Introduction
Lynch syndrome (LS) is a hereditary cancer syndrome caused by germline mutations in one of the mismatch repair (MMR) genes (MSH2, MLH1, MSH6, PMS2), which predispose to multiple types of cancers including colorectal cancer (CRC), endometrial, and urothelial cancers. Deletions of the 3'end of the EPCAM gene also cause LS because of epigenetic silencing of the MSH2 gene. LS represents the most frequent cause of inherited CRC, accounting for 2-4% of all CRC cases (1). Surveillance is beneficial in reducing cancer occurrence as well as mortality (2); therefore, identifying individuals with LS and their subsequent life-long management is of importance for the prevention of cancer-related death in the affected families.
Identification of LS has traditionally relied on assembling a personal and family history of CRC and other related cancers. After additional information such as the age of onset and histological findings are taken into account, those who meet the Amsterdam criteria II (3) and/or the revised Bethesda guidelines (4) are considered to have a higher risk of LS, and are subject to further investigation by microsatellite instability (MSI) testing and/or immunohistochemistry (IHC) for MMR proteins using cancer specimen (1). Recently, an alternative strategy of universal screening has been introduced, in which all patients for whom cancer tissue is available are evaluated by MSI and/or IHC for MMR proteins. Universal screening is reported to be more sensitive in identifying individuals with LS (5,6) at a feasible cost (7,8). After additional studies confirmed its efficacy (9-12), the latest guidelines recommended universal screening for LS in patients with CRC and endometrial cancer (13-17).
In light of these recommendations, universal screening has become widespread; however, the implementation of universal screening in clinical practice is still debated. Brennann et al reported that the yield of LS probands detected is limited in real-world because of infrequent referrals and genetic testing (18). The outcome of LS screening may also be affected by the prevalence of LS in particular ethnic groups (5,6,9,19-21) and the accessibility to genetic counseling and genetic testing (18). Some authors have proposed an age-restricted screening strategy because of the low efficiency of LS screening in older patients (20,21). Hence, despite its advantages of straightforwardness and sensitivity compared with a selective strategy based on Amsterdam criteria II and/or revised Bethesda guidelines, the merits of universal screening might vary depending on the circumstances in individual institutions.
In the current study, we retrospectively analyzed the results of universal screening in patients with CRC that is performed as usual clinical practice in our institution. Further, we investigated the frequencies of deficient MMR (dMMR) tumors and genetic testing candidates (GTCs) in an independent cohort of patients with young-onset (<50 years) CRC. The aims of this study were a) to elucidate the real-world outcomes of universal screening of patients with CRC in terms of the detection frequencies of dMMR tumors, GTCs, and confirmed LS cases, and b) to clarify the rate of dMMR tumors and GTCs in patients with young-onset CRC in a referral hospital in Japan.
Materials and methods
Patients and clinical information
This was a retrospective study conducted at a single referral hospital. The results of universal screening in consecutive patients with CRC who underwent endoscopic and/or surgical resection at Kyoto University Hospital from 2016 until 2018 were analyzed. To evaluate MMR deficiency in an independent cohort of patients with young-onset CRC, we included patients with CRC who were younger than 50 years of age, whose tumor tissues were obtained at Kyoto University Hospital between 2006 and 2015 and for whom formalin-fixed paraffin-embedded (FFPE) samples were available. We defined young-onset CRCs as tumors diagnosed before age 50, in accordance with the Amsterdam criteria II and the revised Bethesda guidelines (3,4). Medical records were reviewed and clinicopathological information and data related to universal screening were collected. Information on genetic testing was also obtained when applicable. The study protocol was approved by the Ethics Committee of Kyoto University Graduate School and Faculty of Medicine (R0821 and R1978).
Immunohistochemistry
Immunohistochemical staining was performed using FFPE blocks. We used antibodies against MLH1 (clone; M1, Roche 518-110215; Roche Diagnostics), MSH2 (clone; G219-1129, Roche 518-101947; Roche Diagnostics), MSH6 (clone; EPR3945, Abcam ab92471; Abcam), PMS2 (clone; EPR3947, Roche 518-110857; Roche Diagnostics), and BRAF V600E (clone; VE1, Abcam ab228461; Abcam). Slides were stained using a Ventana BenchMark ULTRA instrument (Ventana Medical Systems) according to the manufacturer's protocol. Total negativity for nuclear expression of MLH1, MSH2, MSH6, and PMS2 was considered as loss. BRAFV600E was evaluated as positive if diffuse and nearly uniform cytoplasmic staining of tumor cells was present with or without membranous accentuation.
Universal screening for LS
As the first step, IHC for MSH6 and PMS2 was performed, and cases with retained expression of both proteins were regarded as proficient MMR (pMMR). Tumors with loss of expression of either protein were designated as dMMR, and immunostaining for MSH2 and/or MLH1 was added. IHC for BRAF V600E was also performed when a tumor showed loss of MLH1 expression. Tumors showing: i) A combined loss of MSH6 and MSH2, ii) loss of MSH6 alone, iii) loss of PMS2 alone, or iv) a combined loss of PMS2 and MLH1 in conjunction with negative BRAF V600E IHC were considered to indicate possible LS. Tumors with a combined loss of PMS2 and MLH1 with positive BRAF V600E IHC were considered to indicate sporadic dMMR. For patients with multiple synchronous CRCs, the most advanced tumor for each individual was designated as the index tumor if IHC results were concordant. Because patients who have both pMMR and dMMR CRCs are less likely to carry LS, a pMMR CRC was assigned as the index tumor. For patients who had metachronous CRCs during the study period, the first resected tumor was chosen as the index tumor.
Patients with tumors showing a possible LS pattern were designated as GTCs. GTCs were informed of their IHC results at a regular follow-up visit, and those who opted to participate were referred to a specialist for hereditary tumor and genetic counseling, and were asked to consider genetic testing for MMR genes and EPCAM. After retrospective MLH1 methylation analysis, GTCs were divided into suspected sporadic cases (sp-GTC), whose tumors were positive for MLH1 methylation, and bona fide GTCs (bf-GTC) for whom MLH1 methylation was negative (eight cases) or undetermined (two cases; insufficient tissue).
Analyses of BRAF V600E mutation and MLH1 promoter methylation
Genomic DNA was extracted from FFPE samples of tumors and from matched normal epithelial tissues using GeneRead DNA FFPE (Qiagen GmbH). To examine BRAF V600E mutations, DNA was processed using AmpliSeq for Illumina Library Plus (Illumina Inc.), and analyzed using MiniSeq (Illumina). The sequencing data were analyzed using the Local Run Manager software and VariantStudio (both from Illumina Inc.). To investigate the methylation status of CpG sites in the MLH1 promoter, a pair of bisulfite sequencing primers (BSP) was designed to amplify the genomic sequence including 11 CpG sites (Fig. S1A) using Methyl Primer Express software v1.0 (Thermo Fisher Scientific, Inc.). Genomic DNA was analyzed by targeted bisulfite sequencing as follows: Bisulfite conversion of DNA was performed using EZ DNA Methylation kit (Zymo Research). Then bisulfite-treated DNA was amplified using BSPs and a KAPA HiFi Uracil+ kit (Roche Sequencing and Life Science KAPA Biosystems) and was processed using an Ion Plus Fragment Library kit (Thermo Fisher Scientific, Inc.) to prepare libraries for the Ion PGM sequencing system (Thermo Fisher Scientific, Inc.). Ion Torrent sequencing data were analyzed using TABSAT (http://github.com/tadkeys/tabsat) (22). The percentage of methylation in each CpG site was calculated as 100x (reference C reads)/[(C>T converted reads) + (reference C reads)] and the average of 11 CpG sites was determined to represent the methylation level for each sample. We used in vitro methylated and unmethylated control DNA samples to validate the results of this bisulfite sequencing assay and confirmed that methylation levels in the MLH1 promoter region was detected in quantitative manner (Fig. S1B). Because all the normal epithelial tissues and the known nonmethylated colon cancer cells (LoVo) showed methylation levels ≤2% (data not shown), tumor samples showing methylation levels >10% were considered methylated.
Statistical analysis
The Fisher's exact test or the chi-squared test was used to analyze categorical data. All P-values were two-sided and P<0.05 was considered to indicate a statistically significant difference. All analyses were carried out using JMP 15.2 (SAS Institute, Inc.)
Results
Universal screening of patients with CRC
The results of the universal screening are summarized in Fig. 1. A total of 540 CRCs from 503 patients were treated by either surgical or endoscopic resection. Three patients who had either known familial adenomatous polyposis, Crohn's disease, or ulcerative colitis were excluded from the LS screening. An additional 37 CRCs were excluded because IHC was not performed for undetermined reasons. For the 32 patients who possessed multiple CRCs, only the index tumors were included for further analyses.
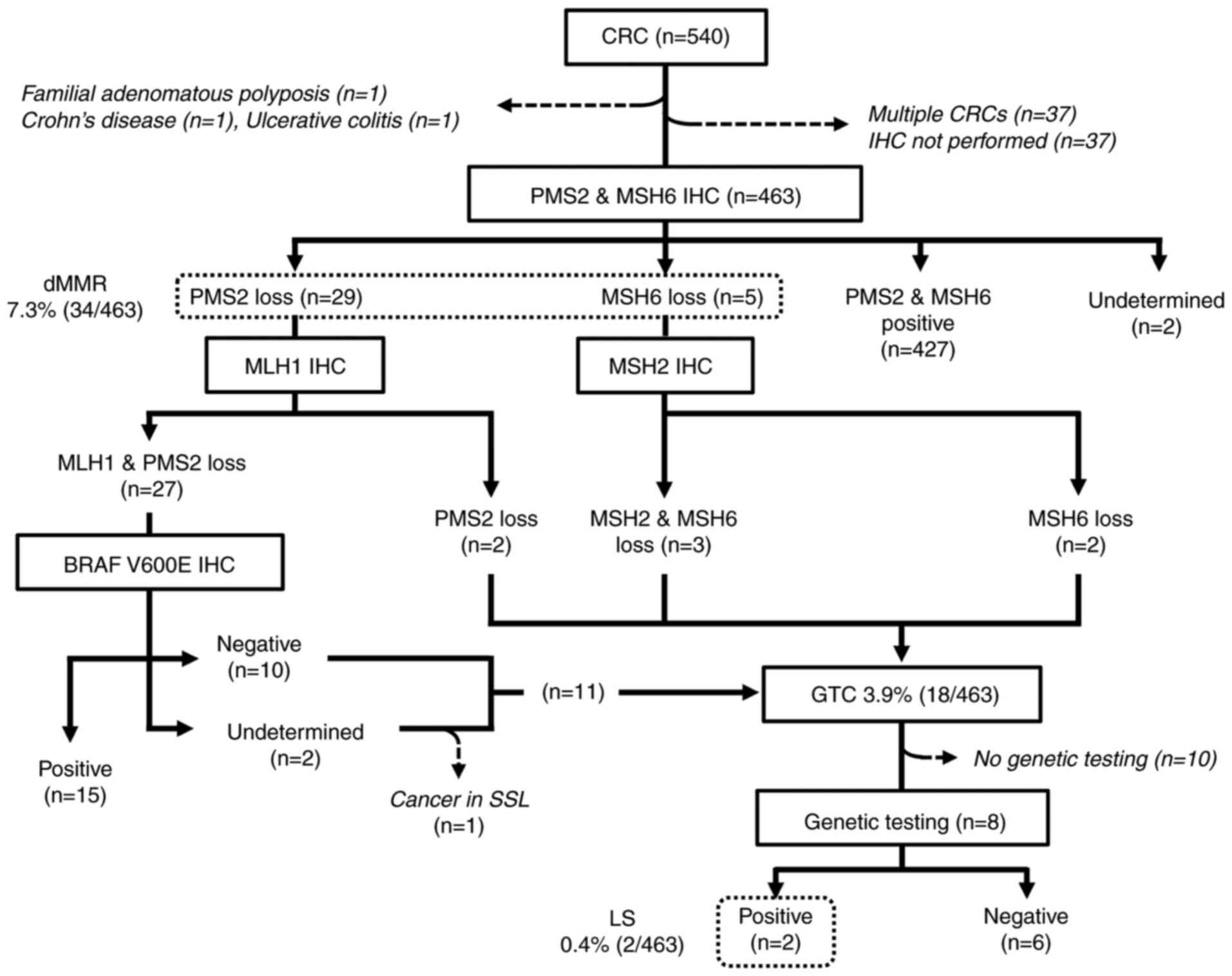 |
Figure 1
Flow diagram and results of the universal screening in patients with CRC. CRC, colorectal cancer; IHC, immunohistochemistry; dMMR, deficient mismatch repair; SSL, sessile serrated lesion; GTC, genetic testing candidate; MSH6, DNA mismatch repair protein MSH6; PMS2, mismatch repair endonuclease PMS2; dMMR, deficient mismatch repair; MLH1, DNA mismatch repair protein Mlh1; MSH2, DNA mismatch repair protein Msh2; LS, Lynch syndrome.
|
Thus, the universal screening procedure was performed in 463 patients with CRC. The patients' characteristics are shown in Table I. Representative images of IHC are shown in Fig. S2A-E. There were 427 pMMR and 34 dMMR tumors; the expression status of PMS2 and/or MSH6 was undetermined in two cases. No tumors showed a combined loss of PMS2 and MSH6. After additional IHC for MLH1, MSH2, and BRAF V600E, 15 dMMR tumors showed the sporadic pattern. Another tumor was considered to be sporadic despite showing combined MLH1 and PMS2 loss with negative BRAF V600E IHC, because it arose within a sessile serrated lesion, which is known to be a major precursor of sporadic dMMR CRC (23). The remaining 18 patients had dMMR tumors with a possible LS pattern and were designated as GTCs. After they were informed of their IHC results at a regular follow-up visit, 11 GTCs opted to consult hereditary tumor specialists and/or the clinical genetics unit, and eight of them subsequently underwent genetic testing. Germline analyses revealed two pathogenic mutations in MMR genes, NM_00251.2(MSH2):c.84_85(p.Lys29X) and NM_00249.3(MLH1):c.1692delC:(p.Ile565PhefsX26), leading to identification of two LS probands (0.4%) (Fig. 2A).
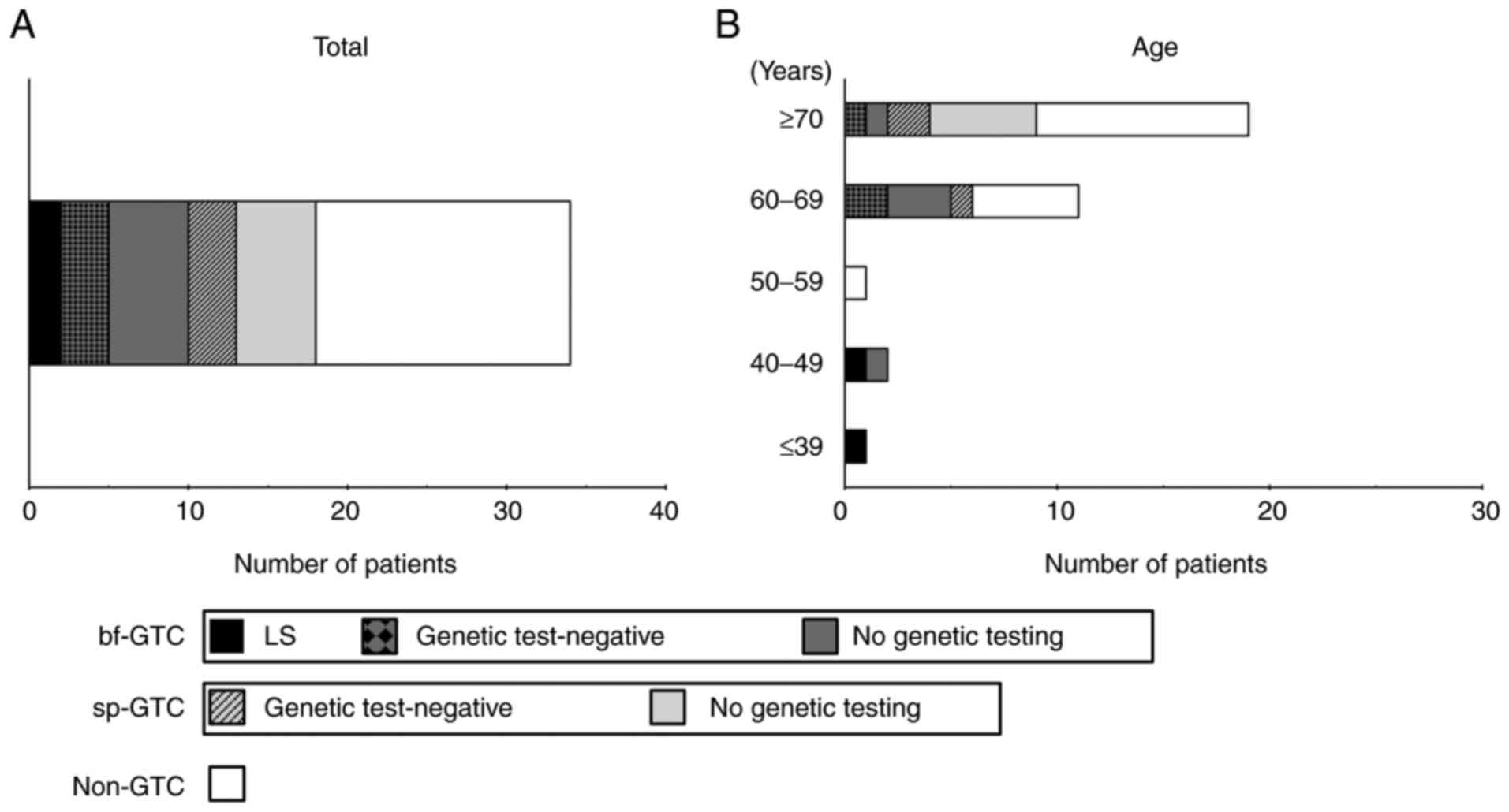 |
Figure 2
Rates of dMMR tumors. Rates of dMMR tumors for (A) all patients with colorectal cancer and (B) in relation to age. In each bar graph, dMMR tumors are divided into LS (black), bf-GTC with genetic test-negative (black and dark grey checkered pattern), bf-GTC with no genetic testing (dark grey), sp-GTC with genetic test-negative (light grey with black diagonal lines), sp-GTC with no genetic testing (light grey) and non-GTC (white). dMMR, deficient mismatch repair; bf-GTC, bona fide genetic testing candidate; sp-GTC, sporadic genetic testing candidate; GTC, genetic testing candidate; LS, Lynch syndrome.
|
 |
Table I
Patient characteristics.
|
Table I
Patient characteristics.
| Patient characteristics |
Number of patients |
| Age, years, median (range) |
69 (28-92) |
| Sex, n |
|
| Female |
198 |
| Male |
265 |
| Treatment modality, n |
|
| Endoscopic resection |
122 |
| Surgery |
341 |
| Tumor location, n |
|
| Right |
157 |
| Left |
306 |
| Tumor histology, n |
|
| High-grade adenocarcinoma |
24 |
| Low-grade adenocarcinoma |
411 |
| Mucinous/othersa |
28 |
| UICC stageb, n |
|
| 0 |
89 |
| I |
101 |
| II |
109 |
| III |
111 |
| IV |
53 |
Proportion of dMMR tumors and clinicopathological findings of CRC
Of the 461 index tumors for which MMR status was successfully determined by IHC, 427 were pMMR and 34 were dMMR (Table II). We then evaluated the proportion of dMMR tumors according to various clinicopathological findings. The dMMR CRCs displayed a bimodal distribution with age with peaks in younger (<50 years old) and older (≥60 years old) patients. In addition, dMMR tumors were significantly enriched within right-sided tumors, and high-grade and mucinous adenocarcinoma, and other histology compared with low-grade adenocarcinoma. Detailed information about the dMMR CRCs is presented in Table SI.
 |
Table II
Rates of dMMR in relation to clinicopathological findings of patients with CRC.
|
Table II
Rates of dMMR in relation to clinicopathological findings of patients with CRC.
| Patient characteristics |
pMMR, n (%) |
dMMR, n (%) |
P-value |
| Total |
427 (92.6) |
34 (7.4) |
|
| Age, years |
|
|
N/Aa |
| ≤39 |
6 (85.7) |
1 (16.7) |
|
| 40-49 |
24 (92.3) |
2 (7.7) |
|
| 50-59 |
64 (98.5) |
1 (1.5) |
|
| 60-69 |
122 (91.7) |
11 (8.3) |
|
| ≥70 |
211 (91.7) |
19 (8.3) |
|
| Personal history of CRC and/or other LS-related cancer |
|
|
0.12 |
| Present |
78 (88.6) |
10 (11.4) |
|
| Absent |
349 (93.6) |
24 (6.4) |
|
| Family history of CRC and/or other LS-related cancer |
|
|
0.48 |
| Present |
186 (91.6) |
17 (8.4) |
|
| Absent |
241 (93.4) |
17 (6.6) |
|
| Mode of treatment |
|
|
0.84 |
| Endoscopic resection |
114 (93.4) |
8 (6.6) |
|
| Surgery |
313 (92.3) |
26 (7.7) |
|
| Tumor location |
|
|
<0.01 |
| Right |
130 (83.3) |
26 (16.7) |
|
| Left |
297 (97.4) |
8 (2.6) |
|
| Tumor histology |
|
|
<0.01 |
| High-grade adenocarcinoma/mucinous/other |
41 (78.8) |
11 (21.2) |
|
| Low-grade adenocarcinoma |
386 (94.4) |
23 (5.6) |
|
| UICC stage |
|
|
0.13 |
| 0 |
84 (94.4) |
5 (5.6) |
|
| I/II |
188 (90.0) |
21 (10.0) |
|
| III/IV |
155 (95.1) |
8 (4.9) |
|
BRAF V600E mutation and MLH1 methylation analyses
Although we did not incorporate assays for BRAF V600E mutations and MLH1 methylation assays into the universal screening process, we retrospectively analyzed them to evaluate the validity of our procedure. Our data showed that BRAF V600E IHC was able to determine BRAF V600E mutational status with a sensitivity and a specificity of 86.7 and 76.5%, respectively (Table SII). MLH1 promoter methylation analysis showed that, among the 32 dMMR tumors tested, all 16 non-GTCs with the sporadic IHC pattern were positive for MLH1 methylation. Eight of 10 dMMR tumors in the GTC group that showed MLH1 protein loss and negative or undetermined BRAF V600E IHC were also positive for MLH1 methylation, indicating that they were more likely sporadic than LS (Table SIII).
Yields of universal and age-restricted screening strategies
After the retrospective MLH1 methylation analysis demonstrated positive MLH1 methylation in the majority of dMMR tumors, we divided the GTCs into two groups: sp-GTC, whose tumors showed MLH1 methylation indicative of a sporadic pattern and bf-GTC without positive-MLH1 methylation. As shown in Table III, universal screening identified dMMR tumors, bf-GTCs, and LS probands in 7.3, 2.2, and 0.4%, respectively, of patients with CRC. Given the low rate of detection of bf-GTC and LS, we then tested a range of age-restricted strategies. Screening of patients with CRC who were <50 years old reduced the required number of tests to 33 (7.1% of all CRC), and enabled a remarkable concentration of bf-GTCs and LS probands. However, seven of 10 bf-GTCs remained untested. Screening of patients with CRC who were <70 years old required 232 tests (50.1% of all CRC), and enabled to detect eight of 10 bf-GTCs and both LS probands identified by universal strategy. In contrast, screening of 231 patients with CRC who were ≥70 years old led to detection of only two bf-GTCs and no confirmed LS cases. (Table III).
 |
Table III
Yields of universal and age-restricted screening strategies.
|
Table III
Yields of universal and age-restricted screening strategies.
| Screening strategies |
Total, n |
dMMR tumor, n (%) |
bf-GTC, n (%) |
LS, n (%) |
| Universal screening |
463 |
34 (7.3) |
10 (2.2) |
2 (0.4) |
| ≤49 years |
33 |
3 (9.1) |
3 (9.1) |
2 (6.1) |
| ≤69 years |
232 |
15 (6.5) |
8 (3.4) |
2 (0.9) |
| ≥70 years |
231 |
19 (8.2) |
2 (0.9) |
0 (0.0) |
Frequency of genetic testing in GTCs
Next, we investigated the frequencies of those who underwent genetic testing in GTCs. Included in the 19 patients with dMMR CRC who were ≥70 years of age were nine GTCs and two bf-GTCs. Of these, three GTCs (3/9, 33.3%) underwent genetic testing, all of whom were negative. In contrast, all three patients <50 years of age who had dMMR CRC were bf-GTCs. Two of these underwent genetic testing, leading to the identification of two LS probands (Fig. 2B). Another woman <50 years old had both colon and endometrial cancers with loss of MSH2/MSH6 expression, and the multiple occurrences of CRCs in her family fulfilled the Amsterdam criteria II. Despite the high level of suspicion of LS, she has not undergone genetic testing.
MMR status in patients with young-onset CRC
To test the enrichment of dMMR tumors and GTCs in an independent cohort of patients with young-onset CRCs, we utilized historical cases from before the start of universal screening in our hospital. As shown in Table IV, evaluation of MMR status in a total of 111 patients with young-onset CRCs (<50 years old) revealed 10 (9.0%) dMMR tumors including nine GTCs and one non-GTC. There was greater enrichment of dMMR tumors among patients <40 years of age. Detailed information about the dMMR CRCs is presented in Table SIV.
 |
Table IV
Rates of dMMR tumors among patients with young-onset CRC.
|
Table IV
Rates of dMMR tumors among patients with young-onset CRC.
| Patient characteristics |
pMMR, n (%) |
dMMR, n (%) |
| Total |
101 (91.0) |
10 (9.0) |
| Age, years |
|
|
| ≤39 |
20 (83.3) |
4 (16.7) |
| 40-49 |
81 (93.1) |
6 (6.9) |
Discussion
In the current study, we analyzed the real-world data from universal screening for LS in patients with CRC in a referral hospital in Japan and identified two LS probands, who accounted for 0.4% of 463 patients with CRC. In real-world, infrequent referrals and access to genetic testing limit the detection of LS (18). Indeed, only eight of 18 GTCs (44.4%) in our series underwent genetic testing. Notably, one of the GTCs was strongly suspected to carry LS but genetic testing is pending awaiting her decision. Such real-world factors would limit the detection of LS in clinical practice, and although the rate of LS in the current study seems low compared with those previously reported (5,6,9,20,21), it appears to be reasonable real-world data considering that a previous study from Japan reported that the prevalence of LS in CRC was 0.7% (19).
As the first step of our universal screening, we utilized IHC for PMS2 and MSH6. This two-protein method was reported to be as effective for detection of dMMR as a four-protein panel including PMS2, MSH6, MLH1, and MSH2 and is less costly; its use for LS screening has been proposed (24). However, Pearlman et al noted that the two-protein method could miss some MSH2-mutated tumors, based on the observation that more than half of MSH2-negative tumors had ambiguous or convincing MSH6 expression (25). In the current study, two-protein IHC demonstrated that 7.3% of all examined CRCs were dMMR. Because we did not perform MSH2 IHC in tumors showing intact MSH6 immunostaining, we may have missed a small number of MSH2-mutated tumors. Nevertheless, because the rates of dMMR and of loss of MSH2 expression in the current series were similar to those reported previously from Japan using four-protein immunostaining (19), two-protein IHC for PMS2 and MSH6 may be acceptable because of the cost and effort saved compared with the four-protein method.
According to the latest guidelines, BRAF V600E mutation and/or MLH1 methylation analyses are recommended for dMMR CRCs with MLH1 loss, because positive BRAF V600E and/or MLH1 methylation results indicate a high likelihood of sporadic tumors (13,15-17) Because some studies have shown a high concordance between BRAF V600E IHC and BRAF V600E mutations in CRC (26,27), in the current series, we chose BRAF V600E IHC rather than BRAF mutational or MLH1 methylation analysis as a cheaper and more straightforward option. Our data demonstrated a moderate concordance of BRAF V600E IHC with BRAF mutational analysis. Of 10 dMMR CRCs in GTC group with loss of MLH1 protein and wild-type BRAF, eight tumors were positive for MLH1 methylation (Table SI and SIII) suggesting that they are more likely to be sporadic rather than LS. Thus, in accordance with previous reports (28-30), MLH1 methylation analysis would be able to identify more sporadic cases and reduce the number of genetic tests required compared with BRAF V600E analysis. However, because the availability of the MLH1 methylation assay is limited, we consider that BRAF V600E IHC could serve as an affordable option for LS screening in daily practice.
The low rate of identification of LS by universal screening prompted us to explore more efficient strategies. In this respect, some authors have proposed age-restricted screening as a simple and efficient alternative (20,21). Our data showed that screening in 33 patients <50 years of age with CRC led to identification of three (9.1%) dMMR CRCs, all of which were bf-GTCs: two confirmed and one suspected LS cases. Furthermore, immunostaining of tumors from an independent historical cohort confirmed the enrichment of GTCs in young-onset CRCs, especially in patients <40 years of age. In contrast, we identified 19 dMMR tumors (8.2%) but only two bf-GTCs (0.87%) in 231 patients with CRC who were aged ≥70 years, and no confirmed LS cases. In accordance with a previous report (18), the infrequent pursuit of genetic testing by older GTCs also affects the low rate of identification of LS in this population, hence limiting the efficacy of universal screening. These data suggest that because of the high rate of sporadic dMMR tumors and infrequent genetic testing in older patients, age restricted strategies might be an efficient practical alternative to universal screening in real-world. Because the outcome of LS screening is affected by various population factors including the prevalence of LS and the accessibility to genetic counseling and genetic testing, the optimal screening strategy for implementation in clinical practice may vary depending on individual institution.
This study had several limitations. First, it was a retrospective study conducted in a single institution. Because the choice of whether to undergo genetic testing was up to the patients, the detection rate demonstrated in this study may underestimate the actual prevalence of LS. Nevertheless, our data reflect the practical outcome of universal screening in real-world, which is important to allow identification of the most efficient screening strategy in daily practice. Second, although we identified dMMR tumors in a second cohort of patients with young-onset CRCs, no data were available on the results of their genetic tests.
In conclusion, we report the real-world outcome of universal screening for LS in patients with CRC using immunostaining for two MMR proteins and BRAF V600E. The low detection rate of LS demonstrated in this study questions the implementation of routine universal screening and urges the reevaluation of optimal screening strategy in daily practice. Considering the enrichment of GTCs in young-onset CRCs combined with the frequent sporadic dMMR tumors and infrequent pursuit of genetic testing among older patients, age-restricted strategies might be simple and efficient practical alternatives to universal screening in real-world, especially in regions with low LS prevalence.
Supplementary Material
MLH1 promoter methylation analysis. (A) Primers used to amplify CpG sites at MLH1 promoter. Primer sequences were as follows: F, TTGTTTTTATTGGTTGGATATT (Tm=56.39) and R, AAATACCAATCAAATTTCTCAA (Tm=56.59). (B) Bisulfite targeted sequencing could determine the levels of MLH1 methylation in methylated control DNA samples in vitro in a quantitative manner. M100, M50, M15, M5 and UM represent 100, 50, 15 and 5% methylated and unmethylated control samples, respectively. F, forward; R, reverse; MLH1, DNA mismatch repair protein Mlh1; Tm, melting temperature.
Representative images of immunohistochemistry in deficient mismatch repair colorectal cancers. (A, B) Histology of a rectal cancer sample from a patient with Lynch syndrome with an MSH2 germline mutation. (A) MSH2 and (B) MSH6 immunohistochemistry. The neoplastic epithelium shows loss of nuclear expression of MSH2 and MSH6, whereas the stroma and lymphocytes show expression of both. (C-E) A transverse colon cancer sample from a patient with positive MLH1 methylation and BRAF V600E mutation. Total loss of (C) MLH1 and (D) PMS2 expression were observed in the tumor cells. (E) BRAF V600E immunohistochemistry shows diffuse cytoplasmic staining of the tumor cells. Magnification, x200. MSH2, DNA mismatch repair protein Msh2; MSH6, DNA mismatch repair protein MSH6; MLH1, DNA mismatch repair protein Mlh1; PMS2, mismatch repair endonuclease PMS2.
Characteristics of patients with dMMR CRC.
Association between BRAF V600E IHC and mutational analyses.
MLH1 promoter methylation in dMMR CRCs.
Characteristics of patients with dMMR tumors in an independent cohort of young-onset CRCs.
Acknowledgements
The authors would like to thank Professor Kenjiro Kosaki (Center for Medical Genetics, Keio University School of Medicine, Tokyo, Japan).
Funding
The present study was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI (grant no. JP16K09309), the Japanese Foundation for Research and Promotion of Endoscopy Grant and in part by The National Cancer Center Research and Development Fund 31-A-2 and AMED (grant nos. JP19ck0106268 and JP20ck0106554).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' contributions
AY designed the present study and wrote the manuscript. YM and SM performed and analyzed the immunohistochemistry experiments. YY performed the mutational and methylation analyses. TK, TS, TH and KK analyzed the clinical data. MT, HM, TY and SK performed genetic counseling and acquired the data related to genetic testing and family medical history. KS performed the genetic testing. HS and MM contributed to the interpretation of data. AY and SM confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The study protocol was approved by the Ethics Committee of Kyoto University Graduate School and Faculty of Medicine (approval nos. R0821 and R1978; Kyoto, Japan).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
|
1
|
Kastrinos F and Stoffel EM: History, genetics, and strategies for cancer prevention in Lynch syndrome. Clin Gastroenterol Hepatol. 12:715–727. 2014.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Järvinen HJ, Aarnio M, Mustonen H, Aktan-Collan K, Aaltonen LA, Peltomäki P, De La Chapelle A and Mecklin JP: Controlled 15-year trial on screening for colorectal cancer in families with hereditary nonpolyposis colorectal cancer. Gastroenterology. 118:829–834. 2000.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Vasen HF, Watson P, Mecklin JP and Lynch HT: New clinical criteria for hereditary nonpolyposis colorectal cancer (HNPCC, Lynch syndrome) proposed by the International Collaborative group on HNPCC. Gastroenterology. 116:1453–1456. 1999.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al: Revised bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 96:261–268. 2004.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Clendenning M, Sotamaa K, Prior T, Westman JA, et al: Feasibility of screening for Lynch syndrome among patients with colorectal cancer. J Clin Oncol. 26:5783–5788. 2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Moreira L, Balaguer F, Lindor N, de la Chapelle A, Hampel H, Aaltonen LA, Hopper JL, Le Marchand L, Gallinger S, Newcomb PA, et al: Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 308:1555–1565. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Di Marco M, DAndrea E, Panic N, Baccolini V, Migliara G, Marzuillo C, De Vito C, Pastorino R, Boccia S and Villari P: Which Lynch syndrome screening programs could be implemented in the ‘Real World’? A systematic review of economic evaluations. Genet Med. 20:1131–1144. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Mvundura M, Grosse SD, Hampel H and Palomaki GE: The cost-effectiveness of genetic testing strategies for Lynch syndrome among newly diagnosed patients with colorectal cancer. Genet Med. 12:93–104. 2010.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Adar T, Rodgers LH, Shannon KM, Yoshida M, Ma T, Mattia A, Lauwers GY, Iafrate AJ, Hartford NM, Oliva E and Chung DC: Universal screening of both endometrial and colon cancers increases the detection of Lynch syndrome. Cancer. 124:3145–3153. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Dillon JL, Gonzalez JL, DeMars L, Bloch KJ and Tafe LJ: Universal screening for Lynch syndrome in endometrial cancers: Frequency of germline mutations and identification of patients with Lynch-like syndrome. Hum Pathol. 70:121–128. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Egoavil C, Alenda C, Castillejo A, Paya A, Peiro G, Sánchez-Heras AB, Castillejo MI, Rojas E, Barberá VM, Cigüenza S, et al: Prevalence of Lynch syndrome among patients with newly diagnosed endometrial cancers. PLoS One. 8(e79737)2013.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ryan NAJ, Glaire MA, Blake D, Cabrera-Dandy M, Evans DG and Crosbie EJ: The proportion of endometrial cancers associated with Lynch syndrome: A systematic review of the literature and meta-analysis. Genet Med. 21:2167–2180. 2019.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Gupta S, Provenzale D, Llor X, Halverson AL, Grady W, Chung DC, Haraldsdottir S, Markowitz AJ, Slavin TP Jr, Hampel H, et al: NCCN Guidelines Insights: Genetic/familial high-risk assessment: Colorectal, version 2.2019. J Natl Compr Cancer Netw. 17:1032–1041. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Crosbie EJ, Ryan NAJ, Arends MJ, Bosse T, Burn J, Cornes JM, Crawford R, Eccles D, Frayling IM, Ghaem-Maghami S, et al: The manchester international consensus group recommendations for the management of gynecological cancers in Lynch syndrome. Genet Med. 21:2390–2400. 2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Giardiello FM, Allen JI, Axilbund JE, Boland CR, Burke CA, Burt RW, Church JM, Dominitz JA, Johnson DA, Kaltenbach T, et al: Guidelines on genetic evaluation and management of Lynch syndrome: A consensus statement by the US Multi-society Task Force on colorectal cancer. Am J Gastroenterol. 109:1159–1179. 2014.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Rubenstein JH, Enns R, Heidelbaugh J and Barkun A: Clinical Guidelines Committee. American gastroenterological association institute guideline on the diagnosis and management of lynch syndrome. Gastroenterology. 149:777–782. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Vangala DB, Cauchin E, Balmaña J, Wyrwicz L, van Cutsem E, Güller U, Castells A, Carneiro F, Hammel P, Ducreux M, et al: Screening and surveillance in hereditary gastrointestinal cancers: Recommendations from the European Society of Digestive Oncology (ESDO) expert discussion at the 20th European Society for Medical Oncology (ESMO)/World Congress on Gastrointestinal Cancer, Barcelona, June 2018. Eur J Cancer. 104:91–103. 2018.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Brennan B, Hemmings CT, Clark I, Yip D, Fadia M and Taupin DR: Universal molecular screening does not effectively detect Lynch syndrome in clinical practice. Therap Adv Gastroenterol. 10:361–371. 2017.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Chika N, Eguchi H, Kumamoto K, Suzuki O, Ishibashi K, Tachikawa T, Akagi K, Tamaru JI, Okazaki Y and Ishida H: Prevalence of Lynch syndrome and Lynch-like syndrome among patients with colorectal cancer in a Japanese hospital-based population. Jpn J Clin Oncol. 47:108–117. 2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Jiang W, Cai MY, Li SY, Bei JX, Wang F, Hampel H, Ling YH, Frayling IM, Sinicrope FA, Rodriguez-Bigas MA, et al: Universal screening for Lynch syndrome in a large consecutive cohort of Chinese colorectal cancer patients: High prevalence and unique molecular features. Int J Cancer. 144:2161–2168. 2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Li D, Hoodfar E, Jiang SF, Udaltsova N, Pham NP, Jodesty Y, Armstrong MA, Hung YY, Baker RJ, Postlethwaite D, et al: Comparison of universal versus age-restricted screening of colorectal tumors for lynch syndrome using mismatch repair immunohistochemistry: A cohort study. Ann Intern Med. 171:19–26. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Pabinger S, Ernst K, Pulverer W, Kallmeyer R, Valdes AM, Metrustry S, Katic D, Nuzzo A, Kriegner A, Vierlinger K and Weinhaeusel A: Analysis and visualization tool for targeted amplicon bisulfite sequencing on ion torrent sequencers. PLoS One. 11(e0160227)2016.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Crockett SD and Nagtegaal ID: Terminology, molecular features, epidemiology, and management of serrated colorectal neoplasia. Gastroenterology. 157:949–966.e4. 2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shia J, Tang LH, Vakiani E, Guillem JG, Stadler ZK, Soslow RA, Katabi N, Weiser MR, Paty PB, Temple LK, et al: Immunohistochemistry as first-line screening for detecting colorectal cancer patients at risk for hereditary nonpolyposis colorectal cancer syndrome: A 2-antibody panel may be as predictive as a 4-antibody panel. Am J Surg Pathol. 33:1639–1645. 2009.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Pearlman R, Markow M, Knight D, Chen W, Arnold CA, Pritchard CC, Hampel H and Frankel WL: Two-stain immunohistochemical screening for Lynch syndrome in colorectal cancer may fail to detect mismatch repair deficiency. Mod Pathol. 31:1891–1900. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Capper D, Voigt A, Bozukova G, Ahadova A, Kickingereder P, von Deimling A, von Knebel Doeberitz M and Kloor M: BRAF V600E-specific immunohistochemistry for the exclusion of Lynch syndrome in MSI-H colorectal cancer. Int J Cancer. 133:1624–1630. 2013.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Toon CW, Walsh MD, Chou A, Capper D, Clarkson A, Sioson L, Clarke S, Mead S, Walters RJ, Clendenning M, et al: BRAFV600E immunohistochemistry facilitates universal screening of colorectal cancers for Lynch syndrome. Am J Surg Pathol. 37:1592–1602. 2013.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Adar T, Rodgers LH, Shannon KM, Yoshida M, Ma T, Mattia A, Lauwers GY, Iafrate AJ and Chung DC: A tailored approach to BRAF and MLH1 methylation testing in a universal screening program for Lynch syndrome. Mod Pathol. 30:440–447. 2017.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Newton K, Jorgensen NM, Wallace AJ, Buchanan DD, Lalloo F, McMahon RF, Hill J and Evans DG: Tumour MLH1 promoter region methylation testing is an effective prescreen for Lynch Syndrome (HNPCC). J Med Genet. 51:789–796. 2014.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Pérez-Carbonell L, Alenda C, Payá A, Castillejo A, Barberá VM, Guillén C, Rojas E, Acame N, Gutiérrez-Aviñó FJ, Castells A, et al: Methylation analysis of MLH1 improves the selection of patients for genetic testing in Lynch syndrome. J Mol Diagn. 12:498–504. 2010.PubMed/NCBI View Article : Google Scholar
|
















