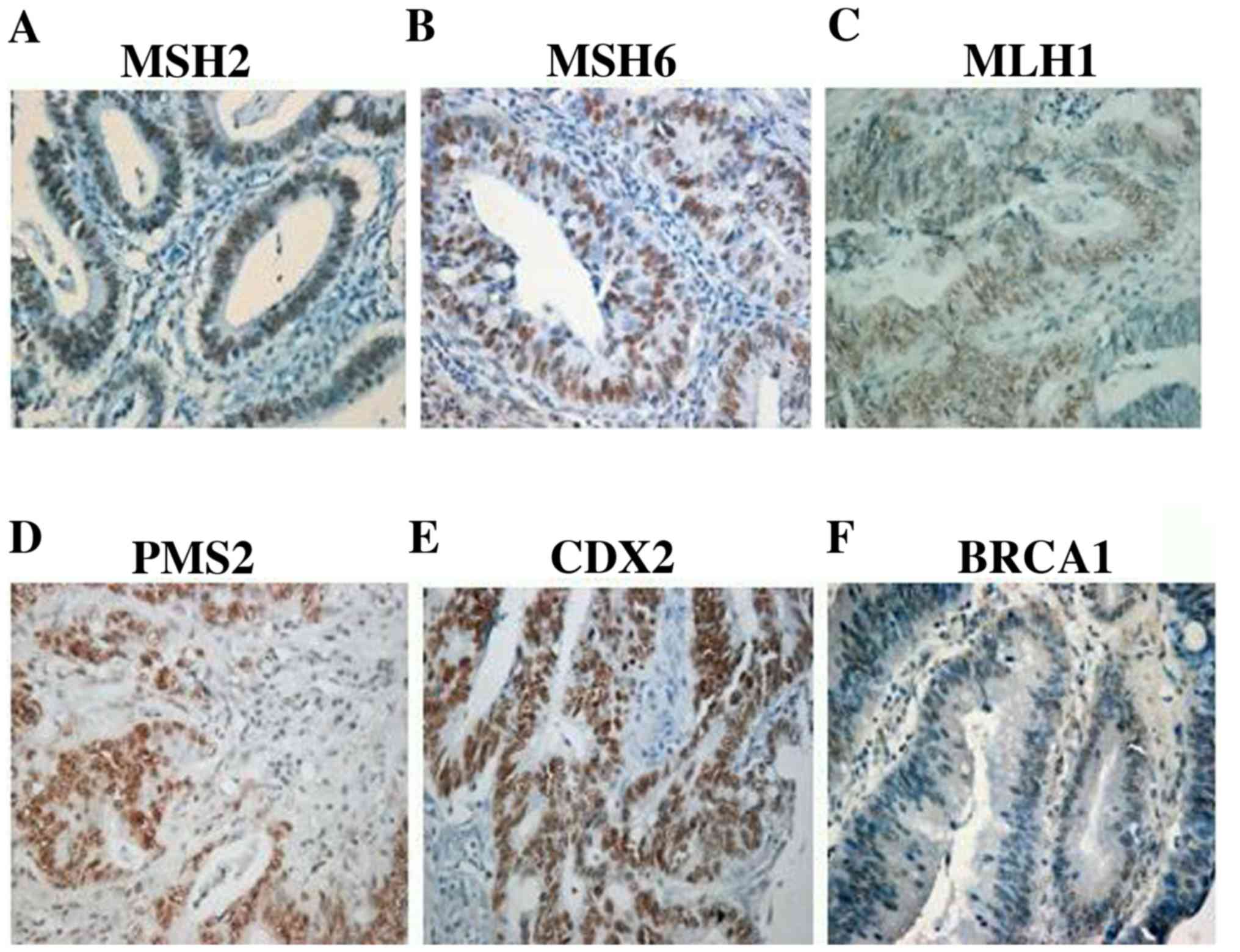 | Figure 1Representative microscopic images of
MMR proteins, CDX2 and BRCA1. Nuclear staining of MMR proteins (A)
MSH2, (B) MSH6, (C) MLH1 and (D) PMS2 in the presence of internal
positive controls (lymphocytes and stromal cells). Nuclear staining
of (E) CDX2 (H score, 100x3-300) and (F) BRCA1 (H score, 50x2-100)
in tumour cells. Magnification, x20. MMR, mismatch repair; DNA
mismatch repair protein Msh2; MSH6, DNA mismatch repair protein
Msh6; MLH1, DNA mismatch repair protein Mlh1; PMS2, mismatch repair
endonuclease PMS2; CDX2, caudal- type homeobox protein 2. |
 | Table IDistribution of MMR protein expression
in colorectal tumours. |
Table I
Distribution of MMR protein expression
in colorectal tumours.
| IHC marker | Loss of
expression | Intact
expression | Focal expression |
|---|
| MLH1 | 42(58) | 22(31) | 8(11) |
| PMS2 | 8(11) | 49(68) | 15(21) |
| MSH2 | 8(11) | 57(79) | 7(10) |
| MSH6 | 29(40) | 30(42) | 13(18) |
| MLH1 and PMS2 dual
loss | 7(10) | - | - |
| MSH2 andMSH6 dual
loss | 8(11) | - | - |
| Isolated PMS2
loss | 1(1) | - | - |
| dMMR | 16(22) | - | - |
| pMMR | 56(78) | - | - |
 | Table IIDistribution of CDX2, BRCA1 and MMR
proteins amongst clinical variables. |
Table II
Distribution of CDX2, BRCA1 and MMR
proteins amongst clinical variables.
| Marker | CDX2 | BRCA1 | MMR |
| Staining | Positive, n-51
(71%) | Negative, n-21
(29%) | Positive, n-15
(21%) | Negative, n-57
(79%) | Proficient, n-56
(78%) | Deficient, n-16
(22%) |
|---|
| Variable | | | | | | |
| Age |
|
Mean | 55.8 | 52.9 | 55.6 | 54.8 | 54.7 | 55.7 |
|
Median | 56 | 55 | 60 | 55 | 55 | 58 |
| Sex |
|
Male | 28(55) | 14(67) | 8(53) | 34(60) | 30(54) | 12(75) |
|
Female | 23(45) | 7(33) | 7(47) | 23(40) | 26(46) | 4(25) |
| Tumor site |
|
Right | 23(45) | 13(62) | 8(53) | 28(49) | 27(48) | 9(56) |
|
Left | 28(55) | 8(38) | 7(47) | 29(51) | 29(52) | 7(44) |
| Grade |
|
I | 2(5) | 1(10) | 0 (0) | 3(7) | 2(5) | 1(10) |
|
II | 36(88) | 7(70) | 9(90) | 34(83) | 37(90) | 6(60) |
|
III | 3(7) | 2(20) | 1(10) | 4(10) | 2(5) | 3(30) |
| LN status |
|
N0 | 29(57) | 10(48) | 9(60) | 30(53) | 34(61) | 5(31)a |
|
N1 | 14(27) | 4(19) | 4(27) | 14(25) | 14(25) | 4(25) |
|
N2 | 8(16) | 7(33) | 2(13) | 13(23) | 8(14) | 7(44) |
| Stage |
|
I | 7(14) | 3(14) | 2(13) | 8(14) | 8(14) | 2(13) |
|
II | 20(39) | 7(33) | 7(47) | 20(35) | 24(43) | 3(19) |
|
III | 21(41) | 10(48) | 6(40) | 25(44) | 20(36) | 11(69) |
|
IV | 3(6) | 1(5) | 0 (0) | 4(7) | 4(7) | 0 (0) |
CDX2 and BRCA1 expression
An intense nuclear staining of CDX2 protein in
>10% of the tumour nuclei was considered positive and observed
in 71% (51/72) of tumours (Fig. 1E;
H score, 100x3-300). Clinical variables, such as age, sex, grade or
stage did not differ between CDX2-positive and -negative tumours
(Table II), although a higher
proportion of right sided tumours was CDX2-negative (62% in right
tumours vs. 45% in left tumours; P-0.2). Two-thirds (67%) of the
CDX2-negative tumours belonged to males and more than half of the
CDX-positive tumours were lymph node-negative.
BRCA1 nuclear expression was observed in only 21%
(15/72) of the tumour samples (Fig.
1F; H score, 50x2-100). Cytoplasmic expression of BRCA1 was
observed in an additional 14% (10/72) of tumours; however, they
were considered negative. No significant differences were seen for
any clinical variables between BRCA1-positive or -negative tumours,
although a higher proportion of BRCA1-positive tumours (60%) was
lymph-node negative (Table
II).
Association of MMR proteins with CDX2
and BRCA1 expression
Association of CDX2 loss is often reported to be
high in MMR-deficient tumours (10,11).
In the present study, 44% (7/16) of dMMR tumours showed loss of
CDX2, compared with 25% (14/56) observed in pMMR tumours (P-0.1).
However, BRCA1 and MMR were inversely correlated, with 38% (6/16)
of the dMMR group expressing BRCA1 protein, compared with only 16%
(9/56) in the pMMR subgroup (P-0.06). When the expressional pattern
of BRCA1 and CDX2 were compared as H scores, a negative correlation
was found between the two proteins (Pearson's correlation
coefficient, -0.13; P-0.2), as expected.
Combined loss of BRCA1 and MMR
proteins show a higher expression of DDR miRNAs
Subsequently, the tumours were divided into two
separate classes based on the dual presence and absence of BRCA1
and MMR status. While two-thirds of the tumours (53/72) showed
either BRCA1 or MMR protein expression, only a small proportion of
tumours (12.5%) had the combined presence of both. A small subset
(10/72; 14%) of tumours was both BRCA1 negative and MMR deficient.
When the clinical variables were compared between these groups
(Table III), tumours with the
combined loss of BRCA1 and MMR proteins showed aggressive features,
such as a younger age, male preponderance and a higher proportion
of grade III and stage III tumours. There was no difference in CDX2
expression between the two groups.
 | Table IIIComparison of clinical variables
between tumors with dual loss of BRCA1 and MMR with other
tumors. |
Table III
Comparison of clinical variables
between tumors with dual loss of BRCA1 and MMR with other
tumors.
| Variable | Dual loss of BRCA1
and MMR, n-10 (14%) | Othersa, n-62 (86%) | P-value |
|---|
| Age | | | NS |
|
Mean | 50.9 | 55.6 | |
|
Median | 50.5 | 55.5 | |
| Sex | | | NS |
|
Male | 8(80) | 34(55) | |
|
Female | 2(20) | 28(45) | |
| Tumor site | | | NS |
|
Right | 5(50) | 31(50) | |
|
Left | 5(50) | 31(50) | |
| Grade | | | 0.072 |
|
I | 1(14) | 2(5) | |
|
II | 3(43) | 40(91) | |
|
III | 3(43) | 2(5) | |
| LN status | | | 0.036b |
|
N0 | 3(30) | 36(58) | |
|
N1 | 2(20) | 16(26) | |
|
N2 | 5(50) | 10(16) | |
| Stage | | | NS |
|
I | 2(20) | 8(13) | |
|
II | 1(10) | 26(42) | |
|
III | 7(70) | 24(39) | |
|
IV | 0 (0) | 4(6) | |
| CDX2 | | | NS |
|
Positive | 4(40) | 17(27) | |
|
Negative | 6(60) | 45(73) | |
| miR-183 | | | |
|
Mean | 8.3 | 7.7 | |
| miR-182 | | | 0.01b |
|
Mean | 10.2 | 9.3 | |
| miR-96 | | | |
|
Mean | 7.4 | 6.8 | |
When the distribution of the miRs implicated in
defective DDR (miR-183, miR-96 and miR-182) was examined in the two
groups, tumours with the combined loss of BRCA1 and MMR showed
trends of higher expression of all the three miRs (P-0.01,
miR-182), as shown in Table III,
indicating defective pathways for DNA damage repair in these
tumours.
Discussion
CRC is the most common malignancy in the western
world and is associated with a significant morbidity. Among the
different molecular pathways involved, although MSI contributes to
less than one quarter of CRC cases, defects in MMR proteins have
been implicated both in carcinogenesis and CRC progression
(27). Although 90% of hereditary
CRC present with MSI, it is limited to 15-20% of sporadic tumours
(28). The present study showed
that 22% of tumours presented with MMR deficiency, consistent with
previously published studies (21,29).
An earlier study on a cohort from a similar setting reported a
deficient MMR prevalence of 22.9% (21). The present study observed loss of
MLH1 protein in more than half of tumours of the cohort. Weaker
staining patterns of MLH1, patchy and heterogenous staining pattern
of MMR proteins, inactivation by promoter methylation and mutations
are considered as multiple reasons for loss of MLH1(30). The results of the present study are
consistent with observations found in previous studies (30,31).
CDX2, a nuclear transcription factor implicated in
CRC prognosis, was also found to be lowly expressed (56%) in MMR
deficient tumours and a higher proportion of CDX2 negative tumours
was right-sided. While some reports (32) have shown similar presence of CDX2
positivity in 50% of dMMR tumours, other reports indicated that the
low/loss of CDX2 expression was significantly associated with MMR
deficiency and with right-sided tumours (10,33).
With the advent of therapeutics that can target
tumours with BRCA1/2 mutations, CRC tumours are also investigated
for mutations in these genes. Although MMR and BRCA1 status is
investigated independently, their combined loss has been reported
in very few studies. A recent study (34) has shown that BRCA1 mutations are
present in 1.1% of CRC tumours using next-generation sequencing on
a 592-gene panel, which was the first study to show that BRCA1/2
mutations are more frequent in MSI high (MSI-H) tumours, and
independently associated with higher tumour mutational burden.
However, other studies have performed combined testing for BRCA1/2
mutational screening, along with MMR proteins, while screening for
hereditary cancers (35). Other
studies have reported IHC detection of BRCA1 in CRC (12,13). A
higher proportion of BRCA1 staining in these reports may be due to
both cytoplasmic and nuclear staining being considered as positive.
The present study considered nuclear presence in >10% of cells
as BRCA1 positive, which perhaps is the reason for the low BRCA1
positivity observed.
The tumours in the present study, with dual loss of
BRCA1 and MMR, showed a high expression of miRs (miR-183-96-182
cluster) implicated in DDR, in addition to other aggressive
features. This cluster is proven to be highly expressed in CRC
tumours, promoting tumorigenesis, cancer progression and metastasis
(36). These DDR miRs impair the
homologous recombination mediated DNA repair by acting as negative
regulators of the genes involved in the DDR pathway and may serve
as predictive biomarkers for prognosis and potential therapeutic
targets in CRC treatment (37). The
present study, a retrospective CRC collected from pathology
archives, has several limitations. A small sample size and TMA
design restricting the evaluation of MMR proteins, which are known
to be heterogenous in expression, may have an impact on the MMR and
BRCA1 status. Moreover, although BRCA1 antibody (clone-MS110) is
the most widely used, it has issues with specificity and
sensitivity (38). While other
studies have shown association of favourable prognostic factors,
such as young age, well-differentiated tumours and low lymph node
positivity with dMMR status, the present study observed a
significant association with a higher lymph node positive status,
which could be due to a higher proportion of stage III disease
(43%) in the present cohort. Although the present study showed that
the incidence of MSI differs between stage II and stage III
diseases, stage-specific analysis was not performed due to the
small sample size. A lack of prognostic information on the present
CRC series has limited ability of validating the effect of dual
loss on clinical progression and requires validation in a larger
series with a clinical outcome.
Newer modalities of treatment with immune checkpoint
inhibitors are approved for treatment of colorectal cancers with
MSI-H (6). However, the approval is
limited to refractory mismatch deficient colorectal tumours in a
metastatic setting (20). Although
extensive efforts to explore additional combination therapies are
ongoing, not all patients uniformly benefit from newer therapeutic
modalities, which showcases the need to identify predictive
biomarkers for a better selection of the patients (2). The present approach of using IHC-based
markers to identify prognostic subtypes shows an easy, novel, and
adaptable approach for subtyping colorectal cancer and should be
further investigated in a larger series to confirm its
relevance.
Supplementary Material
Antibody details.
Control miRNA details.
DNA damage repair miRNA details.
Acknowledgements
Not applicable.
Funding
This study was supported by the Department of
Biotechnology (DBT), India (grant no. BT/MED/30/SP11263/2015).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SR performed miRNA experiments and data analysis.
ACE was involved in the construction of TMA, performing IHC and
sample collection. MC reported the histological details of tumors.
BJ identified the tumor blocks and collated clinical information.
SS conceived and designed the study. JSP was involved in
interpretation of all markers performing histological examination,
analysis of data, conception and design of the study and drafting
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All procedures performed in this study involving
human participants were in accordance with the ethical standards of
the Institutional Ethical Committee, St John's Medical College and
Hospital, Bangalore and approved by the same institute. Waiver of
consent has been approved for this study since this was a
retrospective study that was planned on specimens received for
routine diagnostic purpose in the Department of Pathology, St
John's Medical College and Hospital, Bangalore. All details of
personal information regarding patients were blinded to the
investigators. There was no direct contact between the researcher
and participant.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ferlay J, Colombet M, Soerjomataram I,
Mathers C, Parkin DM, Piñeros M, Znaor A and Bray F: Estimating the
global cancer incidence and mortality in 2018: GLOBOCAN sources and
methods. Int J Cancer. 144:1941–1953. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Hagan S, Orr MC and Doyle B: Targeted
therapies in colorectal cancer-an integrative view by PPPM. EPMA J.
4(3)2013.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kocarnik JM, Shiovitz S and Phipps AI:
Molecular phenotypes of colorectal cancer and potential clinical
applications. Gastroenterol Rep (Oxf). 3:269–276. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Kerr DJ and Midgley R: Defective mismatch
repair in colon cancer: A prognostic or predictive biomarker? J
Clin Oncol. 28:3210–3212. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Nojadeh JN, Behrouz Sharif S and Sakhinia
E: Microsatellite instability in colorectal cancer. Excli J.
17:159–168. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Le DT, Uram JN, Wang H, Bartlett BR,
Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et
al: PD-1 blockade in tumors with mismatch-repair deficiency. N Engl
J Med. 372:2509–2520. 2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Guinney J, Dienstmann R, Wang X, de
Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda
G, Angelino P, et al: The consensus molecular subtypes of
colorectal cancer. Nat Med. 21:1350–1356. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Lenz HJ, Ou FS, Venook AP, Hochster HS,
Niedzwiecki D, Goldberg RM, Mayer RJ, Bertagnolli MM, Blanke CD,
Zemla T, et al: Impact of consensus molecular subtype on survival
in patients with metastatic colorectal cancer: Results From
CALGB/SWOG 80405 (Alliance). J Clin Oncol. 37:1876–1885.
2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ragulan C, Eason K, Fontana E, Nyamundanda
G, Tarazona N, Patil Y, Poudel P, Lawlor RT, Del Rio M, Koo SL, et
al: Analytical validation of multiplex biomarker assay to stratify
colorectal cancer into molecular subtypes. Sci Rep.
9(7665)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Tóth C, Sükösd F, Valicsek E, Herpel E,
Schirmacher P and Tiszlavicz L: Loss of CDX2 gene expression is
associated with DNA repair proteins and is a crucial member of the
Wnt signaling pathway in liver metastasis of colorectal cancer.
Oncol Lett. 15:3586–3593. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Olsen J, Eiholm S, Kirkeby LT, Espersen
ML, Jess P, Gögenür I, Olsen J and Troelsen JT: CDX2 downregulation
is associated with poor differentiation and MMR deficiency in colon
cancer. Exp Mol Pathol. 100:59–66. 2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Grabsch H, Dattani M, Barker L, Maughan N,
Maude K, Hansen O, Gabbert HE, Quirke P and Mueller W: Expression
of DNA double-strand break repair proteins ATM and BRCA1 predicts
survival in colorectal cancer. Clin Cancer Res. 12:1494–1500.
2006.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Wang GH, Zhao CM, Huang Y, Wang W, Zhang S
and Wang X: BRCA1 and BRCA2 expression patterns and prognostic
significance in digestive system cancers. Hum Pathol. 71:135–144.
2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Krajewska M, Fehrmann RS, de Vries EG and
van Vugt MA: Regulators of homologous recombination repair as novel
targets for cancer treatment. Front Genet. 6(96)2015.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Petrovic N, Davidovic R, Bajic V,
Obradovic M and Isenovic RE: MicroRNA in breast cancer: The
association with BRCA1/2. Cancer Biomark. 19:119–128.
2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Wang Y, Huang JW, Calses P, Kemp CJ and
Taniguchi T: miR-96 downregulates REV1 and RAD51 to promote
cellular sensitivity to cisplatin and PARP inhibition. Cancer Res.
72:4037–4046. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Moskwa P, Buffa FM, Pan Y, Panchakshari R,
Gottipati P, Muschel RJ, Beech J, Kulshrestha R, Abdelmohsen K,
Weinstock DM, et al: miR-182-mediated downregulation of BRCA1
impacts DNA repair and sensitivity to PARP inhibitors. Mol Cell.
41:210–220. 2011.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Wahner Hendrickson AE, Menefee ME,
Hartmann LC, Long HJ, Northfelt DW, Reid JM, Boakye-Agyeman F,
Kayode O, Flatten KS, Harrell MI, et al: A Phase I Clinical Trial
of the Poly(ADP-ribose) polymerase inhibitor veliparib and weekly
topotecan in patients with solid tumors. Clin Cancer Res.
24:744–752. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Svensson MC, Warfvinge CF, Fristedt R,
Hedner C, Borg D, Eberhard J, Micke P, Nodin B, Leandersson K and
Jirström K: The integrative clinical impact of tumor-infiltrating T
lymphocytes and NK cells in relation to B lymphocyte and plasma
cell density in esophageal and gastric adenocarcinoma. Oncotarget.
8:72108–72126. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Byrne M and Saif MW: Selecting treatment
options in refractory metastatic colorectal cancer. Onco Targets
Ther. 12:2271–2278. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Nayak SS, Roy P, Arora N, Arun I, Roy MK,
Banerjee S, Mallick I and Mallath MK: Prevalence estimation of
microsatellite instability in colorectal cancers using tissue
microarray based methods-A tertiary care center experience. Indian
J Pathol Microbiol. 61:520–525. 2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mazières J, Brugger W, Cappuzzo F, Middel
P, Frosch A, Bara I, Klingelschmitt G and Klughammer B: Evaluation
of EGFR protein expression by immunohistochemistry using H-score
and the magnification rule: Re-analysis of the SATURN study. Lung
Cancer. 82:231–237. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Bartley AN, Hamilton SR, Alsabeh R,
Ambinder EP, Berman M, Collins E, Fitzgibbons PL, Gress DM, Nowak
JA, Samowitz WS, et al: Members of the Cancer Biomarker Reporting
Workgroup, College of American Pathologists Template for reporting
results of biomarker testing of specimens from patients with
carcinoma of the colon and rectum. Arch Pathol Lab Med. 138:166–70.
2014.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Jørgensen AS, Rasmussen AM, Andersen NKM,
Andersen SK, Emborg J, Røge R and Østergaard LR: Using cell nuclei
features to detect colon cancer tissue in hematoxylin and eosin
stained slides. Cytometry A. 91:785–793. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Korlimarla A, Prabhu JS, Anupama CE,
Remacle J, Wahi K and Sridhar TS: Separate quality-control measures
are necessary for estimation of RNA and methylated DNA from
formalin-fixed, paraffin-embedded specimens by quantitative PCR. J
Mol Diagn. 16:253–260. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Korlimarla A, Prabhu JS, Remacle J,
Rajarajan S, Raja U, C E A, Srinath BS, Manjunath S, K S G, Correa
M, et al: Identification of BRCA1 deficiency using multi-analyte
estimation of BRCA1 and its repressors in FFPE tumor samples from
patients with triple negative breast cancer. PLoS One.
11(e0153113)2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ryan E, Sheahan K, Creavin B, Mohan HM and
Winter DC: The current value of determining the mismatch repair
status of colorectal cancer: A rationale for routine testing. Crit
Rev Oncol Hematol. 116:38–57. 2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Boland CR and Goel A: Microsatellite
instability in colorectal cancer. Gastroenterology.
138:2073–2087.e3. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kumar A, Jain M, Yadav A, Kumari N and
Krishnani N: Pattern of mismatch repair protein loss and its
clinicopathological correlation in colorectal cancer in North
India. S Afr J Surg. 56:25–29. 2018.PubMed/NCBI
|
|
30
|
Chen W, Swanson BJ and Frankel WL:
Molecular genetics of microsatellite-unstable colorectal cancer for
pathologists. Diagn Pathol. 12(24)2017.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Öhrling K, Edler D, Hallström M and
Ragnhammar P: Mismatch repair protein expression is an independent
prognostic factor in sporadic colorectal cancer. Acta Oncol.
49:797–804. 2010.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Neumann J, Heinemann V, Engel J, Kirchner
T and Stintzing S: The prognostic impact of CDX2 correlates with
the underlying mismatch repair status and BRAF mutational status
but not with distant metastasis in colorectal cancer. Virchows
Arch. 473:199–207. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Baba Y, Nosho K, Shima K, Freed E, Irahara
N, Philips J, Meyerhardt JA, Hornick JL, Shivdasani RA, Fuchs CS,
et al: Relationship of CDX2 loss with molecular features and
prognosis in colorectal cancer. Clin Cancer Res. 15:4665–4673.
2009.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Naseem M, Xiu J, Salem ME, Goldberg RM,
Vanderwalde AM, Grothey A, Philip PA, Seeber A, Puccini A, Tokunaga
R, et al: Characteristics of colorectal cancer (CRC) patients with
BRCA1 and BRCA2 mutations. J Clin Oncol. 37 (Suppl 4):S606.
2019.
|
|
35
|
Feliubadaló L, López-Fernández A, Pineda
M, Díez O, Del Valle J, Gutiérrez-Enríquez S, Teulé A, González S,
Stjepanovic N, Salinas M, et al: Opportunistic testing of BRCA1,
BRCA2 and mismatch repair genes improves the yield of phenotype
driven hereditary cancer gene panels. Int J Cancer. 145:2682–2691.
2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ma Y, Liang AJ, Fan YP, Huang YR, Zhao XM,
Sun Y and Chen XF: Dysregulation and functional roles of
miR-183-96-182 cluster in cancer cell proliferation, invasion and
metastasis. Oncotarget. 7:42805–42825. 2016.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Dambal S, Shah M, Mihelich B and Nonn L:
The microRNA-183 cluster: The family that plays together stays
together. Nucleic Acids Res. 43:7173–7188. 2015.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Milner R, Wombwell H, Eckersley S, Barnes
D, Warwicker J, Van Dorp E, Rowlinson R, Dearden S, Hughes G,
Harbron C, et al: Validation of the BRCA1 antibody MS110 and the
utility of BRCA1 as a patient selection biomarker in
immunohistochemical analysis of breast and ovarian tumours.
Virchows Arch. 462:269–279. 2013.PubMed/NCBI View Article : Google Scholar
|















