1. Introduction
RNA modifications are used to fine-tune the
structural features of infrastructural RNAs. In recent years, RNA
modifications have been found to be reversible and involved in
important biological processes, through continued efforts to map
and quantify various RNA modifications in a transcriptome-wide
manner (1). The
N6-methyladenosine (m6A) methylation
modification is the most prevalent internal modification of
eukaryotic mRNA. The latest discoveries of the locations,
functions, and mechanisms of m6A provide new insights
into the regulation mechanism of RNA expression (2). Evidence supports the involvement of
m6A modifications in precursor mRNA (pre-mRNA) splicing,
mRNA stability, RNA structure, translation, and processing of
primary transcripts of microRNAs (miRNAs) (3). The m6A sites are enriched
near the stop codons and 3'-untranslated regions (UTRs), and an
association exists between the m6A residues and the
mRNA-binding site in the 3'-UTR (4,5).
The m6A modification appears to be
reversible under the combined action of the enzymes involved
(6,7). The m6A modification is
mediated, removed, and recognized by methyltransferases,
demethylases, and m6A-binding proteins, respectively
(8). Methyltransferase-like 3
(METTL3) (9),
methyltransferase-like 14 (METTL14) (10), methyltransferase-like 16 (METTL16)
(11), Wilms tumor 1-associating
protein (WTAP) (12), RNA-binding
motif protein 15/15B (RBM15/15B) (13), and vir-like m6A
methyltransferase-associated protein (VIRMA/KIAA1429) (14) are considered to be the components of
‘writers’ that catalyze the formation of m6A; ‘erasers’
such as the obesity-associated protein (FTO) and alkB homolog 5
(ALKBH5) (15,16) remove the methyl code from target
mRNAs; ‘readers’ such as the YTH domain protein families (YTHDF)
and heterogeneous nuclear ribonucleoprotein (HNRNP) families
(17,18) are capable of recognizing
m6A methylation and generating a functional signal
(19). YTH domain proteins read
m6A through a conserved aromatic cage (20) and two other proteins, FMRP
translational regulator 1 (FMR1) and leucine-rich pentatricopeptide
repeat-containing (LRPPRC), can also recognize this modification
(21,22). Therefore, the m6A
modification is a highly dynamic and reversible process (Fig. 1) (23).
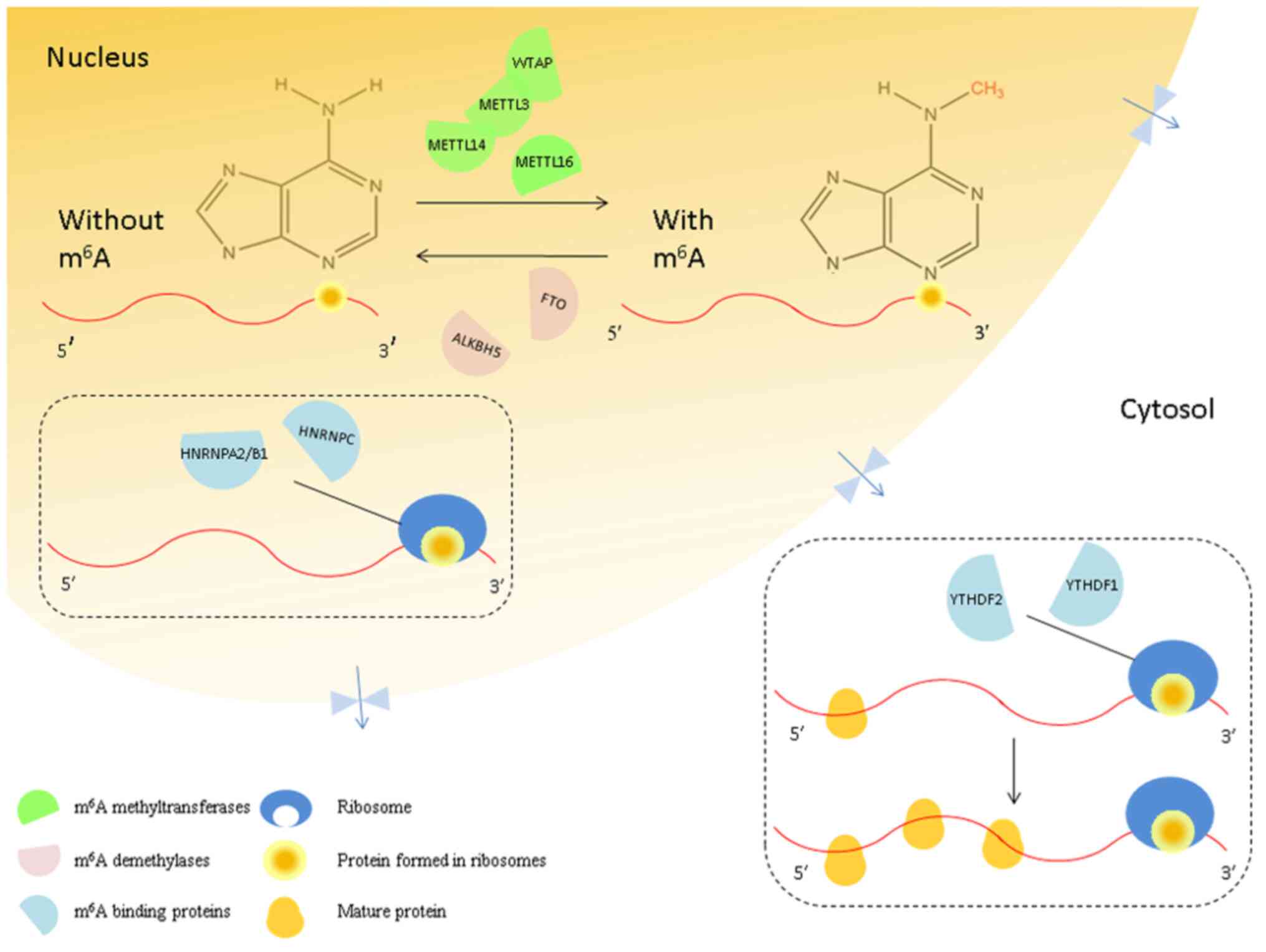 | Figure 1Interaction between the mRNA
methylation ‘writers,’ ‘erasers’ and ‘readers.’ In the nucleus,
m6A methyltransferases (green semicircles) are named
‘writers’. Their binding proteins (blue semicircles) are named
‘readers,’ and the demethylases (pink semicircles) are named
‘erasers.’ The m6A ‘writers’ include METTL3/14/16 and
WTAP, and the ‘erasers’ that can eliminate the m6A from
mRNAs include FTO and ALKBH5. Furthermore, m6A is
recognized by the ‘readers,’ such as YTHDF1/2/3, HNRNPA2/B1, and
HNRNPC. YTHDF1/2/3 (yellow circles) promotes the translation of
m6A-modified mRNAs in the cytosol. m6A,
N6-methyladenosine; METTL, methyltransferase-like; WTAP,
Wilms tumor 1-associating protein; FTO, Fat mass and
obesity-associated protein; ALKBH5, alkB homolog 5; YTHDF, YTH
domain protein families; HNRNP A2/B1, heterogeneous nuclear
ribonucleoprotein A2/B1; HNRNPC, heterogeneous nuclear
ribonucleoprotein C. |
Lung cancer is the most common malignant tumor with
high morbidity and mortality rates worldwide. Non-small cell lung
cancer (NSCLC) accounts for 85-90% of all cases of lung cancer,
including lung squamous cell carcinoma (LUSC), lung adenocarcinoma
(LUAD), and large cell anaplastic carcinoma (LCAC) (24). According to the latest data on
cancer incidence, the 5-year survival rate of patients with NSCLC
is as low as approximately 15%, which can be attributed to atypical
symptoms in the early phase of the disease and lack of effective
treatment (25,26). The epigenetic m6A
modifications are involved in the progression, auxiliary diagnosis,
and prognosis of lung cancer (27).
Moreover, m6A modification is an important factor
affecting the growth, survival, and invasion of cancer cells
(28,29). Here, we review and summarize the
molecular mechanisms and functions of m6A RNA
modification in lung cancer. Further, we discuss the role of
m6A modification in lung cancer to provide a new
theoretical basis for m6A research.
2. m6A writers in lung
cancer
The m6A methyltransferase writer complex,
which catalyzes the m6A mRNA methylation in lung cancer,
primarily consists of METTL3, METTL14, METTL16, and WTAP. METTL3
(also known as MTA70) is the methyltransferase primarily
responsible for the m6A modification. METTL3 and METTL14
form a stable heterodimer core complex of METTL3-METTL14, which
affects the cellular m6A deposition on mammalian nuclear
RNAs. WTAP does not exhibit methylation activity; however, it
interacts with the METTL3-METTL14 complex to significantly impact
the cellular m6A deposition (10).
METTL3
METTL3 levels are upregulated in lung cancer
tissues, which are higher in advanced stage lung cancer patients
(30). METTL3 increases the
translation of target mRNAs by recruiting the eukaryotic
translation initiation factor (eIF)3 to the translation initiation
complex in H1299 cells (31). It
directly interacts with certain components of the multi-subunit
eIF3 complex. Meanwhile, the METTL3-eIF3H interaction is essential
for promoting translation, formation of densely packed
polyribosomes, and oncogenic transformation of A549 cells (Fig. 2A). Disruption of the METTL3-eIF3H
interaction eliminates the ability of METTL3 to promote
translation, influence polysome conformation, and enhance oncogenic
transformation (32). Furthermore,
Lin et al found that METTL3 enhanced RNA translation without
the aid of methyltransferase and reader protein activity. METTL3
increases RNA translation by directly recruiting translation
initiation factors. METTL3 knockdown inhibits the recruitment of
eIF3 to both the cap-binding protein 80 (CBP80)- and eIF4E-cap
binding proteins (33). Inhibition
of m6A with METTL3 short hairpin RNA (shMETTL3)
significantly decreases the expression of eIFs in lung cancer cells
(34).
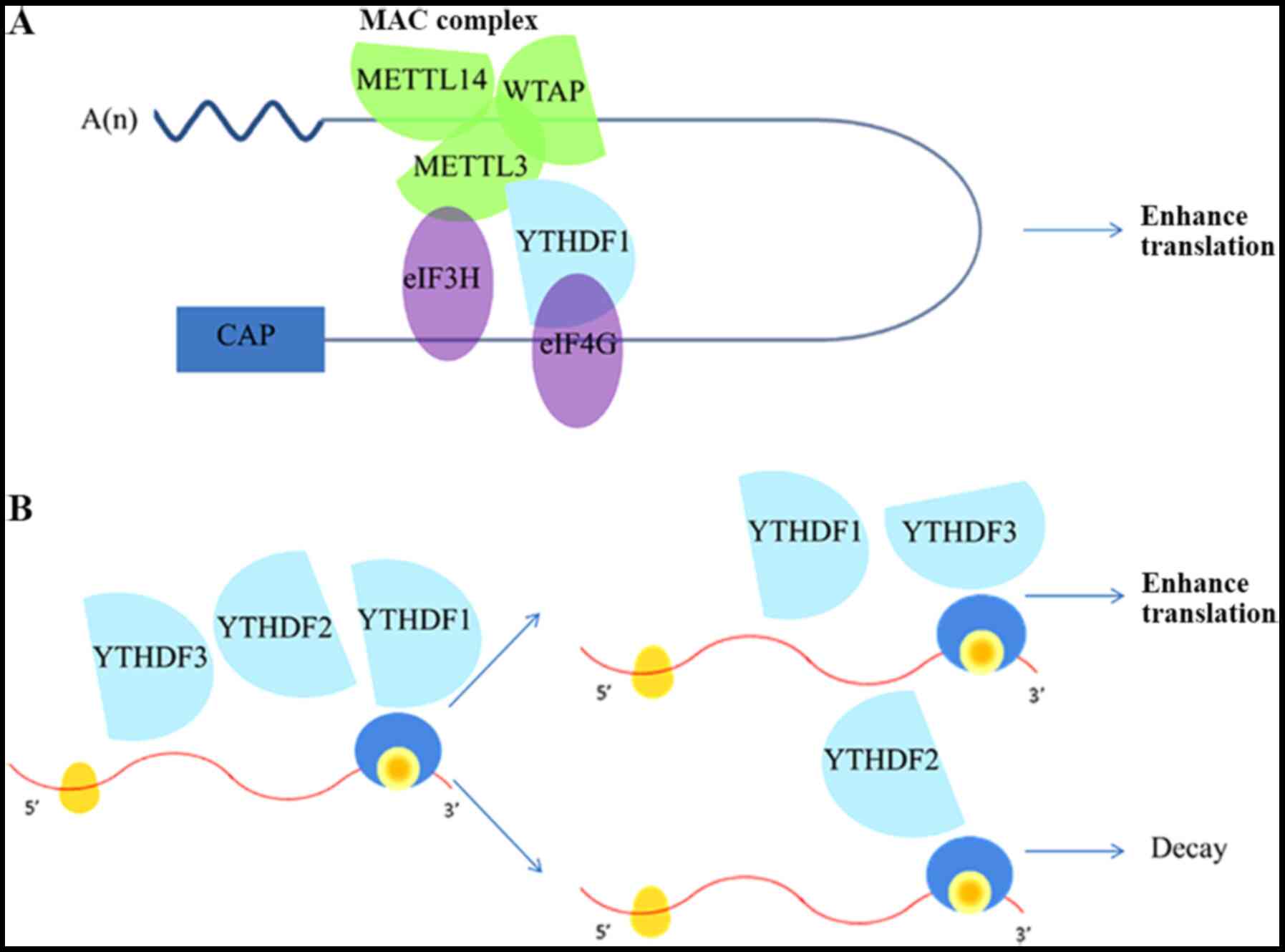 | Figure 2Functional roles of m6A
modification on mRNA expression. (A) RNA circularization. METTL3,
an RNA methyltransferase located at the 3'-UTR, combines with
METTL14 and WTAP to form the MAC complex, which increases the
translation of specific mRNAs independently through its catalytic
activity and recruitment of eIF3H. YTHDF1 binds to eIF4G at 5'-UTR,
which promotes the process of RNA circularization. (B) YTHDF1 and
YTHDF3 synergistically promote protein synthesis, while YTHDF2
affects the attenuation of methylated mRNA. YTHDF1/2/3 serves a key
role in the metabolism of N6-methyladenosine-modified
mRNAs in the cytoplasm. METTL, methyltransferase-like; UTR,
untranslated region; WTAP, Wilms tumor 1-associating protein; MAC,
m6A-METTL complex; eIF3H, eukaryotic translation
initiation factor 3H; YTHDF, YTH domain protein families. |
METTL3 regulates the translation of genes related to
tumor progression and apoptosis. METTL3 knockdown in A549 cells
decreases the expression of bromodomain containing 4 (BRD4) and
other targets, and the cells expressing METTL3 are more sensitive
to pharmacological BRD4 inhibition. METTL3 promotes translation
only when it is tethered to the reporter mRNA at sites close to the
stop codon and assists the mRNA looping mechanism for ribosome
recycling and translational control (32). The mRNAs of several oncogenes, such
as the epidermal growth factor receptor (EGFR), tafazzin (TAZ),
MAPKAPK2 (MK2), and DNA methyltransferase 3 alpha (DNMT3A), have
one or more m6A peaks near the stop codon. The analysis
of m6A levels in EGFR revealed that METTL3 binds to the
EGFR mRNA in A549 cells (33).
METTL3 knockdown significantly increases E-cadherin expression and
decreases the expression of Fibronectin and Vimentin in A549 and
LC-2/ad cells. Moreover, it inhibits the expression changes of
these epithelial-mesenchymal transition (EMT)-relate marker genes
stimulated by transforming growth factor β (TGF-β) treatment. These
results suggest the involvement of endogenous METTL3 in the
transcriptional regulation of TGF-β-induced EMT program (35). Furthermore, the expression of
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), a
transcript related to lung cancer metastasis and prognosis, is
increased due to a higher level of m6A modification
mediated by METTL3. Meanwhile, the METTL3/YTHDF3 complex increases
the stability of MALAT1. METTL3 catalyzes the m6A
methylation modification of the nuclear effector yes-associated
protein (YAP) of the Hippo signaling pathway, promotes its
translation, and mediates the proliferation and metastasis of NSCLC
(31).
The characteristics of METTL3 in activation and
posttranslational modification (such as SUMOylation) of METTL3 may
directly affect the proliferation and xenograft tumor growth of
lung cancer cells. SUMOylation of METTL3 mediates the
m6A mRNA modification and subsequent differences in gene
expression profiles (36).
miRNAs can be differentially expressed and act as
oncogenic or tumor suppressor miRNAs, which are based on the roles
of miRNA-regulated genes (37). The
m6A modification may control arsenite-induced
proliferation and apoptosis of cells by affecting miRNAs (38). Studies have provided new insights
into the mechanism of METTL3 regulation by miRNAs, thus signifying
the potential application of METTL3 as a therapeutic target in
NSCLC. For example, miR-33a inhibits NSCLC cell proliferation by
targeting the 3'-UTR of METTL3 mRNA (39). Moreover, miR-600 attenuates the
METTL3 expression and regulates cell proliferation, metastasis, and
apoptosis by regulating the PKB Protein Kinase (AKT) and β-catenin
signaling pathways (40). METTL3
has been reported to increase the splicing of precursor miR-143-3p,
accelerate the processing and maturation of miR-143-3p. Moreover,
miR-143-3p/VASH1 axis acts as adverse prognosis factors for in
vivo progression and overall survival rate of lung cancer
(41). In summary, miRNA is an
important bridge for m6A to influence the proliferation,
metastasis, invasion, and apoptosis of lung cancer cells.
Other m6A
methyltransferases
WTAP is the target of miRNAs and accelerates the
progression of NSCLC (42). METTL16
is a recently confirmed m6A RNA methyltransferase that
interacts with the 3'-terminal RNA triple helix of MALAT1 in lung
cancer (43). The
three-m6A-regulator signature (KIAA1429, METTL3 and
IGF2BP1) is recognized as an independent prognostic model to
categorize lung cancers into high- and low-risk groups for patient
stratification, prognostic assessment, and personalized treatment
in lung cancer. KIAA1429 and insulin-like growth factor 2
mRNA-binding protein 1 (IGF2BP1) are significantly associated with
multiple biological processes, including proliferation, apoptosis,
metastasis, energy metabolism, drug resistance, and recurrence;
additionally, they target potential genes related to lung cancer
(44). Proteinase
activated-receptor 2 (PAR2) participates in cancer metastasis
promoted by serine proteinases. Knock down of NOP2/sun domain
family, member 2 (NSun2), a new RNA methyltransferase, blocks the
reduction in miR-125b induced by PAR2. NSun2 is shown to interfere
in the mature processing of miR-125b from pri- and pre-miR-125b2 in
A549 cells. Furthermore, PAR2 activation increased the level of
m6A-containing pre-miR-125b in NSun2-dependent manner
(45). Overall, these data reveal
the existence of a complex network of interactions between the
m6A methylases and oncogenes that can regulate the
proliferation, metastasis, invasion, and apoptosis of lung cancer
cells.
3. m6A erasers in lung
cancer
m6A is deposited by the methyltransferase
complex and cleared by the demethylases FTO and ALKBH5. These
demethylases participate in the biological processes of lung cancer
(46,47). m6A in the nuclear RNA is
a substrate of FTO, and FTO causes an enzymatic alteration that may
be related to mRNA transcription (48). ALKBH5, a primary m6A
demethylase, plays important roles in lung cancer by regulating
proliferation, migration, invasion, metastasis, and tumor growth
(49).
FTO
Increased METTL3 and decreased FTO levels
demonstrate that the dysregulated writer and/or eraser may affect
the m6A content in both the cells and tissues of LUAD
patients (34). RNA sequencing
analysis has revealed that some genes are influenced by
m6A demethylation, most of which are associated with
lung cancer, such as laminin γ2, nerve growth factor inducible,
integrin alpha 11, thrombospondin 1, and proprotein convertase
subtilisin/kexin type 9. FTO enhances LUAC cell progression by
activating cell migration (15).
In LUSC, FTO acts as a prognostic factor responsible
for aberrant m6A modifications (50). The proliferation and invasion of
cells in LUSD was found to be effectively decreased by FTO
knockdown. Furthermore, overexpression of FTO rather than its
mutant form promotes the malignant phenotype of cells. Mechanism
analysis demonstrated that FTO decreases the m6A
modification of the myeloid zinc finger 1 (MZF1) transcript and
strengthens its stability, resulting in increased MZF1 expression,
as well as promotion of the occurrence and development of lung
cancer (47).
In addition, FTO represses the m6A levels
and strengthens the mRNA stability of ubiquitin-specific protease 7
(USP7), which relies on the demethylase activity of FTO. FTO
downregulation inhibits proliferation and growth of NSCLC cells by
facilitating the expression of USP7(46). Therefore, overexpression of FTO
promotes the proliferation, migration, and invasion abilities of
lung cancer cells.
ALKBH5
ALKBH5 is upregulated in NSCLC and is closely
associated with a poor prognosis. Functionally, ALKBH5 facilitates
proliferation and inhibits apoptosis of the NSCLC cells in
vitro, whereas ALKBH5 knockdown reduces tumor growth in
vivo (16). The overexpression
of ALKBH5 results in the increase translation efficiency of factor
forkhead box M1 (FOXM1) mRNA by decreasing the level of
m6A in FOXM1, which promotes the growth of LUAD cells
(51). Mechanistically, ALKBH5
knockout inhibits the growth and invasion of A549 and NCI-H566
cells under intermittent hypoxia by downregulating the
m6A modification of FOXM1 and increasing the FOXM1
levels (51). Mechanistically,
methylated RNA immunoprecipitation sequencing revealed that ALKBH5
targets the 3'-UTR of tissue inhibitor of metalloproteinase 3
(TIMP3). ALKBH5 inhibits the TIMP3 transcript stability, thereby
reducing its translation (16). Due
to the upregulation of ALKBH5 in NSCLC, the oncogene ubiquitin
conjugating enzyme E2C (UBE2C) is stabilized epitranscriptionally
with the remaining lower m6A levels within its mature
RNAs. Activation of UBE2C is associated with adverse prognosis and
enhances proliferation, clonogenicity, and invasive growth of NSCLC
cells (52). Furthermore, ALKBH5
restrains tumor growth and metastasis by decreasing the expression
and activity of YAP in a YTHDF- and miR-107/large tumor suppressor
kinase 2 (LATS2)-mediated manner. YAP expression is negatively
correlated with ALKBH5 expression and plays an opposite role in the
regulation of cellular proliferation, invasion, migration, and EMT
of NSCLC cells (53). Collectively,
m6A demethylases affect the proliferation, invasion, and
apoptosis of lung cancer cells by downregulating the m6A
modification of mRNA.
4. m6A readers in lung
cancer
m6A also affects biological processes by
recruiting reader proteins that specifically recognize
m6A RNA methylation and affect the downstream functions
(54,55). YTH N6-methyladenosine
RNA-binding protein (YTHDF)1, YTHDF2, YTHDF3, YTH domain containing
1 (YTHDC)1, and YTHDC2 read mRNA with the m6A
modification specifically in the cytoplasm (56). In the cytoplasm, the
m6A-binding protein YTHDF1 promotes the translation of
m6A-modified mRNAs, and YTHDF2 facilitates the decay of
m6A-modified transcripts. YTHDF3 accelerates protein
synthesis in synergy with YTHDF1 and impacts the methylated mRNA
decay mediated by YTHDF2. Cells deficient in all three YTHDFs show
the most dramatic accumulation of m6A-modified
transcripts (8). In addition to the
YTH domain m6A readers, other readers such as the
heterogeneous nuclear ribonucleoprotein A2/B1 (HNRNP A2/B1) and
heterogeneous nuclear ribonucleoprotein C (HNRNPC) do not directly
interact with m6A but bind to the transcripts containing
m6A (57).
YTH domain family
The expression of YTHDF1 and YTHDF3 is higher, but
that of YTHDF2 is lower, in human lung cancer tissues compared to
adjacent normal lung tissues. These alterations are related to the
functions of mRNA translation and decay of the pre-mRNA target
genes (Fig. 2B). Similarly, YTHDF1
knockdown partly blocks the pre-mRNA processing factor 6 (PRPF6)
expression and cell growth (34).
YTHDF1 positively facilitates protein synthesis by interacting with
the translation machinery (58).
YTHDF1 knockdown restrains NSCLC cell proliferation and xenograft
tumor formation by regulating the translational efficiency of
cyclin-dependent kinase (CDK)2, CDK4, and cyclin D1, whereas YTHDF1
depletion inhibits lung cancer progression. High expression of
YTHDF1 is related to better clinical outcomes, with its depletion
rendering cancer cells resistant to cisplatin treatment.
Mechanistic studies identified the Keap1-Nrf2 axis as the
downstream mediator of YTHDF1(59).
YTHDF2 is upregulated in lung cancer tissues, promotes lung cancer
cell growth and enhances the pentose phosphate pathway (PPP) flux,
which is important for tumor growth. Mechanistically, YTHDF2
directly binds to the m6A modification site of
6-phosphogluconate dehydrogenase(6-PGD) 3'-UTR to promote 6-PGD
mRNA translation in lung cancer cells (60). YTHDF3 with METTL3, YTHDF1, and eIF3b
directly promotes YAP translation via interaction with the
translation initiation machinery in NSCLC. Meanwhile, MALAT1
stability is increased by the METTL3/YTHDF3 complex. Both YAP and
MALAT1 promote carcinogenic activity and are associated with the
occurrence of lung cancer (31).
YTHDF1 and YTHDF2 competitively interacted with YTHDF3 in an
m6A-independent manner to regulate YAP expression.
YTHDF2 facilitated YAP mRNA decay via the AGO2 system, whereas
YTHDF1 promoted YAP mRNA translation by interacting with eIF3a;
both these activities are regulated by m6A modification
(53).
In addition, the m6A-binding protein
YTHDC2 is associated with tumor progression in lung cancer
(27). YTHDC2 is frequently
suppressed in LUAD. Downregulation of YTHDC2 is associated with
poor clinical outcome of LUAD. Moreover, YTHDC2 exhibits antitumor
activity in human LUAD cells. Mechanistically, YTHDC2, through its
m6A-recognizing YTH domain, inhibits cystine uptake and
blocked the downstream antioxidant program. Furthermore, solute
carrier 7A11 (SLC7A11), the catalytic subunit of cystine
transporter system Xc-, is identified to be the direct target of
YTHDC2. YTHDC2 destabilizes SLC7A11 mRNA in an
m6A-dependent manner because YTHDC2 preferentially bound
to m6A-modified SLC7A11 mRNA and thereafter promotes its
decay (61). In summary, these YTH
domain family proteins may influence the fundamental biological
processes in an integrated and cooperative manner in lung
cancer.
HNRNP family
HNRNP A2/B1 is overexpressed in lung cancer and in
other cancers, such as liver cancer, breast cancer, and pancreatic
cancer (62,63). This overexpression is not associated
with the histological classification of lung cancer but with the
clonal expansion of the tumor in NSCLC patients (64,65).
He et al demonstrated that hnRNP A2/B1 formed complexes with
the transcripts of many of the verified downstream genes,
suggesting that HNRNP A2/B1 contributes to the regulation of these
genes (66). The expression of
HNRNP A2/B1 protein is correlated with the expression of anexeleto
(AXL). The expression of HNRNP A2/B1 and AXL affects the prognosis
of patients with NSCLC (67).
CACNA1G-AS1 (CAS1) increases the level of HNRNP A2/B1, which
enhances cancer cell invasion and migration in NSCLC (68). In addition, expression of HNRNP
A2/B1 may affect the function of EMT by regulating the E-cadherin
expression in non-epithelial lung cancer cell lines (62). These results reinforce the
conclusion that HNRNP A2/B1 is associated with cellular processes
that affect the cell cycle and proliferation.
HNRNPC is another RNA-binding protein ‘reader’ of
m6A methylation, and is related to the progression of
various cancers. HNRNPC is upregulated in progressed lung cancer
(27). HNRNPC expression is
significantly related to poor overall survival in patients with
LUAD, indicating that HNRNPC may be a cancer-promoting factor and a
potential prognostic biomarker in LUAD (69). Overexpression of HNRNPC
significantly enhances lung cancer cell proliferation, migration,
and invasion both in vitro and in vivo. In NSCLC cell
lines, HNRNPC interacts with KH-type splicing regulatory protein
(KHSRP), which is considered to be a metastasis-associated
candidate molecule. KHSRP and HNRNPC are significantly associated
with advanced tumor progression and metastasis (both lymph node and
distant) and may induce invasion and metastasis in human lung
cancer (70).
The other m6A binding
proteins
IGF2BPs, a class of RNA-binding proteins, including
IGF2BP1, IGF2BP2, and IGF2BP3, are considered to be the ‘reader’ of
m6A methylation and remarkably affects cancer occurrence
and development. Studies have proved that IGF2BP3 has prognostic
potential in multiple public databases compared with other members
of the IGF2BPs family. IGF2BP3 is abnormally highly expressed in
LUAD tissue, and can lead to worse overall survival. IGF2BP3
expression could serve for independently predicting the prognosis
of LUAD patients. In summary, IGF2BP3 may be an oncogene and
potential prognostic biomarker of LUAD (71).
m6A regulators regulate the expression of
the downstream target genes by mediating the mRNA stability,
translation efficiency, and mRNA decay to affect the proliferation,
migration, and invasion of lung cancer cells (Table I). Knowledge about the mechanism of
m6A methylation is limited, and the supplementary
discoveries of regulatory patterns mediated by m6A in
lung cancer are worth verifying in future studies.
 | Table IPotential mechanisms and target genes
of m6A regulators in lung cancer. |
Table I
Potential mechanisms and target genes
of m6A regulators in lung cancer.
| A, Writer
m6A components |
|---|
| Proteins | Related
targets | Roles in lung
cancer | (Refs.) |
|---|
| METTL3 | EGFR, TAZ, eIF,
CBP80, BRD4, DNMT3A, JUNB, EZH2, ATG7, LC3B, SQSTM1 | Promotes growth,
translation, survival and invasion of lung cancer cells | (28,30,25,49) |
| METTL14 | Unknown | Forms complex with
METTL3 | (10) |
|
METTL16 | MALAT | Combines with
metastasis-related lung adenocarcinoma transcripts to promote lung
cancer activity | (85) |
| WTAP | Unknown | Forms complex with
METTL3 | (86) |
| B, Eraser
m6A components |
| Proteins | Related
targets | Roles in lung
cancer | (Refs.) |
| FTO | USP7, MZF1 | Promotes growth of
lung cancer cells | (46,47) |
| ALKBH5 | FOXM1, YAP UBE2C,
TIMP3 | Promotes growth and
invasion of lung adenocarcinoma cells and stabilize mRNA
transcripts | (16,53) |
| C, Reader
m6A components |
| Proteins | Related
targets | Roles in lung
cancer | (Refs.) |
| YTHDF1 | eIF, G3BP1 | Promotes
translational efficiency | (58) |
| YTHDF2 | 6-PGD | Reduces the
stability of the target transcript | (60) |
| YTHDF3 | YAP | Regulates the
stability of mRNA and cooperates with YTHDF1 to promote protein
synthesis | (53) |
| HNRNP A2/B1 | AXL, COX-2,
PGE2 | Regulates
expression of transcription-related factors and determines cell
fate transition | (64,87,88) |
| HNRNPC | KHSRP, uPAR | Promotes
proliferation, migration, and invasion of lung cancer cells | (70,89) |
5. m6A as a potential therapeutic
target in lung cancer
The treatment options for lung cancer have developed
considerably over the past years; however, most patients are
diagnosed at an advanced stage of the disease due to the insidious
symptoms of early-stage lung cancer. Thus, the survival rate of
lung cancer patients remains low (72). Most RNA methylation regulators had
distinct expressions in tumor tissues and adjacent tissues. The
patients in the high-risk group were more likely to have a higher
stage, more lymph node metastases, and distant metastases, showing
a poor clinical outcome. Different molecular phenotypes constructed
by RNA methylation regulators can be independent ri7sk factors for
the prognosis of LUAD (73). The
expression of m6A methylation regulators between high-
and low-risk LUSC patients is significantly different, and the
high-risk LUSC patients have significantly low levels of ALKBH5,
METTL3, HNRNPC, and KIAA1429. Thus, m6A methylation regulators may
result in a poor prognosis in patients with low-risk LUSC (74).
Recently, cancer immunotherapy has become involved
in treating all forms of cancer and has changed the landscape of
cancer care. LUAD is the most common histological subtype in lung
cancer. LUAD subtypes are identified on the basis of the
immunogenomic profiling of 29 immune signatures. There are three
LUAD subtypes: Immunity High, Immunity Medium, and Immunity Low.
The Immunity High subtype exhibits more sensitivity to
immunotherapy and chemotherapy. Immunity High is significantly
associated with decreased gene expression, such as METTL3, RBM15,
YTHDC1, YTHDF1, and YTHDF2, which are involved in m6A
mRNA methylation. And the level of m6A RNA methylation, associates
with cancer initiation and progression, is reduced in the Immunity
High subtype (75). Interleukin-37
(IL-37) plays a crucial protective role in lung cancer. Treatment
of IL-37 in lung cancer cells induced widespread and dynamic RNA
m6A methylation. It could lead to changes in m6A
methylation level and relates molecule expression level in A546
cells, such as METTL3, METTL14, WTAP, ALKBH5 and FTO, and may
downregulate the proliferation by inhibiting Wnt5a/5b pathway in
A549 cells. It concludes that IL-37 suppresses tumor growth through
regulation of RNA m6A methylation in lung cancer cells
(76). These findings suggest that
m6A RNA methylation is important determinants of
initiation, progression and prognosis in lung cancer and may
provide potential prognostic biomarker or therapeutic target for
immunotherapeutic and chemotherapeutic development.
m6A performs multi-functional roles in
EMT modulation, suggesting the critical roles of m6A in cancer
progression regulation. EMT plays a critical role in lung cancer
progression; thus, it is important to identify the factors that
inhibit EMT in lung cancer treatment (77). METTL3 downregulation in lung cancer
tissues influences EMT via m6A modification of the enhancer of
zeste homolog 2 (EZH2), contributes to the macrophage recruitment,
and reduces the malignant progression of lung cancer (30). Knockdown of METTL3 inhibits the
TGF-β-induced morphological conversion of the cell and increases
the cell migration potential as well as changes in the expression
of EMT-related marker genes (35).
TGF-β1-induced EMT is inhibited in METTL3 knockdown cells. The
expression of TGF-β1 is up-regulated, while self-stimulated
expression of TGF-β1 is suppressed in METTL3 cells. m6A promotes
TGF-β1 mRNA decay, but impairs TGF-β1 translation progress. Besides
this, the autocrine of TGF-β1 is disrupted in METTL3 cells through
interrupting TGF-β1 dimer formation. Snail, which is down-regulated
in METTL3 cells, is a key factor responding to TGF-β1-induced EMT
(78). In addition, YTHDF1 is
positively correlated with the growth, invasion, and EMT of NSCLC
cells, while YTHDF2 plays an opposite role in these cell processes
(53). Recent studies have
demonstrated the inhibitory effect of simvastatin on tumor cell
proliferation. Simvastatin causes METTL3 downregulation in lung
cancer tissues, resulting in EMT via m6A modification of mRNA, thus
restraining the malignant progression of lung cancer (30). These results indicate that m6A
regulators can be potential therapeutic targets for EMT in lung
cancer cells.
In recent years, epigenetics, especially
m6A RNA modification, has been further understood and
explored with the rapid advances in detection methods and
high-throughput sequencing techniques (79). It has been widely illustrated that
the dysregulation of m6A RNA modification is related to
various types of cancers, as well as the drug resistance to
anti-tumor therapy (80). In three
LUAD cell lines, treatment with ammonium tetra thiomolybdate (ATTM)
at high concentrations inhibited the cell growth, while at low ATTM
concentrations the cell growth was promoted. Treatment with ATTM
significantly increased the level of METTL3 but reduced the FTO
levels. Additionally, ATTM upregulates METTL14 expression, which is
not consistent with The Cancer Genome Atlas (TCGA) analysis. This
difference may be due to the uncertainty in gene expression between
the mRNA and protein levels. This unique expression contributes to
ATTM-induced increase in m6A in A549 cells (34). METTL3 promotes the translation of
important oncogenes like EGFR in lung cancer. EGFR inhibitors, such
as gefitinib and erlotinib, have gained approval for the treatment
of patients with NSCLC (81). FTO
inhibitor rhein enhances the antitumor activity of pemetrexed
through influencing autophagy and apoptosis by modulating the p
PI3K-AKT-mTOR pathway and B-cell lymphoma-2 (Bcl-2) family of
proteins in A549 cells. It demonstrates that the potential
application of rhein as a candidate drug in combination with
pemetrexed is promising for treatment of the human lung cancer
(82). m6A
methyltransferase METTL3-mediated autophagy plays an important role
in reversing gefitinib resistance by β-elemene in NSCLC cells.
Mechanistically, β-elemene can reverse gefitinib resistance by
inhibiting the late stage of autophagy in a manner of chloroquine,
which inhibits the maturation of autophagosomes into autolysosomes
through attenuating the lysosomal acidification. In this reversing
process, METTL3 can positively regulate this autophagy process by
targeting autophagy protein (ATG5), ATG7, light chain 3B (LC3B) and
Sequestosome 1 (SQSTM1) (83).
Functional enrichment analysis of the m6A-modified genes
revealed that m6A methylation might modify the cell
cycle to influence the response to afatinib. Furthermore, these
m6A-modified genes are over-represented in the putative
drug resistance-associated genes and FDA-approved drug targets and
have a higher average degree and clustering coefficient than other
genes in the protein–protein interaction network (84). Overall, the action of m6A
regulators may contribute to drug resistance in tumor therapy and
prognosis.
6. Conclusion
m6A methylation has a huge impact on RNA
production/metabolism and is involved in the pathogenesis of many
diseases, including cancer. In the occurrence and development of
lung cancer, m6A-modified mRNA regulates RNA
transcription, splicing, processing, translation, and decay.
Accumulating evidence reveals that m6A regulators and
their mechanisms of action play vital roles in lung cancer. The
m6A modification directly or indirectly affects cell
proliferation, metastasis, invasion, and apoptosis. Systematic
study of the functions and potential molecular mechanisms of
m6A regulators will further improve our understanding of
the complex networks associated with lung cancer. Inhibitors
targeting m6A regulators may have great therapeutic
potential in the treatment of lung cancer. In addition to the known
modulating effects of m6A methylation, the underlying
mechanism of m6A modification in lung cancer needs to be
further investigated.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the project of innovation
plan for graduate students of Beihua University (grant no. 2019021,
2019027), the National Student's Training Program for Innovation
and Entrepreneurship (grant no. 201913706015), the Scientific
Technology Research Project of the Education Department of Jilin
Province (grant nos. JJKH20191068KJ and JJKH20200076KJ), and the
Jilin Science and Technology Innovation Development Program (grant
no. 20190601177).
Availability of data and materials
Not applicable.
Authors' contributions
YW and XS designed the review. MZ, MX, YC, and ZL
were involved in the collection and collation of references. YW
wrote, reviewed, and edited the manuscript. WZ critically revised
the manuscript for intellectual content. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Liu N and Pan T:
N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol.
23:98–102. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jonkhout N, Tran J, Smith M, Schonrock N,
Mattick J and Novoa E: The RNA modification landscape in human
disease. RNA. 23:1754–1769. 2017.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Li L, Fan Y, Leng R, Pan H and Ye D:
Potential link between m6A modification and systemic lupus
erythematosus. Mol Immunol. 93:55–63. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Meyer K, Saletore Y, Zumbo P, Elemento O,
Mason C and Jaffrey S: Comprehensive analysis of mRNA methylation
reveals enrichment in 3'UTRs and near stop codons. Cell.
149:1635–1646. 2012.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang X, Lu Z, Gomez A, Hon G, Yue Y, Han
D, Fu Y, Parisien M, Dai Q, Jia G, et al:
N6-methyladenosine-dependent regulation of messenger RNA stability.
Nature. 505:117–120. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Boccaletto P, Machnicka M, Purta E,
Piatkowski P, Baginski B, Wirecki T, de Crécy-Lagard V, Ross R,
Limbach P, Kotter A, et al: MODOMICS: A database of RNA
modification pathways 2017 update. Nucleic Acids Res. 46:D303–D307.
2018.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Han Z, Niu T, Chang J, Lei X, Zhao M, Wang
Q, Cheng W, Wang J, Feng Y and Chai J: Crystal structure of the FTO
protein reveals basis for its substrate specificity. Nature.
464:1205–1209. 2010.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu
PJ, Liu C and He C: YTHDF3 facilitates translation and decay of
N(6)-methyladenosine-modified RNA. Cell Res. 27:315–328.
2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Schumann U, Shafik A and Preiss T: METTL3
Gains R/W access to the Epitranscriptome. Mol Cell. 62:323–324.
2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang
L, Jia G, Yu M, Lu Z, Deng X, et al: A METTL3-METTL14 complex
mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem
Biol. 10:93–95. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Wu F, Cheng W, Zhao F, Tang M, Diao Y and
Xu R: Association of N6-methyladenosine with viruses and related
diseases. Virol J. 16(133)2019.PubMed/NCBI
|
|
12
|
Ping XL, Sun BF, Wang L, Xiao W, Yang X,
Wang W, Adhikari S, Shi Y, Lv Y, Chen YS, et al: Mammalian WTAP is
a regulatory subunit of the RNA N6-methyladenosine
methyltransferase. Cell Res. 24:177–189. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Meyer K and Jaffrey S: Rethinking m6A
Readers, Writers, and Erasers. Annu Rev Cell Dev Biol. 33:319–342.
2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Schwartz S, Mumbach M, Jovanovic M, Wang
T, Maciag K, Bushkin G, Mertins P, Ter-Ovanesyan D, Habib N,
Cacchiarelli D, et al: Perturbation of m6A writers reveals two
distinct classes of mRNA methylation at internal and 5'sites. Cell
Rep. 8:284–296. 2014.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Ding Y, Qi N, Wang K, Huang Y, Liao J,
Wang H, Tan A, Liu L, Zhang Z, Li J, et al: FTO facilitates lung
adenocarcinoma cell progression by activating cell migration
through mRNA Demethylation. Onco Targets Ther. 13:1461–1470.
2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Zhu Z, Qian Q, Zhao X, Ma L and Chen P: N
6-methyladenosine ALKBH5 promotes non-small cell lung cancer
progress by regulating TIMP3 stability. Gene.
731(144348)2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Han S, Tang Y and Smith R: Functional
diversity of the hnRNPs: Past, present and perspectives. Biochem J.
430:379–392. 2010.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Patil D, Pickering B and Jaffrey S:
Reading m 6 A in the Transcriptome: M 6 A-binding proteins. Trends
Cell Biol. 28:113–127. 2018.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Müller S, Glaß M, Singh A, Haase J, Bley
N, Fuchs T, Lederer M, Dahl A, Huang H, Chen J, et al: IGF2BP1
promotes SRF-dependent transcription in cancer in a m6A- and
miRNA-dependent manner. Nucleic Acids Res. 47:375–390.
2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Xu C, Liu K, Ahmed H, Loppnau P, Schapira
M and Min J: Structural basis for the discriminative recognition of
N6-methyladenosine RNA by the Human YT521-B homology domain family
of proteins. J Biol Chem. 290:24902–24913. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Arguello A, DeLiberto A and Kleiner R: RNA
chemical proteomics reveals the N 6-methyladenosine (m6A)-regulated
protein-RNA Interactome. J Am Chem Soc. 139:17249–17252.
2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Edupuganti RR, Geiger S, Lindeboom RGH,
Shi H, Hsu PJ, Lu ZY, Wang SY, Baltissen MPA, Jansen PWTC, Rossa M,
et al: N 6-methyladenosine (m6A) recruits and repels proteins to
regulate mRNA homeostasis. Nat Struct Mol Biol. 24:870–878.
2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wu D, Li M, Tian W, Wang S, Cui L, Li H,
Wang H, Ji A and Li Y: Hydrogen sulfide acts as a double-edged
sword in human hepatocellular carcinoma cells through
EGFR/ERK/MMP-2 and PTEN/AKT signaling pathways. Sci Rep.
7(5134)2017.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wakelee H, Kelly K and Edelman M: 50 Years
of progress in the systemic therapy of non-small cell lung cancer.
Am Soc Clin Oncol Educ Book: 2014: 177-189 doi:
10.14694/EdBook_AM.2014.34.177.
|
|
25
|
Siegel R, Miller K and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lin JJ, Cardarella S, Lydon CA, Dahlberg
SE, Jackman DM, Jänne PA and Johnson BE: Five-year survival in
EGFR-mutant metastatic lung adenocarcinoma treated with EGFR-TKIs.
J Thorac Oncol. 11:556–565. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhuang Z, Chen L, Mao Y, Zheng Q, Li H,
Huang Y, Hu Z and Jin Y: Diagnostic, progressive and prognostic
performance of m6A methylation RNA regulators in lung
adenocarcinoma. Int J Biol Sci. 16:1785–1797. 2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chen XY, Zhang J and Zhu JS: The role of
m6A RNA methylation in human cancer. Mol Cancer.
18(103)2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Liu Z, Li L, Sun H and Liu S: Link between
m6A modification and cancers. Front Bioeng Biotechnol.
6(89)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen WW, Qi JW, Hang J, Wu JX, Zhou XX,
Chen JZ, Wang J and Wang HH: Simvastatin is beneficial to lung
cancer progression by inducing METTL3-induced m6A modification on
EZH2 mRNA. Eur Rev Med Pharmacol Sci. 24:4263–4270. 2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Jin D, Guo J, Wu Y, Du J, Yang L, Wang X,
Di W, Hu B, An J, Kong L, et al: m(6)A mRNA methylation initiated
by METTL3 directly promotes YAP translation and increases YAP
activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce
NSCLC drug resistance and metastasis. J Hematol Oncol.
12(135)2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Choe J, Lin S, Zhang W, Liu Q, Wang L,
Ramirez-Moya J, Du P, Kim W, Tang S, Sliz P, et al: mRNA
circularization by METTL3-eIF3h enhances translation and promotes
oncogenesis. Nature. 561:556–560. 2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Lin S, Choe J, Du P, Triboulet R and
Gregory RI: The m(6)A methyltransferase METTL3 promotes translation
in human cancer cells. Mol Cell. 62:335–345. 2016.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Li X, Li N, Huang L, Xu S, Zheng X,
Hamsath A, Zhang M, Dai L, Zhang H, Wong JJ, et al: Is hydrogen
sulfide a concern during treatment of lung adenocarcinoma with
ammonium tetrathiomolybdate? Front Oncol. 10(234)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Wanna-Udom S, Terashima M, Lyu H, Ishimura
A, Takino T, Sakari M, Tsukahara T and Suzuki T: The m6A
methyltransferase METTL3 contributes to transforming growth
Factor-beta-induced epithelial-mesenchymal transition of lung
cancer cells through the regulation of JUNB. Biochem Biophys Res
Commun. 524:150–155. 2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Du Y, Hou G, Zhang H, Dou J, He J, Guo Y,
Li L, Chen R, Wang Y, Deng R, et al: SUMOylation of the m6A-RNA
methyltransferase METTL3 modulates its function. Nucleic Acids Res.
46:5195–5208. 2018.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Li L, Chang W and Hsiao M: Aberrant
expression of microRNA clusters in head and neck cancer development
and progression: Current and future translational impacts.
Pharmaceuticals (Basel). 14(194)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Gu S, Sun D, Dai H and Zhang Z: N
6-methyladenosine mediates the cellular proliferation and apoptosis
via microRNAs in Arsenite-transformed cells. Toxicol Lett.
292:1–11. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Du M, Zhang Y, Mao Y, Mou J, Zhao J, Xue
Q, Wang D, Huang J, Gao S and Gao Y: MiR-33a suppresses
proliferation of NSCLC cells via targeting METTL3 mRNA. Biochem
Biophys Res Communs. 482:582–589. 2017.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Wei W, Huo B and Shi X: miR-600 inhibits
lung cancer via downregulating the expression of METTL3. Cancer
Manag Res. 11:1177–1187. 2019.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Wang H, Deng Q, Lv Z, Ling Y, Hou X, Chen
Z, Dinglin X, Ma S, Li D, Wu Y, et al: N6-methyladenosine induced
miR-143-3p promotes the brain metastasis of lung cancer via
regulation of VASH1. Mol Cancer. 18(181)2019.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Wu LS, Qian JY, Wang M and Yang H:
Identifying the role of Wilms tumor 1 associated protein in cancer
prediction using integrative genomic analyses. Mol Med Rep.
14:2823–2831. 2016.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Ruszkowska A, Ruszkowski M, Dauter Z and
Brown J: Structural insights into the RNA methyltransferase domain
of METTL16. Sci Rep. 8(5311)2018.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Li N and Zhan X: Identification of
pathology-specific regulators of m6A RNA modification to
optimize lung cancer management in the context of predictive,
preventive, and personalized medicine. EPMA J. 11:485–504.
2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yang L, Ma Y, Han W, Li W, Cui L, Zhao X,
Tian Y, Zhou Z, Wang W and Wang H: Proteinase-activated receptor 2
promotes cancer cell migration through RNA methylation-mediated
repression of miR-125b. J Biol Chem. 290:26627–26637.
2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Li J, Han Y, Zhang H, Qian Z, Jia W, Gao
Y, Zheng H and Li B: The m6A demethylase FTO promotes the growth of
lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem
Biophys Res Commun. 512:479–485. 2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Liu J, Ren D, Du Z, Wang H, Zhang H and
Jin Y: m6A demethylase FTO facilitates tumor progression
in lung squamous cell carcinoma by regulating MZF1 expression.
Biochem Biophys Res Commun. 502:456–464. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang
Y, Yi C, Lindahl T, Pan T, Yang YG and He C: N6-methyladenosine in
nuclear RNA is a major substrate of the obesity-associated FTO. Nat
Chem Biol. 7:885–887. 2011.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Li F, Wang H, Huang H, Zhang L, Wang D and
Wan Y: m6A RNA methylation regulators participate in the malignant
progression and have clinical prognostic value in lung
adenocarcinoma. Front Genet. 11(994)2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Zhao X, Yang Y, Sun BF, Shi Y, Yang X,
Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al: FTO-dependent
demethylation of N6-methyladenosine regulates mRNA splicing and is
required for adipogenesis. Cell Res. 24:1403–1419. 2014.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Chao Y, Shang J and Ji W:
ALKBH5-m6A-FOXM1 signaling axis promotes proliferation
and invasion of lung adenocarcinoma cells under intermittent
hypoxia. Biochem Biophys Res Commun. 521:499–506. 2020.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Guo J, Wu Y, Du J, Yang L, Chen W, Gong K,
Dai J, Miao S, Jin D and Xi S: Deregulation of UBE2C-mediated
autophagy repression aggravates NSCLC progression. Oncogenesis.
7(49)2018.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Jin D, Guo J, Wu Y, Yang L, Wang X, Du J,
Dai J, Chen W, Gong K, Miao S, et al: m6A demethylase
ALKBH5 inhibits tumor growth and metastasis by reducing
YTHDFs-mediated YAP expression and inhibiting
miR-107/LATS2-mediated YAP activity in NSCLC. Mol Cancer.
19(40)2020.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Deng X, Su R, Weng H, Huang H, Li Z and
Chen J: RNA N(6)-methyladenosine modification in cancers: Current
status and perspectives. Cell Res. 28:507–517. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Yue Y, Liu J and He C: RNA
N6-methyladenosine methylation in post-transcriptional gene
expression regulation. Genes Dev. 29:1343–1355. 2015.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Yang Y, Hsu PJ, Chen YS and Yang YG:
Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers
and functions in RNA metabolism. Cell Res. 28:616–624.
2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Liu N, Dai Q, Zheng G, He C, Parisien M
and Pan T: N(6)-methyladenosine-dependent RNA structural switches
regulate RNA-protein interactions. Nature. 518:560–564.
2015.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Wang X, Zhao BS, Roundtree IA, Lu Z, Han
D, Ma H, Weng X, Chen K, Shi H and He C: N(6)-methyladenosine
modulates messenger RNA translation efficiency. Cell.
161:1388–1399. 2015.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Shi Y, Fan S, Wu M, Zuo Z, Li X, Jiang L,
Shen Q, Xu P, Zeng L, Zhou Y, et al: YTHDF1 links hypoxia
adaptation and non-small cell lung cancer progression. Nat Commun.
10(4892)2019.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Sheng H, Li Z, Su S, Sun W, Zhang X, Li L,
Li J, Liu S, Lu B, Zhang S and Shan C: YTH domain family 2 promotes
lung cancer cell growth by facilitating 6-phosphogluconate
dehydrogenase mRNA translation. Carcinogenesis. 41:541–550.
2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Ma L, Chen T, Zhang X, Miao Y, Tian X, Yu
K, Xu X, Niu Y, Guo S, Zhang C, et al: The m(6)A reader YTHDC2
inhibits lung adenocarcinoma tumorigenesis by suppressing
SLC7A11-dependent antioxidant function. Redox Biol.
38(101801)2021.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Tauler J, Zudaire E, Liu H, Shih J and
Mulshine J: hnRNP A2/B1 modulates epithelial-mesenchymal transition
in lung cancer cell lines. Cancer Res. 70:7137–7147.
2010.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Wu C, Li W, Chen W, Xu D, Wei D and Zhou
Q: Expression of hnRNP A2/B1 in human lung cancer cell lines. Chin
J Lung Cancer. 7:121–124. 2004.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Dai X and Yang H: Cloning of hnRNP A2/B1
gene and detection of its expression in lung cancer tissues. Chin J
Lung Cancer. 8:266–269. 2005.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Zhou J, Nong L, Wloch M, Cantor A,
Mulshine J and Tockman M: Expression of early lung cancer detection
marker: hnRNP-A2/B1 and its relation to microsatellite alteration
in non-small cell lung cancer. Lung Cancer. 34:341–350.
2001.PubMed/NCBI View Article : Google Scholar
|
|
66
|
He Y, Rothnagel J, Epis M, Leedman P and
Smith R: Downstream targets of heterogeneous nuclear
ribonucleoprotein A2 mediate cell proliferation. Mol Carcinog.
48:167–179. 2009.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Qu X, Liu J, Zhong X, Li X and Zhang Q:
Insights into the roles of hnRNP A2/B1 and AXL in non-small cell
lung cancer. Oncol Lett. 10:1677–1685. 2015.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Yu P, Kang A, Jing L and Wang Y: Long
non-coding RNA CACNA1G-AS1 promotes cell migration, invasion and
epithelial-mesenchymal transition by HNRNPA2B1 in non-small cell
lung cancer. Eur Rev Med Pharmacol Sci. 22:993–1002.
2018.PubMed/NCBI View Article : Google Scholar
|
|
69
|
Guo W, Huai Q, Zhang G, Guo L, Song P, Xue
X, Tan F, Xue Q, Gao S and He J: Elevated heterogeneous nuclear
ribonucleoprotein C expression correlates with poor prognosis in
patients with surgically resected lung adenocarcinoma. Front Oncol.
10(598437)2020.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Yan M, Sun L, Li J, Yu H, Lin H, Yu T,
Zhao F, Zhu M, Liu L, Geng Q, et al: RNA-binding protein KHSRP
promotes tumor growth and metastasis in non-small cell lung cancer.
J Exp Clin Cancer Res. 38(478)2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Guo W, Huai Q, Wan H, Guo L, Song P, Gao S
and He J: Prognostic impact of IGF2BP3 expression in patients with
surgically resected lung adenocarcinoma. DNA Cell Biol. 40:316–331.
2021.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Miller K, Nogueira L, Mariotto A, Rowland
J, Yabroff K, Alfano C, Jemal A, Kramer J and Siegel R: Cancer
treatment and survivorship statistics, 2019. CA Cancer J Clin.
69:363–385. 2019.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Sun L, Liu W, Du X, Liu X, Li G, Yao Y,
Han T, Li W and Gu J: Large-scale transcriptome analysis identified
RNA methylation regulators as novel prognostic signatures for lung
adenocarcinoma. Ann Transl Med. 8(751)2020.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Xu F, Zhang H, Chen J, Lin L and Chen Y:
Immune signature of T follicular helper cells predicts clinical
prognostic and therapeutic impact in lung squamous cell carcinoma.
Int Immunopharmacol. 81(105932)2020.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Xu F, Chen J, Yang X, Hong X, Li Z, Lin L
and Chen Y: Analysis of lung adenocarcinoma subtypes based on
immune signatures identifies clinical implications for cancer
therapy. Mol Ther Oncolytics. 17:241–249. 2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Mu X, Zhao Q, Chen W, Zhao Y, Yan Q, Peng
R, Zhu J, Yang C, Lan K, Gu X and Wang Y: IL-37 confers anti-tumor
activity by regulation of m6A methylation. Front Oncol.
10(526866)2020.PubMed/NCBI View Article : Google Scholar
|
|
77
|
Yu L, Kim HJ, Park MK, Byun HJ, Kim EJ,
Kim B, Nguyen MT, Kim JH, Kang GJ, Lee H, et al: Ethacrynic acid, a
loop diuretic, suppresses epithelial-mesenchymal transition of A549
lung cancer cells via blocking of NDP-induced WNT signaling.
Biochem Pharmacol. 183(114339)2020.PubMed/NCBI View Article : Google Scholar
|
|
78
|
Li J, Chen F, Peng Y, Lv Z, Lin X, Chen Z
and Wang H: N6-methyladenosine regulates the expression and
secretion of TGFβ1 to affect the epithelial-mesenchymal transition
of cancer cells. Cells. 9(296)2020.PubMed/NCBI View Article : Google Scholar
|
|
79
|
Gu C, Shi X, Dai C, Shen F, Rocco G, Chen
J, Huang Z, Chen C, He C, Huang T, et al: RNA m6A
modification in cancers: Molecular mechanisms and potential
clinical applications. Innovation. 1(100066)2020.
|
|
80
|
Frye M, Harada B, Behm M and He C: RNA
modifications modulate gene expression during development. Science.
361:1346–1349. 2018.PubMed/NCBI View Article : Google Scholar
|
|
81
|
Modjtahedi H and Essapen S: Epidermal
growth factor receptor inhibitors in cancer treatment: Advances,
challenges and opportunities. Anticancer Drugs. 20:851–855.
2009.PubMed/NCBI View Article : Google Scholar
|
|
82
|
Bu T, Wang C, Jin H, Meng Q, Huo X, Sun H,
Sun P, Wu J, Ma X, Liu Z and Liu K: Organic anion transporters and
PI3K-AKT-mTOR pathway mediate the synergistic anticancer effect of
pemetrexed and rhein. J Cell Physiol. 235:3309–3319.
2020.PubMed/NCBI View Article : Google Scholar
|
|
83
|
Liu S, Li Q, Li G, Zhang Q, Zhuo L, Han X,
Zhang M, Chen X, Pan T, Yan L, et al: The mechanism of
m6A methyltransferase METTL3-mediated autophagy in
reversing gefitinib resistance in NSCLC cells by β-elemene. Cell
Death Dis. 11(969)2020.PubMed/NCBI View Article : Google Scholar
|
|
84
|
Meng Q, Wang S, Zhou S, Liu H, Ma X, Zhou
X, Liu H, Xu C and Jiang W: Dissecting the m6A
methylation affection on afatinib resistance in non-small cell lung
cancer. Pharmacogenomics J. 20:227–234. 2019.PubMed/NCBI View Article : Google Scholar
|
|
85
|
Brown JA, Kinzig CG, DeGregorio SJ and
Steitz JA: Methyltransferase-like protein 16 binds the 3'-terminal
triple helix of MALAT1 long noncoding RNA. Proc Natl Acad Sci USA.
113:14013–14018. 2016.PubMed/NCBI View Article : Google Scholar
|
|
86
|
Fish L, Navickas A, Culbertson B, Xu Y,
Nguyen HCB, Zhang S, Hochman M, Okimoto R, Dill BD, Molina H, et
al: Nuclear TARBP2 drives oncogenic dysregulation of RNA splicing
and decay. Mol Cell. 75:967–981.e9. 2019.PubMed/NCBI View Article : Google Scholar
|
|
87
|
Kwon J, Jo Y, Namgoong S and Kim N:
Functional roles of hnRNPA2/B1 regulated by METTL3 in mammalian
embryonic development. Sci Rep. 9(8640)2019.PubMed/NCBI View Article : Google Scholar
|
|
88
|
Xuan Y, Wang J, Ban L, Lu JJ, Yi C, Li Z,
Yu W, Li M, Xu T, Yang W, et al: hnRNPA2/B1 activates
cyclooxygenase-2 and promotes tumor growth in human lung cancers.
Mol Oncol. 10:610–624. 2016.PubMed/NCBI View Article : Google Scholar
|
|
89
|
Shetty S: Regulation of urokinase receptor
mRNA stability by hnRNP C in lung epithelial cells. Mol Cell
Biochem. 272:107–118. 2005.PubMed/NCBI View Article : Google Scholar
|
















