Introduction
The worldwide incidence of ulcerative colitis (UC)
continues to increase at a significant rate (1). UC is a type of inflammatory bowel
disease characterized by periods of inflammatory recurrence and
remission events, which are accompanied by cell death and
regeneration of the colonic mucosa. It is well known that
long-standing UC leads to dysplasia (i.e., precancerous lesions)
and colorectal cancer (CRC) and is often a threat to the lives of
the patients. The incidence of colorectal dysplasia in patients
with UC has been reported to be 1.9% at 5 years, 5.1% at 15 years,
and 9.2% at 25 years after the onset of UC (2). The risk of developing UC-associated
CRC (UC-CRC) increases 0.5-1% per year in patients who have had UC
for longer than 8-10 years (3). In
addition, the prognosis of CRC is generally poorer in patients with
UC than in patients without UC (4).
Therefore, surveillance colonoscopy is recommended for the
detection of neoplasms in patients with UC. Early detection of
UC-CRC is essential for the successful management of long-standing
UC (4). However, endoscopic and
histologic detection of dysplasia is often difficult due to the
presence of inflammatory and subsequent regenerative changes in the
colonic mucosa (3,5). For the early detection of dysplasia,
it is important to understand the mechanism of CRC development in
patients with UC. While chronic inflammation of the colonic mucosa
is believed to cause UC-CRC (6),
the genetic details of UC-CRC pathogenesis remain unclear (7). In cases of CRC in patients without UC,
p53 mutations are generally considered to be involved in the later
stages of carcinogenesis (8), while
in UC-CRC, p53 mutations have been reported to occur earlier in
tumor development (3,7). p53 immunohistochemistry (IHC) is
therefore often used for the diagnosis of neoplasms in UC.
In both non-UC-CRC and UC-CRC, the expressions of
some genes have different effects on carcinogenesis, but these two
diseases also share many common genes (9).
Previously, our laboratory reported 17 genes
associated with distant metastases extracted from microarrays using
gene expression data (10), as well
as the involvement of the ATF6 in the carcinogenic process
of UC (11). ATF6 was rarely
expressed in normal mucosa but highly expressed in colon adenomas
and CRC. This gene was confirmed to be highly expressed in
dysplasia lesions and UC-related cancers. Therefore, we searched
for genes that may be involved from the early stages of canceration
to the metastatic stage and selected PHLDA1, which encodes
for pleckstrin homology-like domain, family A, member 1, which is
one of the 17 genes (12). In this
study, we focused on the possible involvement of PHLDA1 in
UC carcinogenesis and cancer progression.
Materials and methods
Identification of PHLDA1
The microarray data used was obtained from a
previous study (13). The gene
expression data are deposited in the Gene Expression Omnibus
(http://www.ncbi.nlm.nih.gov/geo/) under
accession number GSE32323.
The gene expression data were analyzed to identify
the candidate genes related to distant recurrence. The criteria to
select candidate genes were as follows: i) A higher expression
level in cancer tissues than in non-cancerous mucosa, and ii) a
significantly higher expression level in cancer cells from the
recurrence group than in the non-recurrence group. A higher
expression was defined as a numerical value 1.5 times greater than
those in another group (10). In
the microarray analysis, 69 genes were identified that fulfilled
the abovementioned criteria. Among 69 genes, 17 genes were found
associated with human malignancies. Among the 17 candidate genes,
we focused on PHLDA1, which is expressed in adenomas in the
early stages of canceration and is also involved in cancer
progression.
Patients and samples for the IHC
study
A total of 49 consecutive patients with UC who
underwent colectomy between January 2004 and December 2017 were
included in this study. Overall, 143 lesions were analyzed in the
IHC study. The diagnosis was based on surgically resected UC
specimens and was made by pathologists who specialize in colon
pathology. The pathologic findings of the UC samples were
categorized into two groups: The absence of neoplasia group
(colitis) and the neoplasia group (dysplasia and UC-CRC).
Samples of non-neoplastic lesions from patients with
or without neoplasia were also selected. Samples of neoplasia were
selected from each neoplastic lesion present. Neoplastic and
non-neoplastic lesions comprised one sample from a single lesion.
Table I summarizes the patients'
characteristics.
 | Table ICharacteristics of patients with
UC. |
Table I
Characteristics of patients with
UC.
| Characteristics | All patients
(n=49) | Patients without
neoplasia (n=30) | Patients with
neoplasia (n=19) |
|---|
| Age, years (mean ±
SD) | 47.6±16.9 | 47.7±19.5 | 47.5±11.2 |
| Sex, n (%) | | | |
|
Male | 30 (61.2) | 17 (56.7) | 13 (68.4) |
|
Female | 19 (38.8) | 13 (43.3) | 6 (31.6) |
| Disease extent, n
(%) | | | |
|
Pancolitis | 49 (100.0) | 30 (61.2) | 19 (38.8) |
|
Left-sided
colitis | 0 (0.0) | 0 (0.0) | 0 (0.0) |
|
Others | 0 (0.0) | 0 (0.0) | 0 (0.0) |
| Past history of 5-ASA
use, n (%) | | | |
|
Yes | 23 (46.9) | 11 (47.8) | 12 (52.2) |
|
No | 26 (53.1) | 19 (73.1) | 7 (26.9) |
| Past history of
steroid use, n (%) | | | |
|
Yes | 32 (65.3) | 24 (75.0) | 8 (25.0) |
|
No | 17 (34.7) | 6 (35.3) | 11 (64.7) |
| Number of samples,
n (%) | | | |
|
Colitis | 90 (62.9) | 64 (71.1) | 26 (28.9) |
|
Dysplasia | 39 (27.3) | 0 (0.0) | 39 (100.0) |
|
UC-CRC | 14 (9.8) | 0 (0.0) | 14 (100.0) |
Immunohistochemistry
IHC analysis was performed on 4-µm-thick sections
cut from formalin-fixed paraffin-embedded tissue blocks obtained
from each patient. PHLDA1 and p53-IHC analysis was performed on all
samples in this study. All sections were scored independently by
two investigators.
IHC for PHLDA1
The streptavidin-biotin method was used for PHLDA1
immunostaining. Xylene is soaked for 10 min five times. In
addition, it is soaked in 100% ethanol three times for 5 min, 95%
ethanol once for 3 min, 90% ethanol once for 3 min, 80% ethanol
once for 3 min, and 70% ethanol once for 3 min. Then it is
rehydrated with double-distilled water for 5 min three times.
Antigen retrieval was performed by heating the
tissue sections in pH 6.0 citrate buffer in a microwave at 98˚C for
25 min. Then, sections were then incubated in a solution of 3%
hydrogen peroxide in 100% methanol for 15 min at room temperature
to quench endogenous peroxidase activity. Next, sections were
incubated in a solution of rabbit polyclonal antibody against
PHLDA1 (1:250; sc-23866; Santa Cruz Biotechnology) for 15 min at
room temperature and then for a further 16 h at 4˚C, after which
they were labeled with a polymer [Histofine Simple Stain MAXPO
(MULTI); Nichirei Bioscience] for 30 min at room temperature.
Staining was visualized after incubation with DAB (0.02%
3,3'-diaminobenzidine tetrahydrochloride; Nichirei Bioscience) for
10 min at room temperature. Finally, the slides were counterstained
in 1% Mayer's hematoxylin, after which they were dehydrated in a
series of increasing alcohol concentrations, which was followed by
xylene immersion, mounting, and coverslipping.
PHLDA1-IHC was evaluated according to a modification
of the method previously described by Krajewska et al
(14). Positive cytoplasmic
staining in colitis, dysplastic, and UC-CRC cells was assessed and
scored on the basis of the immunostaining intensity. The
cytoplasmic immunostaining intensity of the ductal cells was graded
as - (negative), ± (weak), 1+ (strong), or 2+ (very strong)
compared with the stroma cells. The numbers of cells that exhibited
each grade of staining intensity were counted independently and
totaled. Scores corresponding to the percentage (%) of cells of
each grade of staining intensity were calculated relative to the
total number of cells. The score of cytoplasmic staining for PHLDA1
(potentially ranging from 0 to 2) was obtained by summing the
product of each percentage score by the corresponding intensity
score.
IHC for p53
The streptavidin-biotin method was used for mutated
p53 immunostaining. Antigen retrieval was performed by autoclaving
the tissues in pH 6.0 citrate buffer at 121˚C for 15 min.
Endogenous peroxidase activity was quenched using
the same method used for PHLDA1-IHC. The sections were sequentially
incubated with a polyclonal antihuman p53 antibody (1:200;
NCL-L-p53-DO7; Leica Biosystems) for 60 min at room temperature,
MULTI for 30 min at room temperature, DAB for color development,
and 1% Mayer's hematoxylin for the counterstain. Then, the sections
were dehydrated by immersion in a series of alcohol solutions and
xylene according to the method used for PHLDA1-IHC. As Shigaki
et al (15) reported, the
p53 staining pattern in the nucleus was characterized as sporadic,
mosaic, nested, and diffuse. Nested or diffuse patterns were
considered to represent p53-IHC positivity regardless of the
intensity, whereas the sporadic and mosaic patterns were considered
to represent p53-IHC negativity.
Statistical analysis
All statistical analyses were performed using EZR
(Saitama Medical Center, Jichi Medical University, Saitama, Japan),
a graphical user interface for R (The R Foundation for Statistical
Computing, Vienna, Austria). More precisely, it is a modified
version of R commander designed to add statistical functions
frequently used in biostatistics. For categorical data, the
significance of between-group differences was estimated using the
Fisher's exact test and χ2 test, as appropriate. For
continuous variables, descriptive statistics of the mean, median,
and range were calculated, and the significance of between-group
differences was estimated using the Kruskal-Wallis test as
appropriate. The Holm method was used to correct for significant
differences during multiple comparisons. A P value <0.05 was
considered statistically significant.
Results
PHLDA1 expression in UC samples
Of the 49 subjects, 30 exhibited no neoplastic
lesions and 19 had neoplasia. The pathologies of the 143 lesions
were as follows: 90 colitis, 39 dysplasia, and 14 UC-CRC. The
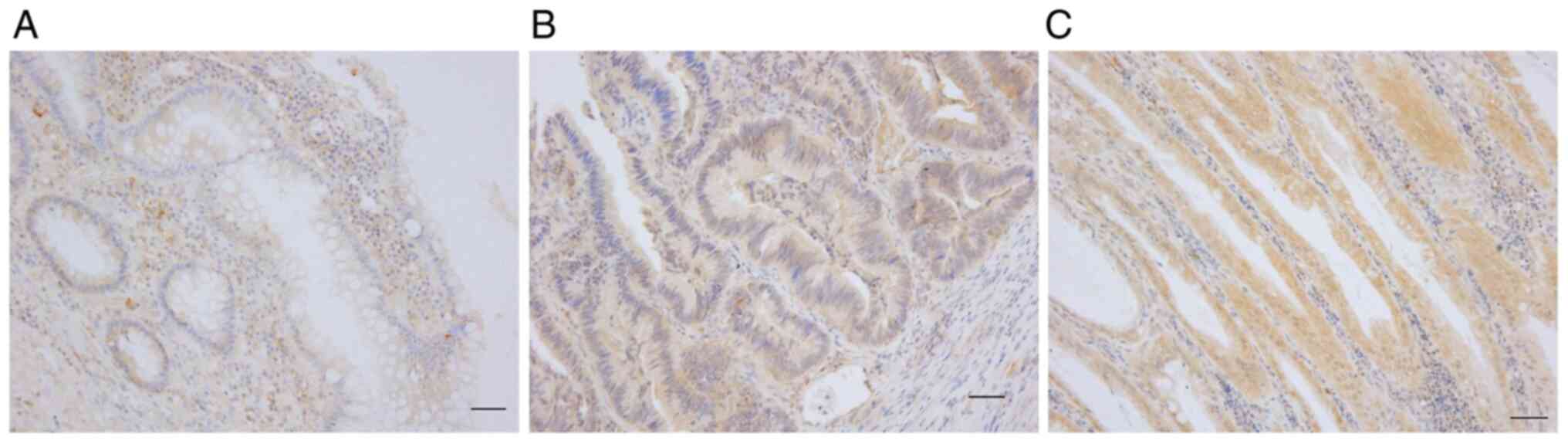
representative results of the PHLDA1-IHC staining in UC samples are
shown in Fig. 1. The median
PHLDA1-IHC score of the 143 UC samples was 0.40 (range, 0.00-1.43).
The median PHLDA1-IHC score in the 39 dysplasia samples was 0.607
(range, 0.03-1.32), which was significantly higher (P<0.001)
than that in the 90 colitis samples (median, 0.29, range, 0-1.29).
The median PHLDA1-IHC score in the 14 UC-CRC samples was 0.865
(range, 0.45-1.42), which was significantly higher (P=0.003) than
that in the dysplasia samples. The PHLDA1-IHC score tended to
increase as the UC cell variant progressed (Fig. 2).
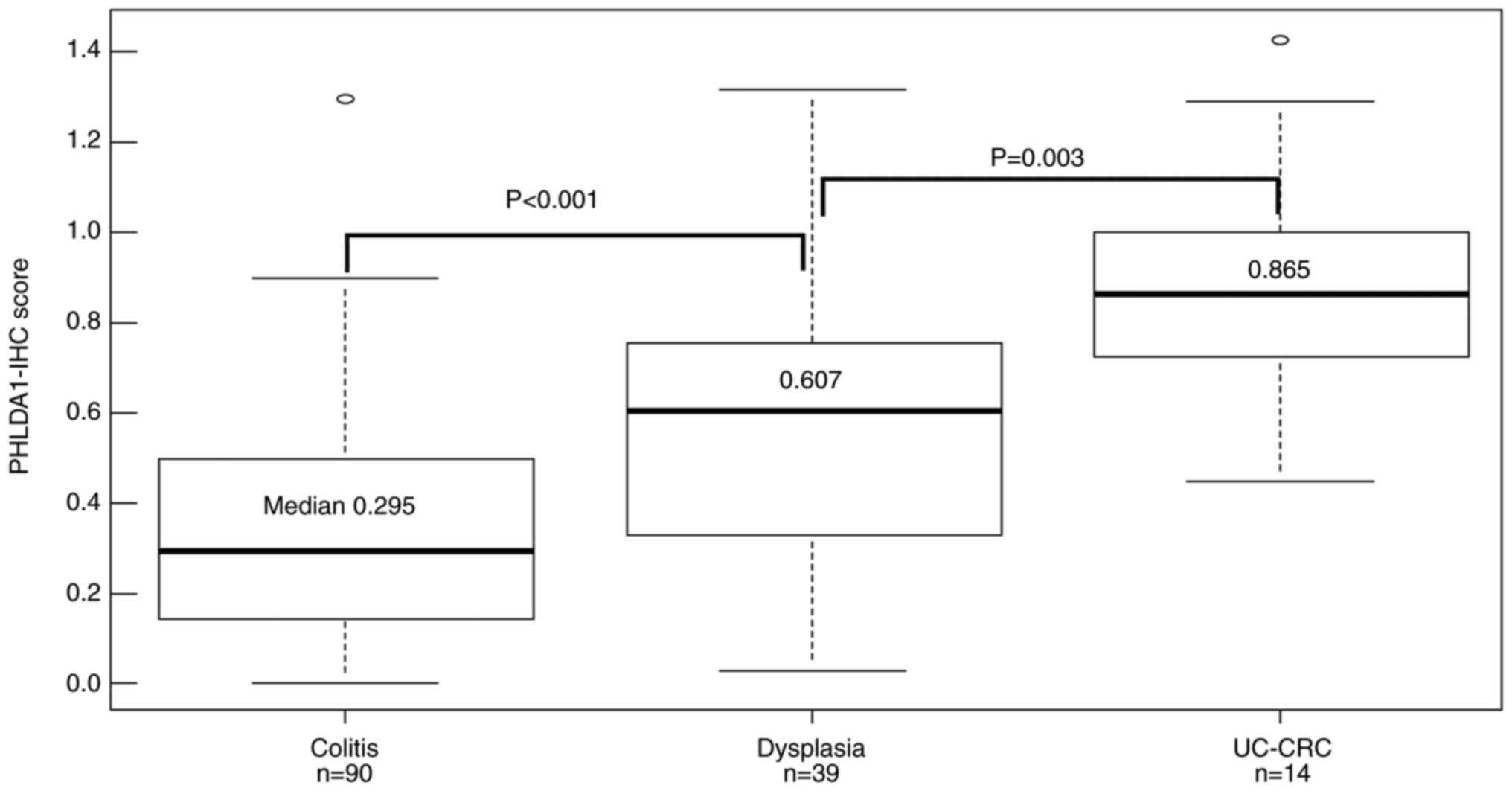 | Figure 2PHLDA1-IHC scores of the UC samples
according to histological category. The median PHLDA1-IHC score of
the 39 dysplasia samples was 0.607 (range, 0.03-1.32), which was
significantly higher (P<0.001) than that of the 90 colitis
samples (median, 0.295; range, 0-1.29). The median PHLDA1-IHC score
of the 14 UC-CRC samples was 0.865 (range, 0.45-1.42), which was
significantly higher (P=0.003) than that of the dysplasia samples.
The Kruskal-Wallis test was performed to compare these three
groups. The Holm method was used to correct for significant
differences during multiple comparisons. PHLDA1, pleckstrin
homology-like domain, family A, member 1; IHC,
immunohistochemistry; UC, ulcerative colitis; UC-CRC, UC-associated
colorectal cancer. |
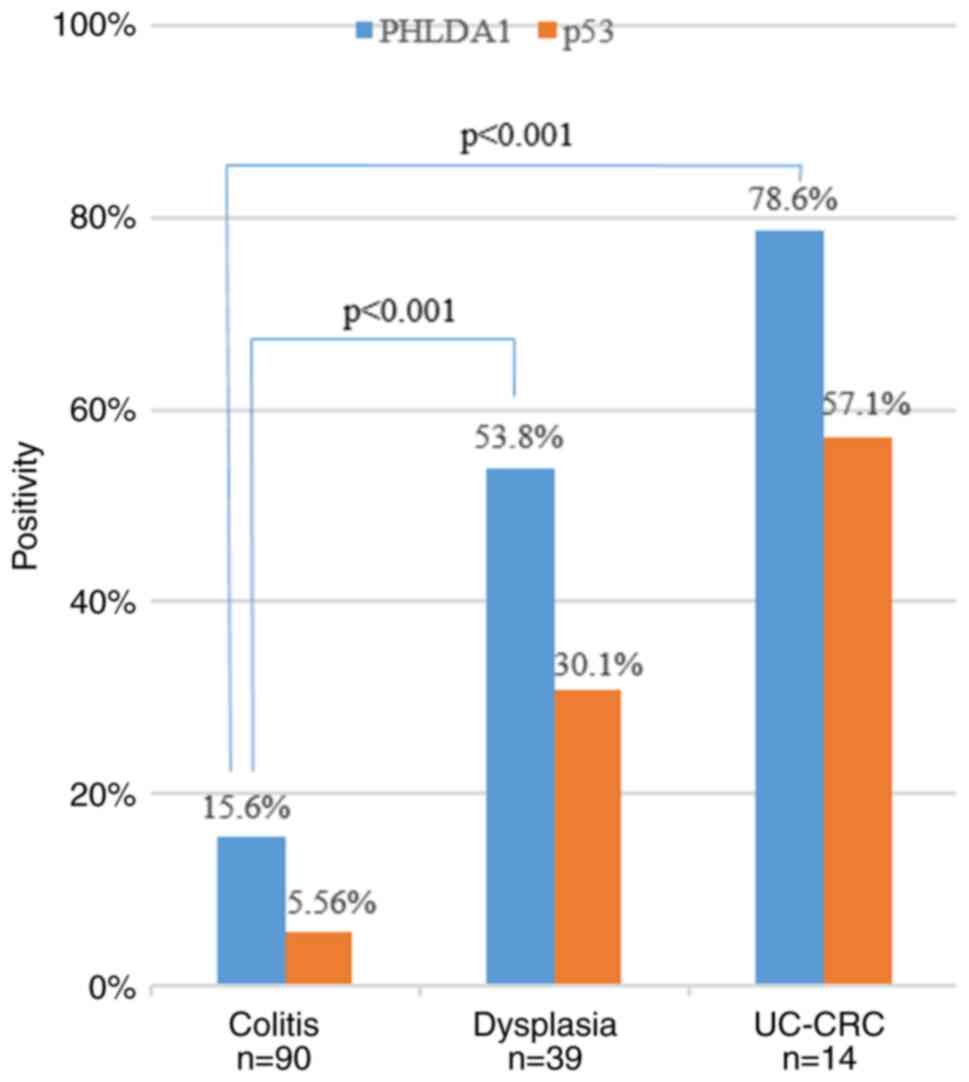
PHLDA1-IHC and p53-IHC positivity
rates in UC samples
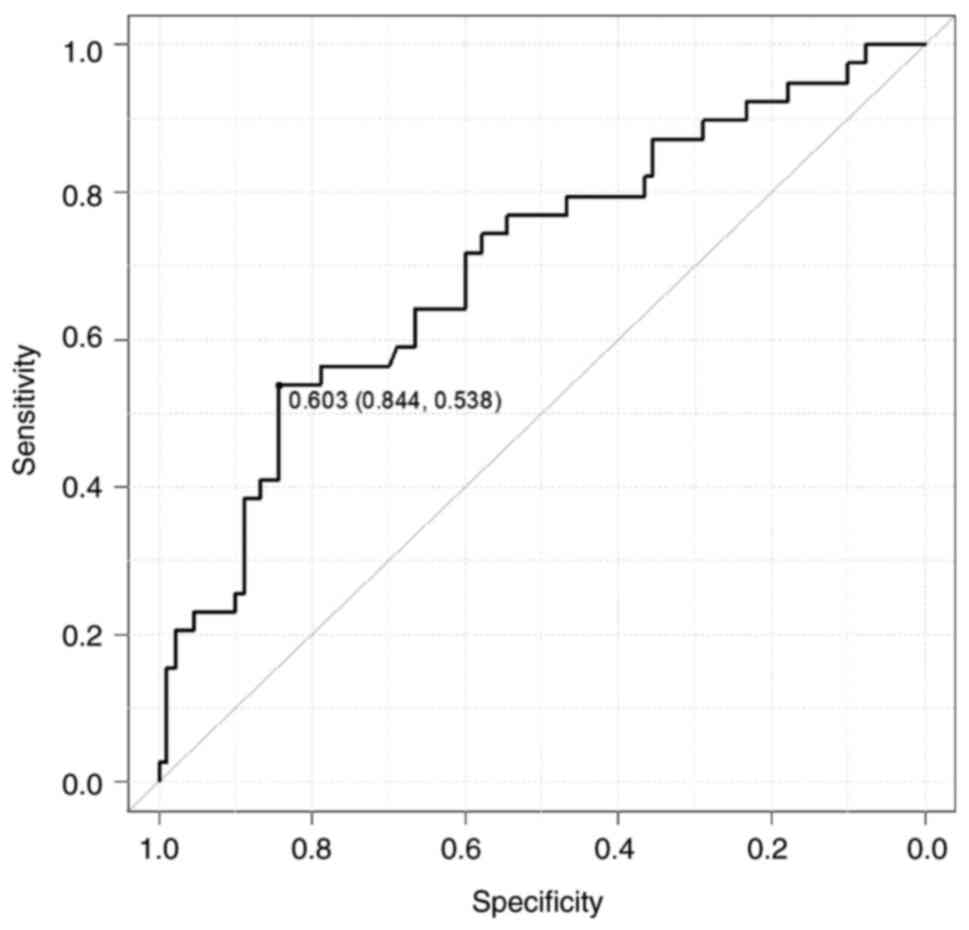
The PHLDA1-IHC scores were divided into two groups
as follows: the PHLDA1-positive (PHLDA1-IHC score ≥0.603 n=47) and
PHLDA1-negative groups (PHLDA1-IHC score ≤0.603, n=96). The cutoff
value using the receiver operating characteristic curve calculated
from colitis and dysplasia was 0.603, which is the threshold at
which the sum of sensitivity and specificity is maximized (Fig. 3). The status of p53-IHC was
classified as either p53-positive (n=25) or p53-negative (n=118)
according to the method reported by Shigaki et al (15). The PHLDA1 and p53 positivity rates
in the UC samples are shown in Fig.
4. The PHLDA1 positivity rates in neoplastic tissues were
higher than that in non-neoplastic tissues. Notably, an obvious
difference was observed in the PHLDA1-IHC positivity rates between
UC-CRC (78.6%) and dysplasia samples (53.8%, P<0.001) and
between UC-CRC and colitis samples (15.6%, P<0.001). On the
contrary, the positivity of PHLDA1 was not statistically
significant (P=0.067) than that of p53 in dysplasia samples.
Accuracy of PHLDA1 and p53-IHC for a
diagnosis of dysplasia
In 129 samples including dysplasia (n=39) and
colitis samples (n=90), we evaluated the accuracy of PHLDA1- and
p53-IHC for the discrimination of dysplasia from the inflammatory
mucosa of UC. The positive predictive value (PPV) of PHLDA1-IHC was
60.0%, which was clearly lower than that of p53 (70.6%). The
negative predictive value (NPV) of PHLDA1-IHC (80.9%) was
relatively higher than that of p53 (75.9%) (Table II). The sensitivity of PHLDA1-IHC
was 53.8%, which was clearly higher than that of p53 (30.8%). The
specificity of PHLDA1-IHC (84.4%) was relatively lower than that of
p53 (94.4%). Twenty-five (17.5%) lesions were PHLDA1 positive and
p53 negative, of which 14 were dysplasia (56.0%). Seven (4.9%)
lesions were PHLDA1 negative and p53 positive, of which five were
dysplasia (71.4%) (Table III). No
significant differences were observed between the PHLDA1 positive
and p53 negative group and PHLDA1 negative and p53 positive group
in age (P=0.65), gender (P=0.64), location (proximal/distal;
P=1.00), and reason for surgery (P=1.00). The UC lesions were
divided into the proximal (cecum, ascending colon and transverse
colon) and distal parts (descending colon, sigmoid colon and
rectum). The proximal parts included 53 lesions, of which 44 were
colitis, and 9 were dysplasia. Ninety lesions were found in the
distant parts, of which 46 were colitis, 30 were dysplasia, and 14
were UC-CRC. PHLDA1 and p53 had no significant differences in
positivity rate in dysplasia in the proximal and distal regions
(Table SI). On the other hand, in
colitis, PHLDA1 showed a significant difference in positivity rate
(Table SII). In the sites in the
proximal region, the PPV of PHLDA1-IHC was 77.8%, which was clearly
higher than that of p53 (66.6%). The NPV of PHLDA1-IHC (95.5%) was
relatively higher than that of p53 (86.0%) (Table SIII). The sensitivity of PHLDA1-IHC
was 77.8%, which was clearly higher than that of p53 (22.2%). The
specificity of PHLDA1-IHC (95.5%) was lower than that of p53
(97.7%). In the sites in the distal region, the PPV of PHLDA1-IHC
was 53.8%, which was clearly lower than that of p53 (71.4%). The
NPV of PHLDA1-IHC (68.0%) was higher than that of p53 (67.7%)
(Table SIV). The sensitivity of
PHLDA1-IHC was 46.7%, which was clearly higher than that of p53
(33.3%). The specificity of PHLDA1-IHC (73.9%) was relatively lower
than that of p53 (91.3%). We found no significant differences in
the positivity rates of PHLDA1 and p53 between the colitis part of
the patients with UC with dysplasia and/or UC-CRC (P=0.24) and
those without dysplasia and UC-CRC (P=0.18) (Table SV).
 | Table IIAccuracy of PHLDA1-IHC and p53-IHC
for diagnosis of dysplasia. |
Table II
Accuracy of PHLDA1-IHC and p53-IHC
for diagnosis of dysplasia.
| Outcome | Dysplasia, n | Colitis, n | PPV, % | NPV, % |
|---|
| PHLDA1 | | | | |
|
Positive | 21 | 14 | 60.0 | 80.9 |
|
Negative | 18 | 76 | | |
| p53 | | | | |
|
Positive | 12 | 5 | 70.6 | 75.9 |
|
Negative | 27 | 85 | | |
 | Table IIIDysplasia rate and combination of
PHLDA1-IHC and p53-IHC status. |
Table III
Dysplasia rate and combination of
PHLDA1-IHC and p53-IHC status.
| Outcome | p53 positive | p53 negative |
|---|
| PHLDA1
positive | 70.0% (7/10) | 56.0% (14/25) |
| PHLDA1
negative | 71.4% (5/7) | 14.9% (13/87) |
Discussion
In this study, we demonstrated that PHLDA1 is highly
expressed in dysplastic and UC-CRC lesions.
PHLDA1 was first identified as a potential
transcription factor that is required for Fas expression and
activation-induced apoptosis in mouse T cell hybridomas (1). The PHLDA1 is located on 12q21.2
and contains PHL domains; these domains interact with membrane
components, which elicit a variety of cellular responses, through
which it participates in cell signaling transduction, vesicular
trafficking, and cytoskeletal rearrangement (16,17).
PHLDA1 is predominantly expressed in the cytoplasm (18-21),
and its expression is induced by different stimuli, such as
estrogens (22), growth factors
(23), differentiation factors
(24), and endoplasmic reticulum
stress-inducing agents (25).
PHLDA1 is also associated with various biological processes, such
as cell apoptosis, cell proliferation, and differentiation.
In terms of the relationship between PHLDA1 and
cancer, it has been reported that an osteosarcoma cell line with a
high metastatic potential had increased PHLDA1 expression (26). It has also been found that PHLDA1
has an apoptosis-suppressing effect in oral cancer (27,28).
In addition, the relationship between breast cancer and PHLDA1 has
been well studied, and E2 and TNF-α are considered promoters of
PHLDA1. However, PHLDA1 in breast cancer is a tumor
suppressor gene (22).
As for the relationship between PHLDA1 and
the colonic mucosa, PHLDA1 is only expressed in
undifferentiated basal cells in the crypts. It is also highly
expressed in colorectal adenoma and carcinoma cells, and high
expression of PHLDA1 has been shown to increase migration ability,
anchorage-independent growth, and cell-matrix adhesion ability
(12).
In this study, we showed that the PHLDA1-IHC score
is significantly elevated as the UC cell variant progresses. Since
no studies have been retrieved showing that the canceration of UC
involves the proliferation of undifferentiated basal cells,
PHLDA1 was presumed to also be involved in the canceration
of UC. However, this study could not clarify whether this gene was
an initiating factor or a promoting factor in the canceration of
UC. Furthermore, the presence of p53 gene mutation suggests the
presence of dysplasia, a precancerous lesion.
p53-IHC is widely used as an auxiliary diagnostic
method, but the correct diagnosis rate of dysplasia is low, and the
sensitivity is believed to be 11-40% (29), which was similar to the findings in
this study. The diagnosis of high p53 protein expression by IHC
does not always correspond to the p53 gene mutation. In other
words, p53 IHC alone may overlook dysplasia. The detection of
promising biomarkers for the diagnosis of dysplasia could impact
the clinical management of patients with UC, who are at a higher
risk for cancer development (3).
This is because if high-grade dysplasia develops, those patients
will be treated by total colectomy, which may significantly reduce
the patient's quality of life. Since p53 IHC has a high PPV
(70.6%), it is probable that dysplasia can be diagnosed in positive
cases. Moreover, since the specificity of p53 IHC is as high as
94.4%, it can be reasonably used as an auxiliary diagnosis.
However, due to its low sensitivity (30.8%), many cases of
dysplasia were present among those who were diagnosed as negative.
Since PHLDA1-IHC is more sensitive than p53-IHC, 56% of dysplasia
cases that were diagnosed as negative by p53-IHC were positive
(Table III). Since PHLDA1-IHC is
more sensitive than p53-IHC, it is more useful as a screening
diagnosis to ensure diagnostic accuracy, and when combined with the
highly sensitive PHLDA1, it may be a useful auxiliary diagnostic
marker. Therefore, it may be possible to include, by PHLDA1-IHC,
those cases that have been determined to be negative by p53-IHC and
that are difficult to determine pathologically. In addition, PHLDA1
had a high PPV, which suggests that it may be more effective,
especially for dysplasia that occurred in the proximal colon
(Table SIII).
However, one limitation of this study includes its
small sample size. This is because we collected multiple samples
(pathological sections) from a single patient. However, as far as
possible, we collected different and individual tissues from that
patient, including UC-CRC, dysplasia, inflammatory, and other
tissues. Moreover, the histological type was as different as
possible in the tissues collected from that patient, e.g., UC-CRC
tissue and cells. We collected and examined dysplasia from the
section that appeared to be strongly atypical as well as from the
section where cell atypia was likely to be weak, the section with
strong inflammation, and the section with weak inflammation.
Furthermore, this study was only verified by immunostaining since
no blood and stool samples were collected, which could otherwise
further verify PHLDA1 expression. A future prospective study
that includes a larger number of endoscopic biopsy samples along
with long-term surveillance with endoscopy is needed.
In conclusion, PHLDA1 was believed to be
involved in UC carcinogenesis, and PHLDA1-IHC has been suggested to
contribute to the diagnosis of dysplasia when used in combination
with p53-IHC.
Supplementary Material
Comparison of positivity of PHLDA1 or
p53 between colitis sites in the proximal and distant parts of
dysplasia.
Comparison of positivity of PHLDA1 or
p53 between colitis sites in the proximal and distant parts.
Accuracy of PHLDA1-IHC and p53-IHC for
a diagnosis of dysplasia in the proximal lesions.
Accuracy of PHLDA1-IHC and p53-IHC for
a diagnosis of dysplasia in the distal lesions.
Comparison of positivity of PHLDA1 or
p53 in the colitis part of a patient with or without colorectal
cancer or dysplasia.
Acknowledgements
The authors would like to thank Mrs. Yoko Takagi and
Mrs. Junko Inoue (Department of Specialized Surgeries, Graduate
School of Medical and Dental Sciences, Tokyo Medical and Dental
University, Bunkyo-ku, Tokyo 113-8519, Japan) for their excellent
technical assistance.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
FO was responsible for the study conception and
design, the data collection and analysis, performing the
experiments, writing the manuscript and the statistical analysis.
TI, MI and SO contributed to the study conception and design, data
analysis, and data interpretation, and provided technical and
material support. SY, AK, TM, MT, HU and YK contributed to the
study conception, data interpretation, and critically revised the
manuscript. FO and TI confirmed the authenticity of all the raw
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was conducted in accordance with
the Declaration of Helsinki and its later amendments or comparable
ethical standards. The study protocol was approved by the
Institutional Review Board of Tokyo Medical and Dental University
(approval. no. D2000-831; date, January 1, 2000 to June 30, 2025;
Tokyo, Japan), and written informed consent was obtained from all
the patients before enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Molodecky NA, Soon IS, Rabi DM, Ghali WA,
Ferris M, Chernoff G, Benchimol EI, Panaccione R, Ghosh S, Barkema
HW and Kaplan GG: Increasing incidence and prevalence of the
inflammatory bowel diseases with time, based on systematic review.
Gastroenterol. 142:46–54. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Jess T, LoftusEV Jr, Velayos FS, Harmsen
WS, Zinsmeister AR, Smyrk TC, Tremaine WJ, Melton LJ 3rd, Munkholm
P and Sandborn WJ: Incidence and prognosis of colorectal dysplasia
in inflammatory bowel disease: A population-based study from
olmsted county, Minnesota. Inflamm Bowel Dis. 12:669–676.
2006.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Chen R, Lai LA, Brentnall TA and Pan S:
Biomarker for colitis-associated colorectal cancer. World J
Gastroenterol. 22:7882–7891. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Watanabe T, Konishi T, Kishimoto J, Kotake
K, Muto T and Sugihara K: Japanese Society for Cancer of the Colon
and Rectum. Ulcerative colitis-associated colorectal cancer shows a
poorer survival than sporadic cancer: A nationwide Japanese study.
Inflamm Bowel Dis. 17:802–808. 2011.PubMed/NCBI View Article : Google Scholar
|
|
5
|
DeRoche TC, Xiao SY and Liu X:
Histological evaluation in ulcerative colitis. Gastroenterol Rep
(Oxf). 2:178–192. 2014.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Okamoto R and Watanabe M: Role of
epithelial cells in the pathogenesis and treatment of inflammatory
bowel disease. J Gastroenterol. 51:11–21. 2016.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Triantafillidis JK, Nasioulas G and
Kosmidis PA: Colorectal cancer and inflammatory bowel disease:
Epidemiology, risk factors, mechanisms of development and
prevention strategies. Anticancer Res. 29:2727–2737.
2009.PubMed/NCBI
|
|
8
|
Vogelstein B, Fearson ER, Hamilton SR,
Kern SE, Preisinger AC, Leppert M, Nakamura Y, White R, Smits AM
and Bos JL: Genetic alterations during colorectal-tumor
development. N Engl J Med. 319:525–532. 1988.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Ullmam TA and Itzkowitz SH: Intestinal
inflammation and cancer. Gastroenterology. 140:1807–1816.
2011.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Takahashi H, Ishikawa T, Ishiguro M,
Okazaki S, Mogushi K, Kobayashi H, Iida S, Mizushima H, Tanaka H,
Uetake H and Sugihara K: Prognostic significance of Traf2- and Nck-
interacting kinase (TNIK) in colorectal cancer. BMC Cancer.
15(794)2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hanaoka M, Ishikawa T, Ishiguro M, Tokura
M, Yamauchi S, Kikuchi A, Uetake H, Yasuno M and Kawano T:
Expression of ATF6 as a marker of pre-cancerous atypical change in
ulcerative colitis-associated colorectal cancer. A potential role
in the management of dysplasia. J Gastroenterol. 53:631–641.
2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sakthianandeswaren A, Christie M,
D'Andreti C, Tsui C, Jorissen RN, Li S, Fleming NI, Gibbs P, Lipton
L, Malaterre J, et al: PHLDA1 expression marks the putative
epithelial stem cells and contributes to intestinal tumorigenesis.
Cancer Res. 71:3709–3719. 2011.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Khamas A, Ishikawa T, Shimokawa K, Mogushi
K, Iida S, Ishiguro M, Mizushima H, Tanaka H, Uetake H and Sugihara
K: Screening for epigenetically masked genes in colorectal cancer
using 5-Aza-2'-deoxycytidine, microarray and gene expression
profile. Cancer Genomics Proteomics. 9:67–75. 2012.PubMed/NCBI
|
|
14
|
Krajewska M, Krajewski S, Epstein JI,
Shabaik A, Sauvageot J, Song K, Kitada S and Reed JC:
Immunohistochemical analysis of bcl-2, bax, bcl-X, and mcl-1
expression in prostate cancers. Am J Pathol. 148:1567–1576.
1996.PubMed/NCBI
|
|
15
|
Shigaki K, Mitomi H, Fujimori T, Ichikawa
K, Tomita S, Imura J, Fujii S, Itabashi M, Kameoka S, Sahara R and
Takenoshita S: Immunohistochemical analysis of chromogranin A and
p53 expressions in ulcerative colitis-associated neoplasia:
Neuroendocrine differentiation as an early event in the
colitis-neoplasia sequence. Hum Pathol. 44:2393–2399.
2013.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Lemmon MA and Ferguson KM:
Signal-dependent membrane targeting by pleckstrin homology (PH)
domains. Biochem J. 15:1–18. 2000.PubMed/NCBI
|
|
17
|
Scheffzek K and Welti S: Pleckstrin
homology (PH) like domains - versatile modules in protein-protein
interaction platforms. FEBS Lett. 586:2662–2673. 2012.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Hinz T, Flindt S, Marx A, Janssen O and
Kabelitz D: Inhibition of protein synthesis by the T cell
receptor-inducible human TDAG51 gene product. Cell Signal.
13:345–352. 2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Neef R, Kuske MA, Pröls E and Johnson JP:
Identification of the human PHLDA1/TDAG51 gene: Down-regulation in
metastatic melanoma contributes to apoptosis resistance and growth
deregulation. Cancer Res. 62:5920–5929. 2002.PubMed/NCBI
|
|
20
|
Nagai MA, Fregnani JH, Netto MM, Brentani
MM and Soares FA: Down-regulation of PHLDA1 gene expression is
associated with breast cancer progression. Breast Cancer Res Treat.
106:49–56. 2007.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Zhao P, Lu Y and Liu L: Correlation of
decreased expression of PHLDA1 protein with malignant phenotype of
gastric adenocarcinoma. Int J Clin Exp Pathol. 8:5230–5235.
2015.PubMed/NCBI
|
|
22
|
Marchiori AC, Casolari DA and Nagai MA:
Transcriptional up-regulation of PHLDA1 by 17beta-estradiol in
MCF-7 breast cancer cells. Braz J Med Biol Res. 41:579–582.
2008.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Toyoshima Y, Karas M, Yakar S, Dupont J,
Helman L and LeRoith D: TDAG51 mediates the effects of insulin-like
growth factor I (IGF-I) on cell survival. J Biol Chem.
279:25898–25904. 2004.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hossain GS, van Thienen JV, Werstuck GH,
Zhou J, Sood SK, Dickhout JG, de Koning AB, Tang D, Wu D, Falk E,
et al: TDAG51 is induced by homocysteine, promotes
detachment-mediated programmed cell death, and contributes to the
cevelopment of atherosclerosis in hyperhomocysteinemia. J Biol
Chem. 278:30317–3027. 2003.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Gomes I, Xiong W, Miki T and Rosner MR: A
proline- and glutamine-rich protein promotes apoptosis in neuronal
cells. J Neurochem. 73:612–622. 1999.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ren L, Mendoza A, Zhu J, Briggs JW, Halsey
C, Hong ES, Burkett SS, Morrow J, Lizardo MM, Osborne T, et al:
Characterization of the metastatic phenotype of a panel of
established osteosarcoma cells. Oncotarget. 6:29469–29481.
2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Murata T, Sato T, Kamoda T, Moriyama H,
Kumazawa Y and Hanada N: Differential susceptibility to hydrogen
sulfide-induced apoptosis between PHLDA1-overexpressing oral cancer
cell lines and oral keratinocytes: Role of PHLDA1 as an apoptosis
suppressor. Exp Cell Res. 320:247–257. 2014.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Coutinho-Camillo CM, Lourenço SV, Nonogaki
S, Vartanian JG, Nagai MA, Kowalski LP and Soares FA: Expression of
PAR-4 and PHLDA1 is prognostic for overall and disease-free
survival in oral squamous cell carcinomas. Virchows Arch.
463:31–39. 2013.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Taylor HW, Boyle M, Smith SC, Bustin S and
Williams NS: Expression of p53 in colorectal cancer and dysplasia
complicating ulcerative colitis. Br J Surg. 80:442–444.
1993.PubMed/NCBI View Article : Google Scholar
|