Introduction
Breast cancer is the second most common cancer
worldwide with regard to incidence. Two million new cases were
reached in 2018, a number that is continually on the increase
(1). Invasive ductal carcinoma
(IDC) is most common type, constituting ~70-80% of all breast
cancer diagnoses (2). Abnormal
cancer cells that form in the milk ducts spread beyond the ducts
into other parts of the breast tissue (2). In particular, tumors with
overexpressed or amplified human epidermal growth factor receptor 2
(HER2) show strong proliferation, migration and invasion. Early
treatment is necessary, most commonly in the form of chemotherapy
and surgery (3). Recently,
HER2-directed targeted therapy using monoclonal antibodies, such as
Trastuzumab and Pertuzumab, has been established as standard of
care. Trastuzumab acts directly against the extracellular domain of
the HER2 receptor and prevents ligand-independent HER2 signaling,
downregulates HER2 expression and reduces the more active p95-HER2
form of HER2. Pertuzumab binds to the extracellular domain II of
HER2 and inhibits ligand-dependent HER2-HER3 dimerization. These
monoclonal antibody therapies are recommended for use together with
radiotherapy and/or chemotherapy (4-8).
However, some tumors are resistant to treatment by antibody
therapy, chemotherapy and radiotherapy. The mechanisms of the
resistance are not known.
In order to gain more insight into the resistance
mechanisms, a metabolome analysis of serum collected repeatedly
from a HER2-positive breast cancer patient who showed resistance to
postoperative adjuvant therapy (PAT) was performed. The focus was
on low-molecular-mass metabolites in blood because they, better
than other ‘omic’ profiles, reflect the patients' biochemical
status or physiopathological condition. Alterations at the
metabolomic level not only reflect the alterations at the genomics
and proteomics levels, but also are influenced by environmental
factors (9).
In recent years, metabolomic studies have been
successfully used to identify biomarkers and altered metabolic
pathways in various cancer systems, including gastric, brain,
breast, and lung cancer (10-13).
In particular, a metabolomics approach was applied to identify
biomarkers potentially associated with pathological complete
response to trastuzumab-paclitaxel neoadjuvant therapy in
HER2-positive breast cancer patients. Consequently, good responders
showed higher levels of spermidine and lower amounts of tryptophan
compared to the poor responders (14).
Patients and methods
Study subject and ethics approval
The PAT-resistant patient was selected from
HER2-positive breast cancer patients (all >40 years) who were
treated by mastectomy at the Mutsu General Hospital (Aomori, Japan)
between February, 2017 and October, 2019. During this research
period, a PAT-resistant case was found and further analysis was
performed. The characteristic treatment flow and investigational
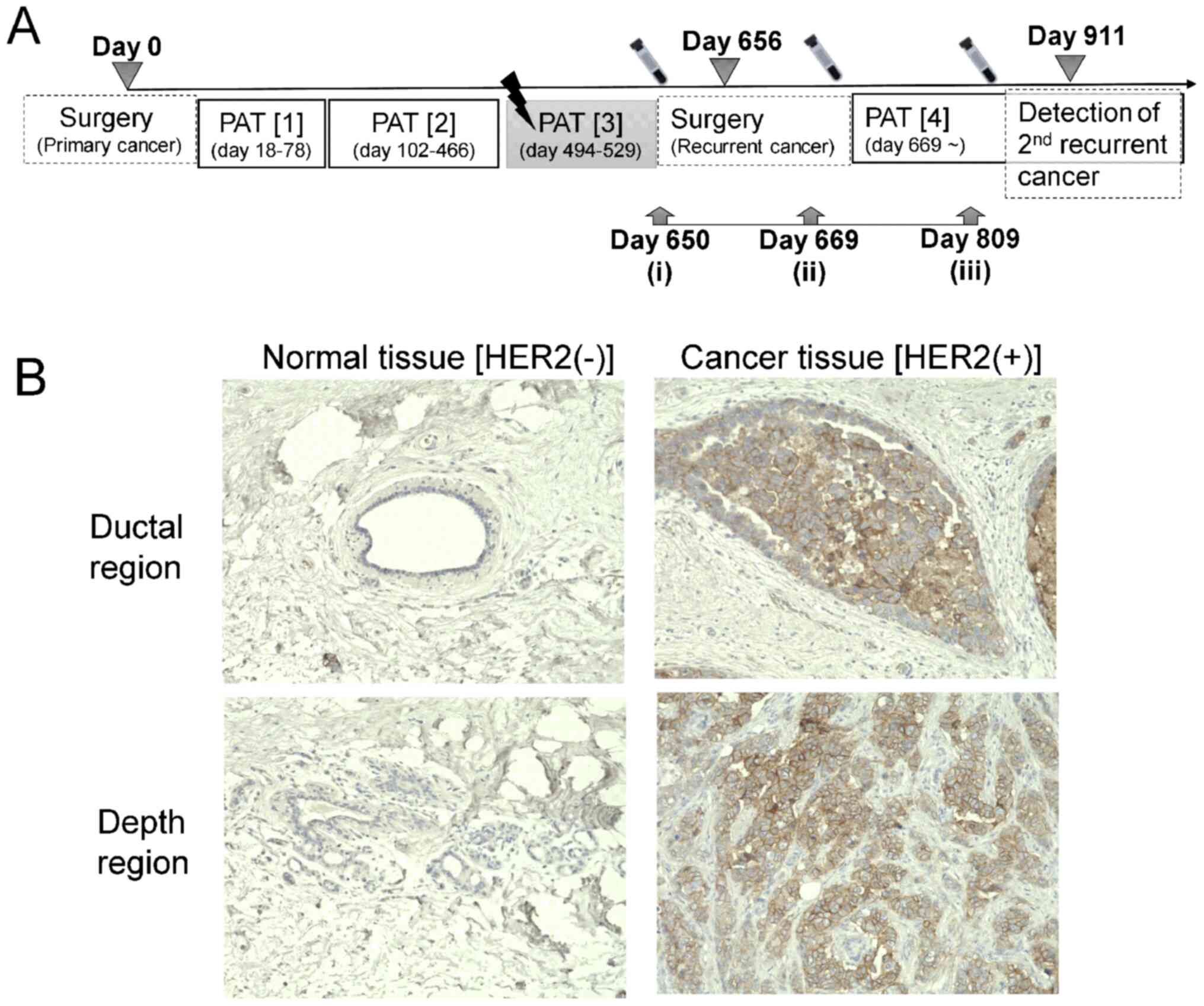
points are summarized in Fig. 1A.
The diagnosis was ductal adenocarcinoma in primary and solid
tubular et scirrhous carcinoma in recurrent,
T4bN2aM0, Stage IIIB. The patient
was treated using Fluorouracil + Epirubicin Hydrochloride +
Cyclophosphamide Hydrate (PAT1; day 18-78), and then with
Trastuzumab + Paclitaxel (PAT2; day 102-466) (Fig. 1A). External radiotherapy (photon
beam, 50 Gy per fraction, 25 fractions) was carried out on day
494-529 (PAT3). After surgery for removal of recurrent cancer
tissue, the patient was treated using Trastuzumab + Pertuzumab +
Docetaxel (PAT4; from day 669).
The study was approved by the Committee of Medical
Ethics of the Hirosaki University Graduate School of Health
Sciences (Hirosaki, Japan; no. 2016-051) and Mutsu General Hospital
(Mutsu, Japan; no. 000-282) to ensure the welfare and privacy of
the donors. Following a detailed verbal explanation regarding the
content of this study, a written informed consent was obtained.
Follow up by blood test
To observe the patient's condition during the
treatment, counts of white blood cells (WBC), red blood cells (RBC)
and platelets (Plt) were recorded and the level of hemoglobin (Hg)
was measured by routine hematological techniques. At the same time,
the concentration of C-reactive protein (CRP), carcinoembryonic
antigen (CEA), cancer antigen 15-3 (CA15-3) and breast cancer
antigen 225 (BCA225) were also quantified.
Collection of peripheral blood (PB)
serum for metabolomics analysis
PB was collected directly into serum separation
tubes (BD Biosciences). The serum was separated and stored at
-80˚C.
Mass spectrometry for
metabolomics
Serum metabolites were measured as part of a
targeted metabolomics panel using the AbsoluteIDQ P180 kit
(Biocrates, Inc.). The kit is a fully automated assay based on
phenylisothiocyanate (PITC) derivatization of the target analytes
using internal standards for quantitation. Human serum sample
preparation was carried out according to the manufacturer's
protocol. Briefly, 10 µl of serum was transferred to the first
96-well plate and dried under a nitrogen stream. Thereafter, 50 µl
of a 5% PITC solution was added to derivatize amino acids and
biogenic amines. After incubation, the filter spots were dried
again before the metabolites were extracted using 5 mM ammonium
acetate in methanol (300 µl) into the second 96-well plate for
analysis after further dilution using the Biocrates MS running
solvent A.
Acylcarnitines, lyso-phosphatidylcholines with acyl
residue, phosphatidylcholine with diacyl residue sum (PC aa), and
phosphatidylcholine with acyl-alkyl residue sum (PC ae),
sphingomyelins, and the sum of hexoses were analyzed by the flow
injection analysis (FIA) method in positive ion mode using the
liquid chromatography-mass spectrometry (LC-MS/MS). The LC-MS/MS
analysis was carried out by an HPLC system (ExionLC™ AD, AB Sciex)
coupled to a QTRAP6500+ triple quadruple ion trap hybrid mass
spectrometer system (AB Sciex) in electrospray ionization (ESI)
mode. The full list of metabolites and their abbreviations are
presented in Table I. The
metabolites were identified and quantified with multiple reaction
monitoring (MRM) with individual transition and parameters of
declustering potential (DP) and collision energy (CE) as listed in
Table S1. Flow rate settings for
FIA were as follows: 0.03 ml/min in 1.6 min; 0.03 to 0.2 ml/min in
0.8 min; 0.2 ml/min in 0.4 min; 0.2 to 0.03 ml/min in 0.2 min. MS
settings for FIA mode were as follows: Curtain gas, 45; ion spray
voltage, 5,500 V; temperature, 175˚C; ion source gas 1, 40 psi; ion
source gas 2, 50 psi; CAD gas, 6 psi; entrance potential, 10 V;
collision cell exit potential, 15 V. Twenty microliters of the
sample extract were used in the FIA. Quantification was carried out
using internal standards and a calibration curve.
 | Table IThe classification of metabolites in
this study using the Biocrates AbsoluteIDQ™ kit. |
Table I
The classification of metabolites in
this study using the Biocrates AbsoluteIDQ™ kit.
| Metabolite class | No. of analytes | Analyte name |
|---|
| Amino acid biogenic
amine | 42 | Ala, Arg, Asn, Asp,
Cit, Gln, Glu, Gly, His, Ile, Leu, Lys, Met, Orn, Phe, Pro, Ser,
Thr, Trp, Tyr, Val, Ac-Orn, ADMA, alpha-AAA, c4-OH-Pro, Carnosine,
Creatinine, DOPA, Dopamine, Histamine, Kynurenine, Met-SO,
Nitro-Tyr, PEA, Putrescine, SDMA, Serotonin, Spermidine, Spermine,
t4-OH-Pro, Taurine, total DMA |
| Carnitine
acylcarnitine | 26 | C0, C2, C3, C3:1,
C4, C4:1, C5, C5:1, C6 (C4:1-DC), C6:1, C8, C9, C10, C10:1, C10:2,
C12, C12:1, C14, C14:1, C14:2, C16, C16:1, C16:2, C18, C18:1,
C18:2 |
| Hydroxy- and
dicarboxyacylcarnitines | 14 | C3-DC (C4-OH),
C3-OH, C5-DC (C6-OH), C5-M-DC, C5-OH (C3-DC-M), C5:1-DC, C7-DC,
C12-DC, C14:1-OH, C14:2-OH, C16-OH, C16:1-OH, C16:2-OH,
C18:1-OH |
| Sphingomyeline,
hydroxysphingomyelins | 15 | SM (OH) C14:1, SM
(OH) C16:1, SM (OH) C22:1, SM (OH) C22:2, SM (OH) C24:1, SM C16:0,
SM C16:1, SM C18:0, SM C18:1, SM C20:2, SM C22:3, SM C24:0, SM
C24:1, SM C26:0, SM C26:1 |
| Diacyl
phosphatidylcholine | 38 | PC aa C24:0, PC aa
C26:0, PC aa C28:1, PC aa C30:0, PC aa C30:2, PC aa C32:0, PC aa
C32:1, PC aa C32:2, PC aa C32:3, PC aa C34:1, PC aa C34:2, PC aa
C34:3, PC aa C34:4, PC aa C36:0, PC aa C36:1, PC aa C36:2, PC aa
C36:3, PC aa C36:4, PC aa C36:5, PC aa C36:6, PC aa C38:0, PC aa
C38:1, PC aa C38:3, PC aa C38:4, PC aa C38:5, PC aa C38:6, PC aa
C40:1, PC aa C40:2, PC aa C40:3, PC aa C40:4, PC aa C40:5, PC aa
C40:6, PC aa C42:0, PC aa C42:1, PC aa C42:2, PC aa C42:4, PC aa
C42:5, PC aa C42:6 |
| Acyl-alkyl
phosphatidylcholine | 38 | PC ae C30:0, PC ae
C30:1, PC ae C30:2, PC ae C32:1, PC ae C32:2, PC ae C34:0, PC ae
C34:1, PC ae C34:2, PC ae C34:3, PC ae C36:0, PC ae C36:1, PC ae
C36:2, PC ae C36:3, PC ae C36:4, PC ae C36:5, PC ae C38:0, PC ae
C38:1, PC ae C38:2, PC ae C38:3, PC ae C38:4, PC ae C38:5, PC ae
C38:6, PC ae C40:1, PC ae C40:2, PC ae C40:3, PC ae C40:4, PC ae
C40:5, PC ae C40:6, PC ae C42:0, PC ae C42:1, PC ae C42:2, PC ae
C42:3, PC ae C42:4, PC ae C42:5, PC ae C44:3, PC ae C44:4, PC ae
C44:5, PC ae C44:6 |
|
Lysophosphatidylcholine | 14 | lysoPC a C14:0,
lysoPC a C16:0, lysoPC a C16:1, lysoPC a C17:0, lysoPC a C18:0,
lysoPC a C18:1, lysoPC a C18:2, lysoPC a C20:3, lysoPC a C20:4,
lysoPC a C24:0, lysoPC a C26:0, lysoPC a C26:1, lysoPC a C28:0,
lysoPC a C28:1 |
| Sugar | 1 | H1 |
| Total | 188 | |
Amino acids and biogenic amines were analyzed via
LC-MS/MS in positive ion mode. Then, 10 µl of the sample extract
were injected onto an HPLC C18 column (Zorbax Eclipse XDB-C18
column, 3x100 mm, 3.5 µm; Agilent) with a guard column (Zorbax
SB-C18, 3x100 mm, 1.85 µm; Agilent) at 40˚C using a 10-min solvent
gradient employing 0.2% formic acid in water (solvent A) and 0.2%
formic acid in acetonitrile (solvent B). Additional LC settings for
LC-MS/MS were as follows: 0% B in 0.5 min; 0-95% B in 5 min; 95% B
in 1 min; 95-0% B in 0.5 min; 0% B in 2.5 min at a flow rate of 0.5
ml/min. MS settings for LC-MS/MS mode were as follows: Curtain gas,
45; ion spray voltage, 5,500 V; temperature, 500˚C; ion source gas
1, 40 psi; ion source gas 2, 50 psi; CAD gas, 6 psi; entrance
potential, 10 V; collision cell exit potential, 15 V. All
metabolites were identified and quantified using
isotopically-labeled internal standards and MRM as optimized and
provided by Biocrates Inc.
Immunological analysis in
HER2-positive cells in cancer tissue sections
Paraffin-embedded sections of the removed cancer
tissues were deparaffinized with xylene and ethanol, washed in
D-PBS (-) and treated with 3% hydrogen peroxide for 5 min. The
slides were treated with 10 mM citrate buffer (pH 6.0) and
incubated for 10 min at 121˚C using an autoclave. Then, the slides
were washed in TBS buffer (25 mM Tris-HCl and 150 mM NaCl, pH 7.2).
To perform blocking, the slides were treated with the blocking
solution (5% normal goat serum in TBS buffer) at room temperature
for 30 min. Next, the slides were incubated with the blocking
solution containing a primary rabbit monoclonal antibody directed
against human HER2/ERBB2 (no. 4290; Cell Signaling Technology Inc.)
at a 1:1,000 dilution at room temperature for 60 min. The slides
were washed three times with TBS buffer and incubated at room
temperature for 60 min with an anti-rabbit IgG horseradish
peroxidase (HRP)-linked antibody (no. 7074, Cell Signaling
Technology) prepared in the blocking solution at a 1:2,000
dilution. The slides were washed three times with TBS buffer at
room temperature for 5 min. Signals were visualized using
3,3'-diaminobenzidine (DAB) (Sigma-Aldrich). The slides were washed
in water and nuclei were stained using hematoxylin solution.
Finally, the slides were dehydrated with ethanol and xylene, and
mounted using a hydrophobic mounting agent. Fluorescent
immunological analysis was performed as in our previous study
(15). Mouse monoclonal antibody
directed against human nSMase2 (sc-166637; Santa Cruz
Biotechnology, Inc.) (1:100 dilution) and rabbit monoclonal
HER2/ERBB2 antibody (1:1,000 dilution) described above was used as
a primary antibody. Anti-rabbit IgG Alexa Fluor 488-conjugated
antibody (no. 4412, Cell Signaling Technology) and Anti-mouse IgG
Alexa Fluor 647-conjugated antibody (no. 4410, Cell Signaling
Technology) were used as secondary antibodies. The cell nuclear
staining for fluorescence analysis DAPI (no. 4083; Cell Signaling
Technology) was used. Fluorescence signals were detected using a
fluorescence microscope (BZ-X700; Keyence).
Results
Patient description
Following diagnosis of right breast adenocarcinoma
with the expression of HER2 as primary cancer, the tumor was
removed by surgery (day 0, Fig.
1A). Three months after completing PAT3 (day 606), locally
recurrent cancer lesion in right breast within radiation field was
detected and removed by surgery on day 656. A second locally
recurrent cancer lesion and metastatic lymph node tumor were
observed on day 911. According to pathological analysis, the
removed primary tumor tissue (day 0) showed a distinctly higher
expression of HER2 as compared to corresponding areas of normal
tissue (Fig. 1B). A transient
increase in WBC (days 1 and 657) and Plt (days 7-102 and 669) after
surgery was observed (Fig. 2A and
B). On the other hand, the number
and concentration of RBC was stable (Fig. 2C and D). The inflammation marker CRP responded
similarly to WBC and Plt (Fig. 2E),
whereas the level of CEA remained stable (Fig. 2F). The levels of CA15-3 and BCA225
gradually increased during the observation period however these
markers were within the normal range (Fig. 2G and H).
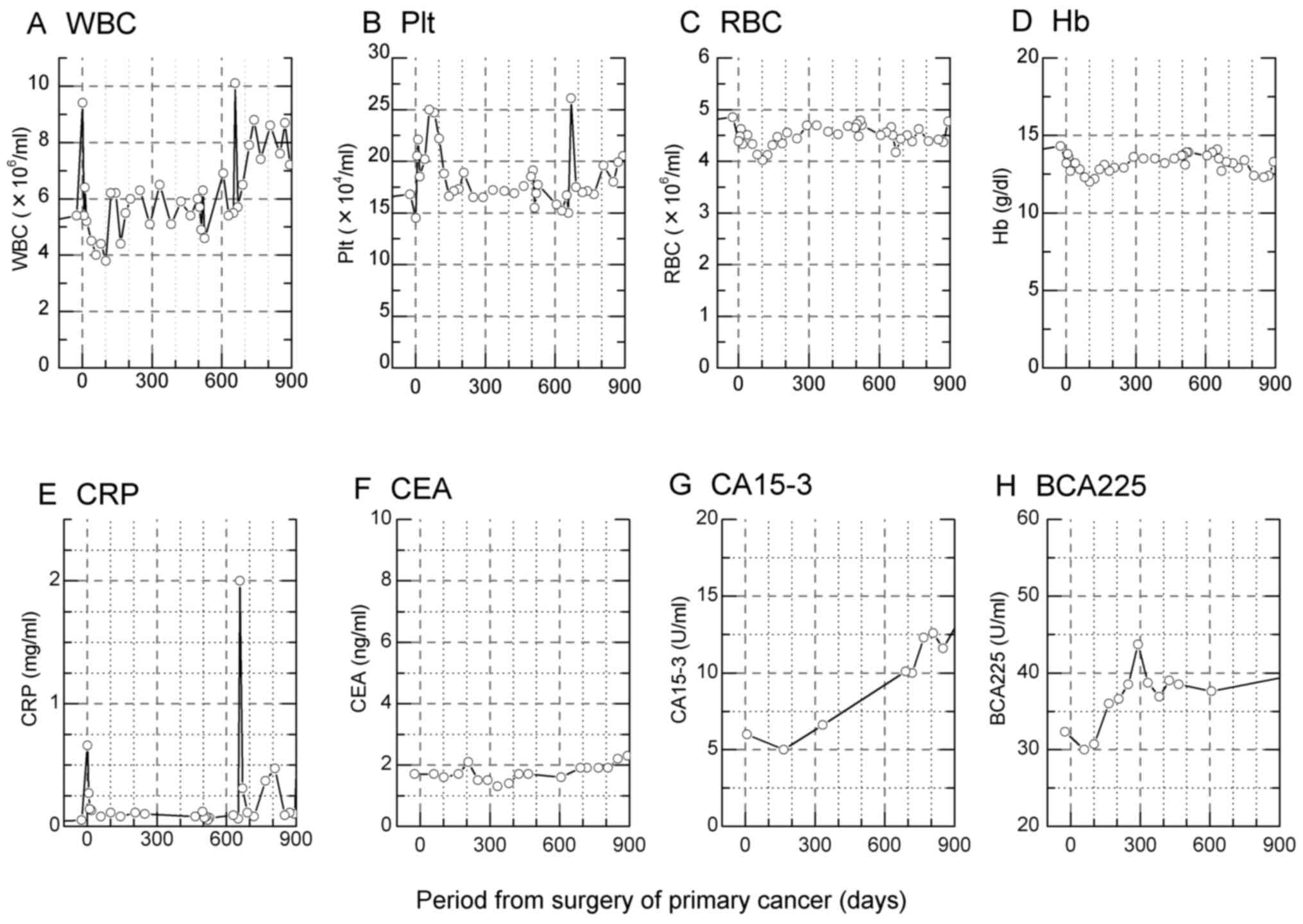 | Figure 2Peripheral blood cell counts,
haemoglobin, and biochemical markers during the course of therapy.
Concentrations of (A) white blood cell (WBC), (B) platelet (Plt),
(C) red blood cell (RBC), (D) level of hemoglobine (Hb), (E)
C-reactive protein (CRP) inflammation marker, and (F, G and H)
breast cancer-specific markers carcinoembryonic antigen (CEA),
cancer antigen 15-3 (CA15-3) and breast cancer antigen 225
(BCA225), respectively. The normal range of WBC, Plt, RBC, Hb, CRP,
CEA, CA15-3 and BCA225 were: 4.5-11.0x106/ml,
1.5-4.5x105/ml, 4.2-5.4x106/ml, 12.5-15.5
g/dl, <3.0 mg/ml, <5.0 ng/ml, 30 U/ml and <160 U/ml,
respectively. |
Repeated tumor recurrence during
PAT
After the first surgery, 4 cycles of PAT were
performed (Fig. 1A). According to
pathological analysis, the staging T1N1M0
(Stage IIA) at the first diagnostic stage (before day 0) changed to
rT4bN2aM0 when the recurrent tumor
was detected in the same original region and with a part invading
into the greater pectoral muscle as well as presenting as lymph
node metastasis (~day 650).
Metabolomic analysis in PB serum
None of the four PAT cycles showed effectiveness. To
investigate whether specific biomarker(s) were released from cancer
tissues to circulating body fluids, PB serum was collected at the
time before surgery for 1st recurrent tissue (day 650, Fig. 1-i), after 13 days from surgery (day
669, Fig. 1-ii) and the day when
the 2nd recurrent cancer was diagnosed (day 809, Fig. 1-iii), and analyzed by mass
spectrometry for wide-targeted metabolomic markers of response. The
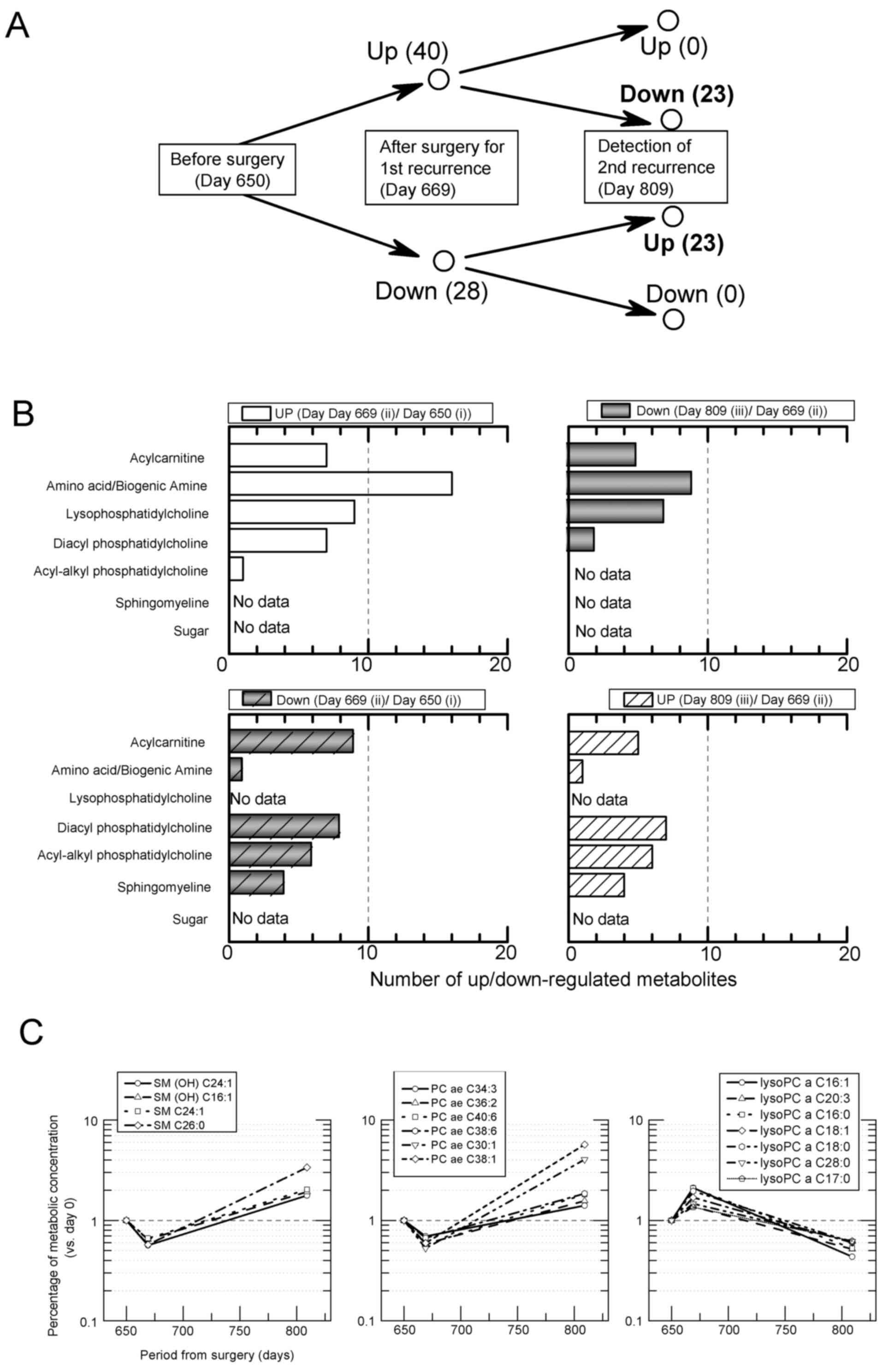
concentration of 188 molecules was analyzed (Table I), and, of these, 40 upregulated
molecules and 28 downregulated molecules were observed on day 669
(Fig. 1A-ii) as compared to day 650
(Fig. 1A-i) (Fig. 3A and B) by merely calculating the molecules in
Table I. In addition, 23 molecules
among the 40 upregulated molecules (Fig. 1A-ii) were downregulated on day 809
(Fig. 1A-iii) in comparison to day
669 (Fig. 1A-ii). On the other
hand, 23 molecules of the 28 downregulated molecules on day 669 (as
compared to day 650) were upregulated on day 809 (as compared to
day 669). These molecules that were up-to-down- and
down-to-up-regulated contained many lipids of three classes:
Acyl-alkyl phosphatidylcholine (6 molecules), sphingomyeline (4
molecules) and lysophosphatidylcholine (lysoPC) (7 molecules)
(Fig. 3C). Interestingly, the
concentrations of molecules in these three classes showed
fluctuations which correlated with the detection of cancer
recurrence (Fig. 3C). LysoPC is
produced from partial hydrolysis of phosphatidylcholines, and is
regarded as an inflammation-resolution lipid mediator (16). In this case study, half of the 14
lysoPC species measured were increased after surgery and decreased
when cancer recurred.
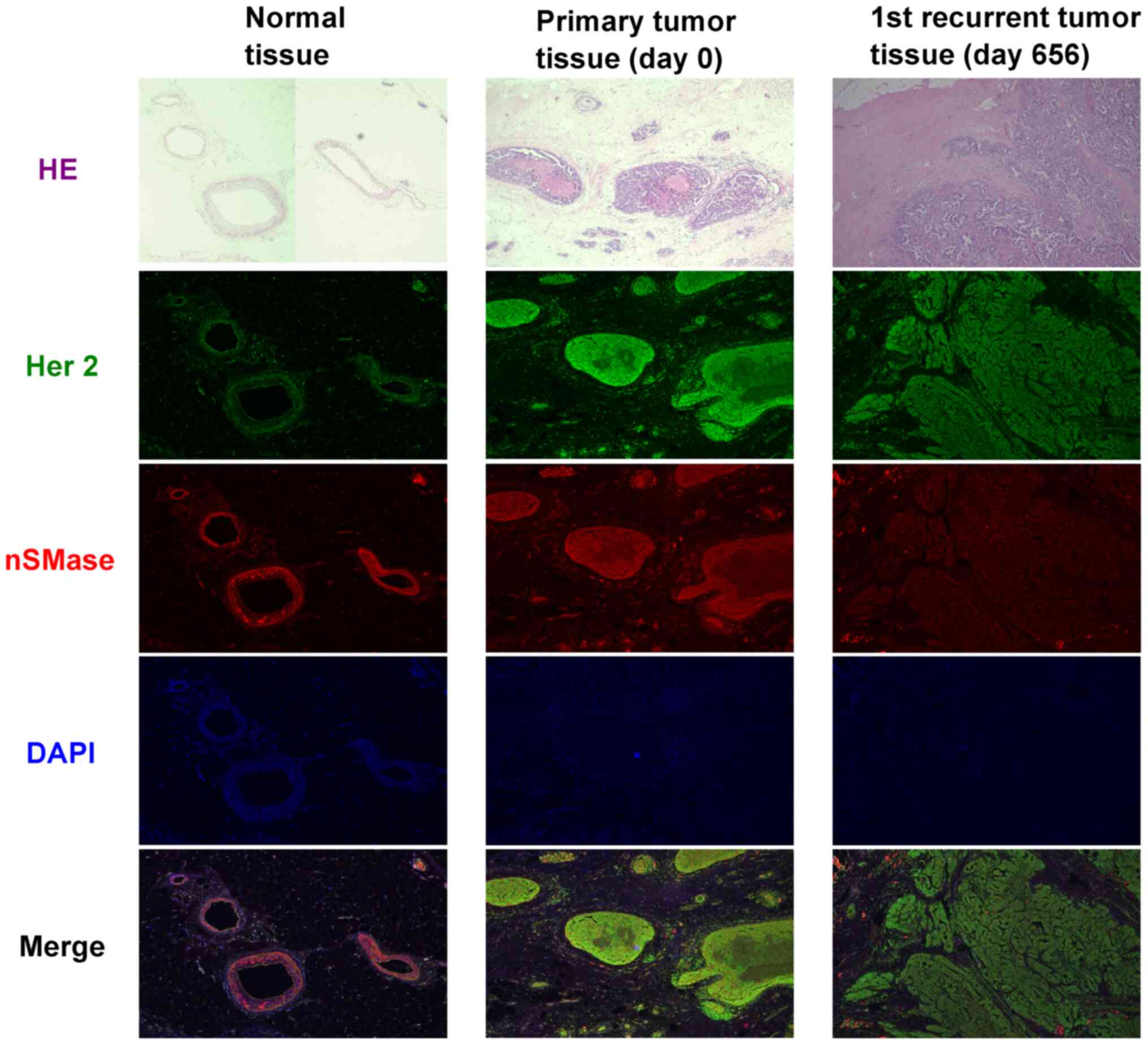
Immunoexpression analysis in cancer
tissue
Phosphatidylcholines and sphingomyelins are known as
components of cell membranes, especially in extracellular vesicles
which are associated with cell-to-cell communication (17-19).
Therefore, we focused on neutral sphingomyelinase-2 (nSMase), which
is one of the important factors for exocytosis or endocytosis. In
primary cancer tissue and 1st recurrent cancer tissue, a higher
expression of Her2 protein was observed (Fig. 4). However, the lowest accumulation
of intracellular nSMase in recurrent tumor was shown in comparison
to normal or primary tumor. This means that the exocytosis or
endocytosis occurs for release as extracellular vesicles (EVs).
Discussion
In the present study, we focused on a single
HER2-positive breast cancer patient with PAT resistance to identify
the characteristics of serum metabolites using metabolome analysis.
The applied PATs were not effective in their anti-tumorigenic,
anti-invasion and anti-lymph node metastatic action. In addition,
the biochemical markers in peripheral blood did not respond to the
PATs in a strong manner. It is known that ~60% of patients with
HER2-positive breast cancer develop resistance to trastuzumab,
partially due to loss of expression of HER2 extracellular domain on
their tumor cells (20). In
clinical sites, this is sometimes diagnosed as a resistant fraction
to administration of chemotherapeutic drugs (i.e., taxans) and/or
external photon radiotherapy in HER2-positive cancer patients.
However, the essential cause or the key factor driving this
resistance remains to be determined. Previous findings have shown
that the PI3K/Akt/mTOR pathway, STAT3-survivin signaling and
expression of mitogen-activated protein kinase phosphatase 1
produce for an environment that promotes resistance (21-23).
Therefore, there may be different subpopulations of cancer cells
that are responsive or non-responsive to cytotoxic drugs or
ionizing radiation in HER2-positive breast cancer.
A poor response of traditional breast cancer
biomarkers in serum (CEA, CA15-3 and BCA225) was observed in the
blood of the studied patient (Fig.
2) and they were of no value for predicting the recurrence of
cancer until the pathological test was performed and diagnosed with
correctly (day 645-650). Therefore, the identification of earlier
specific biomarkers of HER2-positive breast cancer progression and
therapeutic effectiveness is needed. We aimed to identify new serum
biomarker(s) using the metabolomics technique and found that the
concentration of the sphingomyeline family in serum is
inverse-correlated to the accumulation of nSMase in HER2-positive
cancer tissue (Figs. 3 and 4). Sphingomyelin is one of the
sphingolipids, an important structural component of biological
membranes and one of the end-points in the synthesis of
sphingolipids (24). Its hydrolysis
releases ceramide and phosphocholine and several stimuli are known
to activate sphingomyelin hydrolysis (25). Ceramide is an important promoter of
apoptosis (18), but additionally,
ceramide also triggers budding of EVs, such as exosomes into
multivesicular endosomes (26). It
is known that EVs are nanometer-scale particles that are secreted
by cells and mediate intercellular communication by transferring
biomolecules between cells. Thus, their expression relates to
cancer tissue growth or distant metastasis. We have previously
reported that during the active release of EVs to blood flow after
exposure to higher dose of ionizing radiation, the lower
accumulation of EV components in their original tissue cells was
observed in comparison to the other tissues with non-active release
of EVs (27,28). Our case data showed that the
concentration of nSMase co-expressed with HER2 in 1st recurrent
cancer tissue, where the levels of both are lower than in primary
cancer tissue (Fig. 4), and a
higher concentration of sphingomyelin family in serum was detected
when the 2nd recurrent cancer tissue was diagnosed. These responses
suggested that recurrent cancer tissues more easily release EVs to
peripheral blood serum and promote cancer progression and
expansion. Thus, one reported mechanism is the transfer of miRNAs
via exosomes from cancer cells to microenvironmental cells,
promoting angiogenesis (28,29).
It is possible that full resistance to PAT could be
connected to sphingomyelines, which may serve as a serum marker.
The gene sphingomyeline phosphodiesterase 3 (SMPD3) which
codes for nSMase was shown to act as a tumor suppressor gene in
hepatocellular carcinoma, supported by overexpression and knockdown
experiments (30). Of note, a
recent study showed that high levels of sphingomyelin synthase 2
(SGMS2), which controls the synthesis of sphingomyelins from
ceramide and thereby disrupts the ceramide-associated apoptosis
pathway, is correlated with breast cancer aggressiveness and
metastasis (31).
Additionally, we saw a clear opposite pattern for
diacyl and acyl-alkyl phosphatidylcholines, where there was a
decrease at 30 days after surgery, and then an increase, versus
lysoPCs, which was initially increased and then decreased.
Histological damage and infection caused by surgery induce
inflammatory biological responses which are evoked as a result of
exogenous and endogenous mediators secreted not only locally but
also systemically. Of note is that increased levels of the enzyme
lysoPC acyltransferase 1 (LPCAT1), controlling the conversion of
lysoPC to phosphatidylcholine, were linked to poor prognosis and
adverse features such as estrogen and progesterone receptor
negativity, amplification of HER2 and MYC and deletions of PTEN and
CDKNA2 in breast cancer (32,33).
Kim et al also reported a similar statement that lipid
profiles, such as sphingomyelin and phosphatidylcholine, correlated
with various clinicopathological characteristics of HER2-positive
breast cancer (34).
Although the PAT resistance could not be clearly
deciphered, our results provide novel information of a parallel
expression intensity of HER2 and nSMase, which possibly is
connected to cell communication by EVs. Recently, alteration of
gene expression by EV transportation was reported to induce
trastuzumab-resistant HER2-positive metastatic breast cancer
(35). Clearly, additional,
detailed analysis is required on a larger number of patients.
In conclusion, complete therapeutic-resistant cancer
tissue connected to reduced expression of nSMase in HER2-positive
breast cancer are strongly co-expressed with the sphingomyelin
family which can be detected at the peripheral blood serum
level.
Supplementary Material
Metabolite multiple reaction
monitoring transition.
Acknowledgements
The authors are grateful to Miyu Miyazaki at the
Center for Scientific Equipment Management, Hirosaki University
Graduate School of Medicine, for assistance with LC-MS/MS
analysis.
Funding
This study was supported by ‘JSPS KAKENHI,
Grant-in-Aid for Scientific Research (C) (General) (project no.
16K10339, Satoru Monzen)’, ‘JSPS KAKENHI, Fund for the Promotion of
Joint International Research (Fostering Joint International
Research) (project no. 17KK0181, Satoru Monzen)’, and
‘Interdisciplinary Collaborative Research Grant for Young
Scientists, Hirosaki University’.
Availability of data and materials
All data generated or analysed during the present
study are included in this published article.
Authors' contributions
SM and YM designed the study and prepared the
manuscript draft and substantively participated in revising the
manuscript. SM, YT and MC conceived the study, and contributed by
analyzing the patient's data and revised the manuscript. AW and LL
supervised the study and critically reviewed the manuscript, and
gave final approval of the version to be published. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Committee of Medical
Ethics of the Hirosaki University Graduate School of Health
Sciences, (no. 2016-051) and Mutsu General Hospital (no. 000-282)
to ensure the welfare and privacy of the donors. Following a
detailed verbal explanation regarding the content of this study, a
written informed consent was obtained.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tsuchiya SI, Yamaguchi R, Tsuchiya K and
Ohashi R: Characteristics of the Japanese histological
classification for breast cancer: Correlations with imaging and
cytology. Breast Cancer. 23:534–539. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ahmed S, Sami A and Xiang J: HER2-Directed
therapy: Current treatment options for HER2-positive breast cancer.
Breast Cancer. 22:101–116. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Wuerstlein R and Harbeck N: Neoadjuvant
therapy for HER2-positive breast cancer. Rev Recent Clin Trials.
12:81–92. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kubo M, Kawai M, Kumamaru H, Miyata H,
Tamura K, Yoshida M, Ogo E, Nagahashi M, Asaga S, Kojima Y, et al:
A population-based recurrence risk management study of patients
with pT1 node-negative HER2+ breast cancer: A national
clinical database study. Breast Cancer Res Treat. 178:647–656.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Goldvaser H, Korzets Y, Shepshelovich D,
Yerushalmi R, Sarfaty M, Ribnikar D, Thavendiranathan P and Amir E:
Deescalating adjuvant trastuzumab in HER2-positive early-stage
breast cancer: A systemic review and meta-analysis. JNCI Cancer
Spectr. 3(pkz033)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Hirsova P, Ibrahim SH, Krishnan A, Verma
VK, Bronk SF, Werneburg NW, Charlton MR, Shah VH, Malhi H and Gores
GJ: Lipid-Induced signaling causes release of inflammatory
extracellular vesicles from hepatocytes. Gastroenterology.
150:956–967. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Gu G, Dustin D and Fuqua SA: Targeted
therapy for breast cancer and molecular mechanisms of resistance to
treatment. Curr Opin Pharmacol. 31:97–103. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gomez-Casati DF and Busi MV: Clinical
Molecular Medicine. In: Principles and Practice. 1st Edition. Kumar
D (ed.) Academic Press, pp47-55, 2019.
|
|
10
|
Wang D, Li W, Zou Q, Yin L, Du Y, Gu J and
Suo J: Serum metabolomic profiling of human gastric cancer and its
relationship with the prognosis. Oncotarget. 8:110000–110015.
2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Chinnaiyan P, Kensicki E, Bloom G, Prabhu
A, Sarcar B, Kahali S, Eschrich S, Qu X, Forsyth P and Gillies R:
The metabolomic signature of malignant glioma reflects accelerated
anabolic metabolism. Cancer Res. 72:5878–5888. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Hadi NI, Jamal Q, Iqbal A, Shaikh F,
Somroo S and Musharraf SG: Serum metabolomic profiles for breast
cancer diagnosis, grading and staging by gas chromatography-mass
spectrometry. Sci Rep. 7(1715)2017.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Kumar N, Shahjaman M, Mollah MN, Islam SM
and Hoque MA: Serum and plasma metabolomic biomarkers for lung
cancer. Bioinformation. 13:202–208. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Miolo G, Muraro E, Caruso D, Crivellari D,
Ash A, Scalone S, Lombardi D, Rizzolio F, Giordano A and Corona G:
Pharmacometabolomics study identifies circulating spermidine and
tryptophan as potential biomarkers associated with the complete
pathological response to trastuzumab-paclitaxel neoadjuvant therapy
in HER-2 positive breast cancer. Oncotarget. 7:39809–39822.
2016.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chiba M, Kubota S, Sakai A and Monzen S:
Cell-To-Cell communication via extracellular vesicles among human
pancreatic cancer cells derived from the same patient. Mol Med Rep.
18:3989–3996. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yan JJ, Jung JS, Lee JE, Lee J, Huh SO,
Kim HS, Jung KC, Cho JY, Nam JS, Suh HW, et al: Therapeutic effects
of lysophosphatidylcholine in experimental sepsis. Nat Med.
10:161–167. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
17
|
van Meer G, Voelker DR and Feigenson GW:
Membrane lipids: Where they are and how they behave. Nat Rev Mol
Cell Biol. 9:112–124. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Shamseddine AA, Airola MV and Hannun YA:
Roles and regulation of neutral sphingomyelinase-2 in cellular and
pathological processes. Adv Biol Regul. 57:24–41. 2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Andrews NW, Corrotte M and Castro-Gomes T:
Above the fray: Surface remodeling by secreted lysosomal enzymes
leads to endocytosis-mediated plasma membrane repair. Semin Cell
Dev Biol. 45:10–17. 2015.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Nami B and Wang Z: HER2 in breast cancer
stemness: A negative feedback loop towards trastuzumab resistance.
Cancers (Basel). 26(40)2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wilks ST: Potential of overcoming
resistance to HER2-targeted therapies through the PI3K/Akt/mTOR
pathway. Breast. 24:548–555. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Kim JS, Kim HA, Seong MK, Seol H, Oh JS,
Kim EK, Chang JW, Hwang SG and Noh WC: STAT3-Survivin signaling
mediates a poor response to radiotherapy in HER2-positive breast
cancers. Oncotarget. 7:7055–7065. 2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Candas D and Li JJ: MKP1 mediates
resistance to therapy in HER2-positive breast tumors. Mol Cell
Oncol. 2(e997518)2015.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Adada M, Luberto C and Canals D:
Inhibitors of the sphingomyelin cycle: Sphingomyelin synthases and
sphingomyelinases. Chem Phys Lipids. 197:45–59. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Signorelli P and Hannun YA: Analysis and
quantitation of ceramide. Methods Enzymol. 345:275–294.
2002.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Elsherbini A and Bieberich E: Ceramide and
exosomes: A novel target in cancer biology and therapy. Adv Cancer
Res. 140:121–154. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Chiba M, Kubota S, Sato K and Monzen S:
Exosomes released from pancreatic cancer cells enhance angiogenic
activities via dynamin-dependent endocytosis in endothelial cells
in vitro. Sci Rep. 8(11972)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chiba M, Monzen S, Iwaya C, Kashiwagi Y,
Yamada S, Hosokawa Y, Mariya Y, Nakamura T and Wojcik A: Serum
miR-375-3p increase in mice exposed to a high dose of ionizing
radiation. Sci Rep. 8(1302)2018.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Kosaka N, Iguchi H, Hagiwara K, Yoshioka
Y, Takeshita F and Ochiya T: Neutral sphingomyelinase 2
(nSMase2)-dependent exosomal transfer of angiogenic microRNAs
regulate cancer cell metastasis. J Biol Chem. 288:10849–10859.
2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Revill K, Wang T, Lachenmayer A, Kojima K,
Harrington A, Li J, Hoshida Y, Llovet JM and Powers S: Genome-Wide
methylation analysis and epigenetic unmasking identify tumor
suppressor genes in hepatocellular carcinoma. Gastroenterology.
145:1424–1435. 2013.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zheng K, Chen Z, Feng H, Chen Y, Zhang C,
Yu J, Luo Y, Zhao L, Jiang X and Shi F: Sphingomyelin synthase 2
promotes an aggressive breast cancer phenotype by disrupting the
homoeostasis of ceramide and sphingomyelin. Cell Death Dis.
10(157)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Lebok P, von Hassel A, Meiners J,
Hube-Magg C, Simon R, Höflmayer D, Hinsch A, Dum D, Fraune C, Göbel
C, et al: Up-regulation of lysophosphatidylcholine acyltransferase
1 (LPCAT1) is linked to poor prognosis in breast cancer. Aging
(Albany NY). 11:7796–7804. 2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Abdelzaher E and Mostafa MF:
Lysophosphatidylcholine acyltransferase 1 (LPCAT1) upregulation in
breast carcinoma contributes to tumor progression and predicts
early tumor recurrence. Tumor Biol. 36:5473–5483. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kim IC, Lee JH, Bang G, Choi SH, Kim YH,
Kim KP, Kim HK and Ro J: Lipid profiles for HER2-positive breast
cancer. Anticancer Res. 33:2467–2472. 2013.PubMed/NCBI
|
|
35
|
de Oliveira Taveira M, Nabavi S, Wang Y,
Tonellato P, Esteva FJ, Cantley LC and Wulf GM: Genomic
characteristics of trastuzumab-resistant her2-positive metastatic
breast cancer. J Cancer Res Clin Oncol. 143:1255–1262.
2017.PubMed/NCBI View Article : Google Scholar
|