Introduction
The prognosis of patients with colorectal cancer
(CRC) after surgical resection is based on the American Joint
Committee on Cancer (AJCC)/International Union Against Cancer
(UICC)-TNM staging system (1,2).
However, this anatomy-based system gives useful but insufficient
prognostic information, particularly for prognosis assessments in
patients with UICC stage II and III CRC (2-5).
In numerous solid tumor types, such as CRC,
accumulating studies have indicated that the invasion of immune
factors, depending on the type, density and location of
infiltration (6) in the tumor
microenvironment, has a profound impact on the prognosis of cancer
patients (7-10).
Specifically, a high density of adaptive immune cells infiltrating
the core and invasive margin of the tumor was associated with a
favorable prognostic effect for disease-free survival and overall
survival (OS) in patients with early and advanced-stage CRC
(11,12). Thus, an immunoscore based on the
density and location of CD3/CD8+ lymphocytes was used as
a supplementary component for CRC classification (13).
However, the tumor immune microenvironment is
complex and involves multiple immune cell types (14), as well as co-signaling molecules
and immunomodulatory factors (15,16),
which may produce both pro- and antitumor functions. Multiple
immune effector cells, such as CD8+TIL or
CD4+TIL, and immunosuppressive cells, such as regulatory
T (Treg) cells or myeloid-derived suppressor cells (MDSCs), are
reportedly present at the tumor site (14). Immune cell subtypes have
demonstrated differential prognostic value depending on the
histological type of cancer (17).
In melanoma, as well as colorectal and breast cancer, high
CD8+TIL infiltration was strongly correlated with
favorable clinical outcome (8).
However, the role of Treg cells in CRC is controversial, as
Foxp3+Treg infiltration indicated favorable prognosis in
certain studies (18-21).
Furthermore, MDSCs are a heterogeneous collection of immature
myeloid cells that exhibit pathological activation and display
potent immunosuppressive activity in the tumor microenvironment
(22). Recently, the prognostic
value of MDSCs was studied in different types of cancer (23-25),
but the prognostic significance of tumor-infiltrating MDSCs in CRC
has remained to be fully determined.
To promote or suppress T-cell activation,
co-signaling molecules may be classified as co-stimulators or
co-inhibitors (15). Programmed
cell death 1 (PD1)/PD1 ligand 1 (PD-L1), the most extensively
studied co-inhibitors, initiated a new wave of cancer immunotherapy
(26). Within the past several
years, PD1 or PD-L1 inhibitors have been successively approved as
first- or second-line therapies for various solid and hematological
tumors, including metastatic CRC with mismatch-repair-deficiency or
microsatellite instability-high (27,28).
Furthermore, co-inhibitors such as lymphocyte-activating 3 (LAG3)
and T-cell immunoglobulin mucin family member 3 (TIM3), as well as
co-stimulators tumor necrosis factor receptor superfamily, member 4
(OX40) and inducible T-cell costimulator (ICOS), either alone or in
combination with the PD1/PD-L1 pathway, have been tested in
clinical trials for various advanced malignancies, including CRC
(16,29). However, the prognostic value of
these co-signaling molecules remains controversial and has remained
to be fully elucidated. Another factor of interest is indoleamine
2,3-dioxygenase 1 (IDO1), an immunomodulatory factor expressed in
immune cells and various tumor cells (TCs) that potently mediates
immunosuppressive effects in cancer (30). Whether IDO1 expression in different
cells offers prognostic value for CRC warrants investigation.
Recently, a visualized nomogram model was developed
for a variety of cancer types, which involved more parameters and
had better survival prediction ability than TNM staging (24,31,32).
Thus, a wide range of immune markers may be used to establish an
accurate immunoprofile to evaluate the prognosis of patients with
CRC. In the present study, the infiltration of four immune cell
types (CD8+TIL, CD4+TIL,
Foxp3+Treg and CD33+MDSC), as well as the
expression of four co-inhibitors (PD1/PD-L1/TIM3/LAG3), two
co-stimulators (OX40/ICOS) and the immunomodulatory factor IDO1
were evaluated in CRC. Subsequently, the independent prognostic
impact of the above variables was ascertained and a nomogram-based
immunoprofile for CRC was established. The nomogram-based
immunoprofile system provides diagnostic accuracy in the prognosis
of clinical outcomes and is a useful supplement to TNM staging for
patients with stage II/III CRC. It is expected that the present
results will improve the ability of clinicians to distinguish
different clinical outcomes in patients with CRC and the same TNM
stage, particularly for TNM stage II/III patients.
Materials and methods
Study population and selection
criteria
A retrospective study was performed on a primary
cohort of 96 patients who underwent surgical resection of primary
CRC at the First Medical Centre of Chinese PLA General Hospital
(Beijing, China) between April 2010 and November 2013, with a mean
follow-up time of 63.1±23.7 months. The inclusion criteria were as
follows: No history of neoadjuvant treatment, complete resection of
colorectal tumors, histologic diagnosis of colorectal carcinoma and
administration of standardized combination chemotherapy if
relapsed. The exclusion criteria were as follows: Other
malignancies, missing clinicopathological and follow-up
information, and perioperative mortality. Using the same criteria,
from January 2009 to May 2010, an independent validation cohort of
consecutive patients with CRC (n=153), who were followed up for a
mean of 50.4±26.8 months, was evaluated at Qingyang People's
Hospital (Qingyang, China). All patients were classified using the
AJCC 8th TNM staging system. These studies complied with ethical
approval processes and were approved by the institutional review
board the Chinese PLA General Hospital (Beijing, China). The
clinical characteristics of the two cohorts are summarized in
Table I.
 | Table IBasic clinical and pathological
features of the patients from the two cohorts. |
Table I
Basic clinical and pathological
features of the patients from the two cohorts.
| Parameter | Primary cohort
(n=96) | Validation cohort
(n=153) | P-value |
|---|
| Age, years | | | 0.191 |
|
Median | 62.5 | 66 | |
|
Range | 24-90 | 22-95 | |
| Sex | | | 0.240 |
|
Male | 63 (65.6) | 89 (58.2) | |
|
Female | 33 (34.4) | 64 (41.8) | |
| Tumor location | | | 0.108 |
|
Colon | 62 (64.6) | 83 (54.2) | |
|
Rectum | 34 (35.4) | 70 (45.8) | |
| Histological
type | | | 0.270 |
|
Adenocarcinoma | 94 (97.9) | 144 (94.1) | |
|
Othera | 2 (2.1) | 9 (5.9) | |
| Vascular
invasion | | | 0.505 |
|
Yes | 7 (7.3) | 8 (5.2) | |
|
No | 89 (92.7) | 145 (94.8) | |
| Grade | | | 0.705 |
|
G1 | 5 (5.2) | 5 (3.3) | |
|
G2 | 74 (77.1) | 123 (80.4) | |
|
G3 | 17 (17.7) | 25 (16.3) | |
| T stage | | | 0.866 |
|
T1+T2 | 9 (9.4) | 12 (7.8) | |
|
T3 | 76 (79.2) | 121 (79.1) | |
|
T4 | 11 (11.5) | 20 (13.1) | |
| N stage | | | 0.976 |
|
N0 | 58 (60.4) | 91 (59.5) | |
|
N1 | 27 (28.1) | 45 (29.4) | |
|
N2 | 11 (11.5) | 17 (11.1) | |
| TNM stage | | | 0.246 |
|
IA+IB | 7 (7.3) | 12 (7.8) | |
|
IIA+IIB | 45 (46.9) | 76 (49.7) | |
|
IIIA+IIIB | 35 (36.5) | 60 (39.2) | |
|
IVA | 9 (9.3) | 5 (3.3) | |
Immunohistochemistry (IHC) and
pathologic assessment
IHC staining of immune cell markers (CD4, CD8, Foxp3
and CD33) and other immunological markers (PD-L1, PD1, LAG3, TIM3,
OX40, ICOS and IDO1) was performed as described previously
(24), with slight modifications.
In brief, after antigen retrieval and blocking of endogenous
peroxidase, as well as blocking of non-specific binding sites,
3-µm-thick tissue sections were incubated with specific primary
antibodies at 4˚C overnight. Subsequently, the sections were
incubated for 30 min with HRP-labeled rabbit/mouse secondary
antibody (Gene Tech) at 37˚C. The primary antibodies used for IHC
are listed in Table SI.
Immunostaining results were examined by two
pathologists independently blinded to any information on the
clinicopathological features of the patients using a
semi-quantitative score. The relative percentage of positive cells
for each marker was calculated using the mean value of five
randomly selected high-magnification (x200) fields on full slides.
The analysis of CD33 and the immunological markers, such as LAG3,
TIM3, OX40 and ICOS, was performed on the entire tumor region
(e.g., parenchyma and mesenchyme). The other markers
(CD4/CD8/Foxp3/PD-L1/PD1/IDO1) were assessed in both the tumor
parenchyma and mesenchyme regions. The assessment of PD-L1 in the
tumor parenchyma distinguished the TCs and immune cells (ICs). For
statistical analyses, the optimal cutoff value for each marker was
determined using the minimum P-value, which was calibrated using
the X-Tile tool (33). Each marker
was stratified into dichotomous variables (high vs. low) and a
calibrated KM analysis was recorded (Table SII).
Multiplex IHC (mIHC) and evaluation of
staining
Measurement of CD8, CD33 and CK (separated tumor
tissue) was performed by mIHC using the Opal iterative staining
protocol according to the manufacturer's protocol (Akoya
Biosciences) and previously published methods (24,34).
In brief, pretreatment of mIHC sections was performed as in the IHC
assay described above and antigen retrieval was performed by
microwave heating. Subsequently, sections were subjected to CD33,
cytokeratin (CK) and CD8 Opal iterative staining and Opal 520, Opal
570 and Opal 620 (Akoya Biosciences) were applied to each antibody.
Nuclei were counterstained with DAPI (Akoya Biosciences). The
primary antibodies used for mIHC are listed in Table SI.
MIHC slide images were captured using the Vectra
platform (Perkin-Elmer) and analyzed using inForm software version
2.2.1 (Akoya Biosciences). To better achieve the multispectral
unmixing of mIHC fluorescence signals, the images of single stained
slides of each marker were used to create a spectrum library and
those of unstained slides were used to extract tissue
autofluorescence. The quantification of positive cells for each
marker was performed using the minimum region signal threshold in
the inForm software automated counting tool. Measurements were
recorded as the mean value of five randomly selected
high-magnification (x200) fields on full slides. For
CD8+TIL, signals from tumor parenchyma (CK-positive) and
mesenchyme (CK-negative) were counted. For CD33+MDSCs,
the levels were assessed in whole tumor regions (e.g., parenchyma
and mesenchyme). Similarly, the minimum P-value of
CD8+TIL and CD33+MDSC in mIHC was set as the
optimal cutoff value.
Statistical analysis
Values are expressed as the mean ± standard
deviation. Statistical calculations were performed using SPSS
Statistics (version 22; IBM Corporation) and GraphPad Prism
(version 7; GraphPad Software, Inc.). The χ2 test was
used to examine the categorized variables. For continuous
variables, the Mann-Whitney U test was used. The correlation
between the expression levels of each marker was determined using
the Spearman coefficient and the correlation matrix was subjected
to unsupervised hierarchical clustering. OS was defined as the time
from surgical resection to last contact. The Kaplan-Meier method
and log-rank test were used to evaluate OS. The univariate and
multivariate Cox proportional hazards model was used to adjust the
independent risk factors. Statistical significance was set at
P<0.05 (two-tailed).
Using the rms package in R version 3.6.1, a
visualized nomogram based on the results of the Cox model analysis
was developed using data from the primary cohort. The primary
cohort served as the internal validation group for the nomogram,
while external validation of the nomogram was performed using the
validation cohort. The nomogram's model performance was quantified
using Harrell's concordance index (C-index). Internal validation of
the nomogram was performed using the bootstrap method and a
relatively unbiased estimate was acquired through 1,000
repetitions. The predictive accuracy and recognition capacity of
the nomogram using the internal and external validation cohorts was
assessed by calculating the C-index and calibration curve. To
facilitate clinical practice, the nomogram was simplified and an
immunoprofile based on the nomogram was constructed. Finally, the
receiver operating characteristic (ROC) curve of the immunoprofile
and TNM stage system were compared in patients with stage II/III
CRC.
Results
Patient clinical characteristics in
both cohorts
The basic clinical and pathological features of the
patients with CRC from the two cohorts are listed in Table I. No significant differences in
clinical characteristics between the two cohorts were observed
(Table I). At the time of
diagnosis, in the primary cohort, the median age was 62.5 (range,
24-90) years, 63 (65.6%) patients were male, and in the validation
cohort, the median age was 66 (range, 22-95) years, of which 89
(58.2%) patients were male. In the present study, most patients
presented with locally advanced stages of the disease (TNM stage II
and III), accounting for 83.3% (80/96) in the primary cohort and
88.9% (136/153) in the validation cohort. Postoperative adjuvant
therapy was as follows: Patients with TNM stage I entered the
follow-up period directly after curative surgical resection,
whereas most patients with TNM stage II/III received close
surveillance and capecitabine or 5-FU-based standard adjuvant
chemotherapy. Advanced patients were given systemic treatment. The
patients of the two cohorts were followed up for at least 5
years.
CRC immune markers Infiltrating
ICs
Infiltration of CD4+TILs,
CD8+TILs, Foxp3+Tregs and
CD33+MDSCs at the CRC tumor in situ was observed
(Fig. 1, upper panel). In CRC, the
location and density of the four tumor-infiltrating ICs were
heterogeneous. As presented in Fig.
1, the compositions of CD4+TILs, CD8+TILs
and Foxp3+Tregs were different in the tumor parenchyma
and mesenchyme regions. These ICs, particularly CD4+TIL
and CD8+TIL, were markedly higher in the tumor
mesenchyme than in the tumor parenchyma. Immunopositives in the
tumor parenchyma and mesenchyme were described as ‘(P)’ and ‘(M)’,
respectively. In the primary cohort, 85% of patients (82/96; P) and
97% of patients (94/96; M) had at least 1% CD8+TIL
infiltration. A total of 26% of patients (25/96; P) and 100% of
patients (96/96; M) had at least 1% CD4+TIL
infiltration. Furthermore, 21% of patients (20/96; P) and 97% of
patients (94/96; M) had at least 1% Foxp3+Treg
infiltration. Furthermore, the infiltration of
CD33+MDSCs was relatively sparse in the tumor mesenchyme
and parenchyma. Thus, the analysis of CD33+MDSCs was
performed in the entire tumor region and at least 1%
CD33+MDSC infiltration was noted in 48% of patients
(46/96) (Table SII).
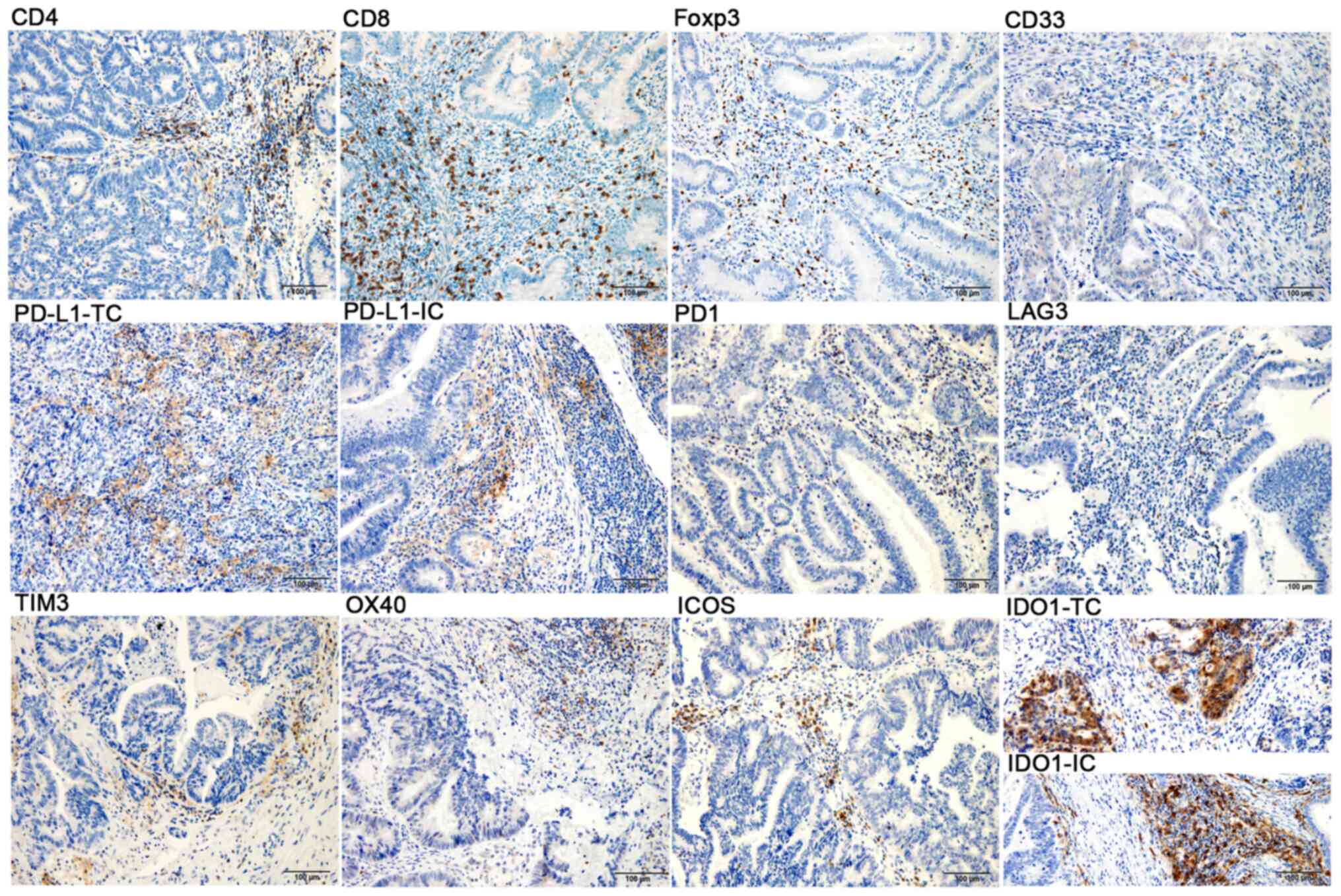 | Figure 1Immunohistochemical staining of CD4,
CD8, Foxp3, CD33, PD-L1, PD1, LAG3, TIM3, OX40, ICOS and IDO1 in
colorectal cancer samples (magnification, x200; scale bar, 100 µm;
blue, nuclear staining; brown, positive staining). TC, tumor cell;
IC, immune cell; Foxp3, forkhead box p3; PD1, programmed cell death
1; PD-L1, PD1 ligand 1; TIM3, T-cell immunoglobulin mucin family
member 3; LAG3, lymphocyte-activating 3; OX40, tumor necrosis
factor receptor superfamily, member 4; ICOS, inducible T-cell
costimulator; IDO1, indoleamine 2,3-dioxygenase 1. |
Expression of co-signaling molecules and
immunomodulatory factors. The expression levels of PD-L1, PD1,
LAG3, TIM3, OX40, ICOS and IDO1 were examined in the CRC tumors
in situ by IHC (Fig. 1,
middle and lower panels). Overall, the expression rate of PD-L1 in
tumor parenchyma was low, with only seven patients (7/96)
expressing ≥1% PD-L1 in TCs, and only one case (1/7) with 5% PD-L1
in ICs, whereas 70% (67/96) of the patients had at least 1% PD-L1
expression in the tumor mesenchyme. At least 1% PD-1 expression was
observed in 21% of patients (20/96; P) and 60% of patients (58/96;
M). The positive expression was defined as ≥1% expression in ICs.
The expression of LAG3, TIM3, OX40 and ICOS was 8% (8/96), 43%
(41/96), 59% (57/96) and 79% (76/96), respectively. Furthermore,
IDO1 was observed in both TCs and ICs. When considering ≥1% to
indicate positive staining, it was observed that 36% of patients
(35/96) exhibited IDO1 expression in TCs, while 89% of patients
(85/96) expressed IDO1 in ICs.
Correlation between tumor-infiltrating ICs and
co-signaling molecules or immunomodulatory factors. According
to the mechanisms governing tumor immunology, ICs may be regulated
by the expression of activated and inhibited molecules, which may
shift the immune response toward anti-inflammatory profiles or
escaped antitumor immunity. Thus, the correlation between
CD8+TIL, CD4+TIL, Foxp3+Treg,
CD33+MDSC and PD-L1, PD-1, LAG3, TIM3, OX40, ICOS and
IDO1 was analyzed (Fig. S1). The
densities of CD8+TIL, both in the tumor parenchyma and
mesenchyme, were significantly correlated with the expression of
PD-L1 (P), PD1 (M), TIM3, OX40 and ICOS (all P<0.001) and
increased along with CD4+TIL (M) and
Foxp3+Treg (M) (all P<0.001). By contrast,
CD33+MDSCs were associated with the infiltration of
CD4+TIL (P<0.001) and expression of TIM3 (P=0.016).
In addition, CD4+TIL (M) and Foxp3+Treg (M)
were also positively correlated with the expression of PD-L1 (P),
PD1 (M), TIM3, OX40, ICOS and IDO1 (all P<0.001). These
correlations may reflect a highly activated immune microenvironment
in CRC.
Association between immune markers and
clinical outcomes Infiltrating ICs and clinical outcomes
The relationship between the patients' OS and
CD4+TIL, CD8+TIL and Foxp3+Treg
expression in the tumor parenchyma and mesenchyme was evaluated.
The survival curves indicated that the favorable prognosis of
patients with high densities of CD8+TIL, both in the
tumor parenchyma and mesenchyme according to IHC and mIHC staining,
was statistically significant compared to those with low
CD8+TIL (Fig. 2A and
B). The densities of
CD8+TIL in both tumor regions were analyzed in
combination, with poor infiltration defined as ≤1%
CD8+TIL in the parenchyma, together with ≤5%
CD8+TIL in the mesenchyme, whereas other combinations
were regarded as rich infiltration (Figs. 2B and S2). In both cohorts, the
CD8+TIL rich groups were significantly associated with
prolonged patient survival (P=0.0002 in the primary cohort and
P<0.001 in the validation cohort; Fig. 2B). The favorable prognostic effect
mentioned above was not observed in the parenchyma, mesenchyme or
whole tumor region for either CD4+TIL or
Foxp3+Tregs (Fig.
S3A). Of note, the analysis of CD33+MDSC revealed
that a high density of CD33+MDSCs resulted in a
significantly unfavorable prognosis compared to low
CD33+MDSC in both cohorts of patients with CRC (P=0.0019
in the primary cohort and P=0.0003 in the validation cohort;
Fig. 2C and D).
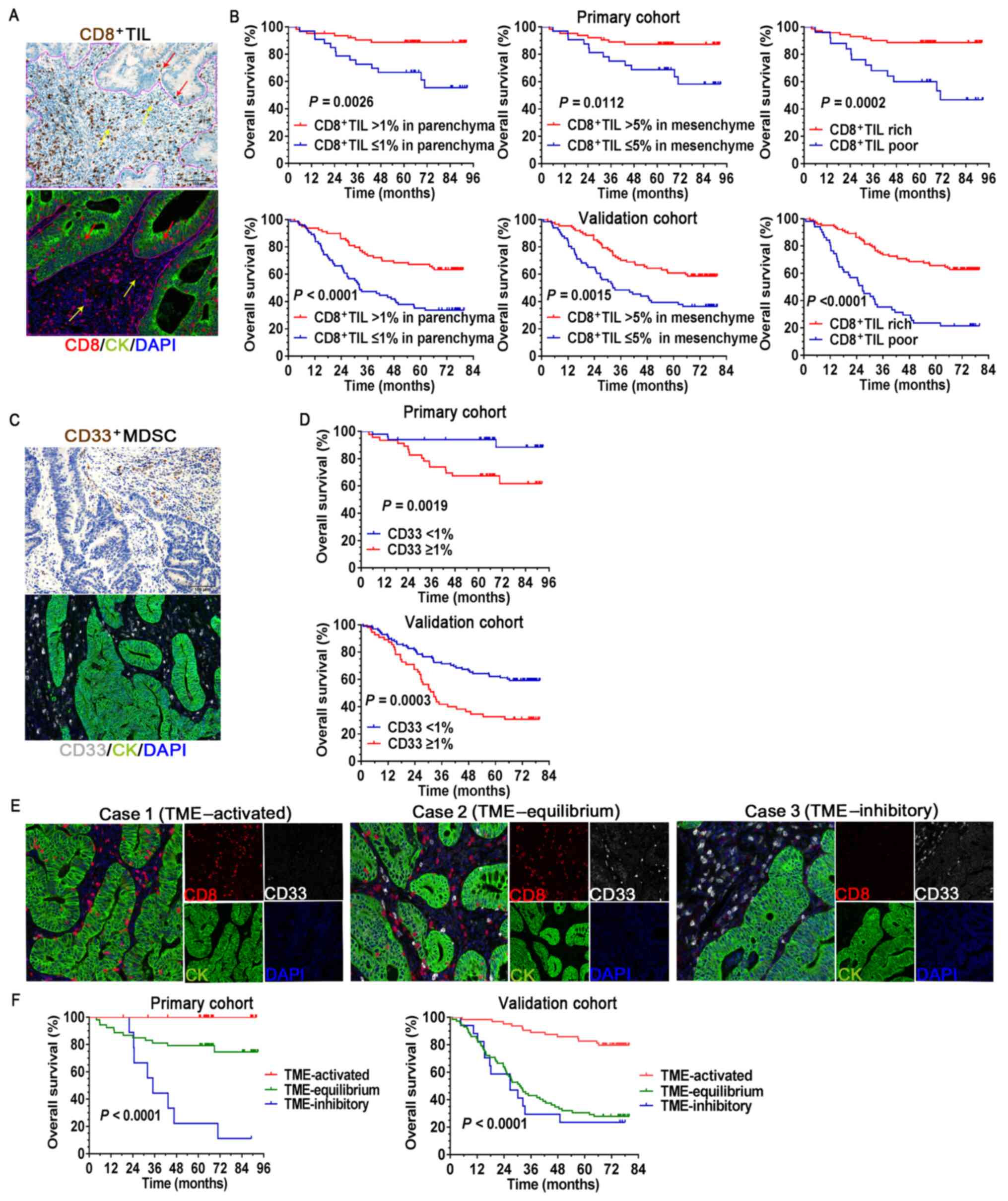 | Figure 2Association between the immune cells
and clinical outcomes. (A) IHC and mIHC staining of
CD8+TIL in the tumor parenchyma (red arrow) and
mesenchyme (yellow arrow), and the solid purple line is the
delimitation of parenchyma/mesenchyme. (B) Survival curves
comparing OS of patients stratified by different CD8+TIL
infiltration status. (C) IHC and mIHC staining of
CD33+MDSCs. The assessment of CD33+MDSCs was
performed in the entire tumor region. (D) Survival curves comparing
OS of patients stratified by different CD33+MDSC
infiltration status. (E) Representative mIHC images for three
different statuses of tumor-infiltrating immune cells in the TME
(magnification, x200; scale bar, 100 µm). (F) Survival curves
comparing the OS of patients stratified by different TME status.
MDSCs, myeloid-derived suppressor cells; OS, overall survival; IHC,
immunohistochemistry; mIHC, multiplex IHC; TME, tumor
microenvironment; TIL, tumor-infiltrating lymphocytes; CK,
cytokeratin. |
Next, the prognostic value of the different
proportions of the CD8+TILs and CD33+MDSCs
when they appeared simultaneously in the CRC microenvironment was
analyzed (Fig. 2E and F). Tumors with rich CD8+TILs
in the TME (TME-activated) were associated with favorable survival,
whereas tumors with high CD33+MDSC (TME-inhibitory) were
associated with poor clinical outcomes. Near-equal infiltration of
CD8+TIL and CD33+MDSCs tended to indicate an
equilibrium TME, in which the patients had inferior survival
compared to those with an activated TME. However, no significant
differences were observed between TME-equilibrium and
TME-inhibitory in the validation cohort. These data highlight the
importance of defining a good scoring system to assess the status
of TME in CRC.
Co-signaling molecules or immunomodulatory
factors and clinical outcomes. Among the six co-signaling
molecules examined, only PD1 (P) and PD1 (M) were significantly
associated with favorable OS (P=0.0421 and P=0.0197, respectively),
whereas PD-L1 (P), PD-L1 (M), LAG3, TIM3, OX40, ICOS, IDO1 (P) and
IDO1 (M) were not significantly associated with any outcome
(Fig. S3B and C). To further assess the prognostic
significance of PD1, PD1 (P) and PD1 (M) were defined, with ≥1% as
PD1+TIL-rich and the others as PD1+TIL-poor.
Similarly, PD1+TIL-rich cells were significantly
associated with prolonged patient survival (P=0.0459; Fig. S3B). IDO1 was observed to be
expressed by TCs and ICs (Fig. 1,
lower panels) and Kaplan-Meier survival analyses suggested that no
statistically significant differences were present between groups
with high and low IDO1 expression levels in the different cells
(Fig. S3C).
Construction and validation of the
nomogram-based immunoprofile Independent prognostic factors
First, the hazard ratio of the clinical parameters
and immune markers was analyzed in the primary cohort using a
univariate regression model. As presented in Table II, among the clinical
characteristics, only pathologic T stage, N stage and TNM stage
were associated with increased risk of death and OS (Fig. S4A-C). Subsequently, TNM stage and
immunological markers with P<0.1 in the univariate analysis were
incorporated into a Cox proportional regression model for
multivariate analysis (Table II).
It was determined that CD8 (HR=0.201; 95% CI: 0.056-0.727;
P=0.014), CD33 (HR=4.565; 95% CI: 1.428-14.592; P=0.010) and TNM
stage (II vs. IV, HR=0.129; 95% CI: 0.034-0.498; P=0.003) exhibited
independent prognostic value for OS in patients with CRC.
 | Table IIUnivariate/multivariate analyses for
overall survival in the primary cohort. |
Table II
Univariate/multivariate analyses for
overall survival in the primary cohort.
| | Univariate
analyses | Multivariate
analyses |
|---|
| Prognostic
factor | Hazard ratio | P-value | Hazard ratio | P-value |
|---|
| Age, <63 vs. ≥63
years | 1.596
(0.652-3.907) | 0.306 | | |
| Male vs. female
sex | 0.465
(0.155-1.392) | 0.171 | | |
| History of smoking,
yes vs. no | 1.807
(0.752-4.344) | 0.186 | | |
| History of
drinking, no vs. yes | 1.517
(0.551-4.178) | 0.420 | | |
| Histological type,
adenocarcinoma vs. othera | 2.643
(0.351-19.889) | 0.345 | | |
| Vascular invasion,
no vs. yes | 2.578
(0.754-8.820) | 0.131 | | |
| Grade, G1 vs.
G3 | 0 (0) | 0.983 | | |
| Grade, G2 vs.
G3 | 0.613
(0.223-1.688) | 0.344 | | |
| T stage, T1+T2 vs.
T4 | 0 (0) | 0.980 | | |
| T stage, T3 vs.
T4 | 0.304
(0.116-0.796) | 0.015 | | |
| N stage, N0 vs.
N2 | 0.193
(0.061-0.609) | 0.005 | | |
| N stage, N1 vs.
N2 | 0.439
(0.138-1.396) | 0.163 | | |
| AJCC TNM stage, Ι
vs. IV | 0 (0) | 0.976 | 0 (0) | 0.983 |
| AJCC TNM stage, II
vs. IV | 0.080
(0.023-0.279) | <0.001 | 0.129
(0.034-0.498) | 0.003 |
| AJCC TNM stage, III
vs. IV | 0.248
(0.086-0.712) | 0.010 | 0.358
(0.119-1.077) | 0.068 |
| CD4+TIL,
poor vs. rich | 1.206
(0.495-2.941) | 0.680 | | |
| CD8+TIL,
poor vs. rich | 0.216
(0.088-0.530) | 0.001 | 0.201
(0.056-0.727) | 0.014 |
|
Foxp3+TIL, poor vs. rich | 1.336
(0.546-3.271) | 0.525 | | |
|
CD33+MDSC, low vs. high | 4.816
(1.609-14.414) | 0.005 | 4.565
(1.428-14.592) | 0.010 |
| PD-L1-parenchyma,
low vs. high | 1.618
(0.374-6.998) | 0.520 | | |
| PD-L1-mesenchyme,
low vs. high | 0.454
(0.188-1.098) | 0.080 | 0.631
(0.222-1.792) | 0.388 |
| PD1+TIL,
poor vs. rich | 0.418
(0.173-1.011) | 0.053 | 0.796
(0.228-2.776) | 0.720 |
| LAG3, low vs.
high | 0.042
(0.000-27.922) | 0.340 | | |
| TIM3, low vs.
high | 0.903
(0.369-2.210) | 0.823 | | |
| OX40, low vs.
high | 0.525
(0.217-1.269) | 0.152 | | |
| ICOS, low vs.
high | 0.427
(0.174-1.045) | 0.062 | 1.407
(0.392-5.054) | 0.600 |
| IDO1-parenchyma,
low vs. high | 1.735
(0.709-4.249) | 0.228 | | |
| IDO1-mesenchyme,
low vs. high | 0.622
(0.226-1.716) | 0.359 | | |
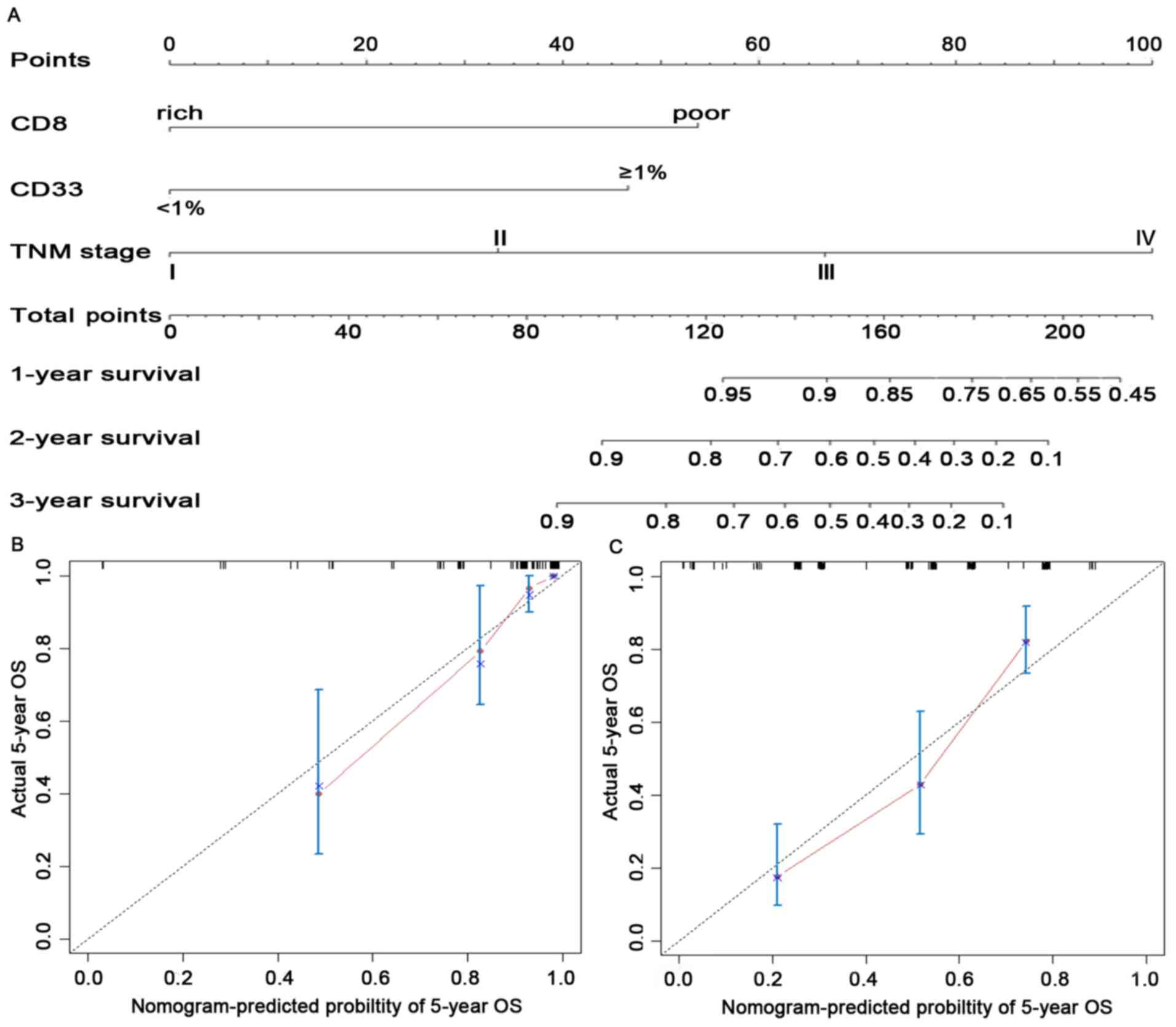
Construction of the nomogram-based immunoprofile
and validation. Although both CD8+TIL and
CD33+MDSC had independent prognostic significance in the
multivariate analyses (Table II),
they were reported to have functionally opposite effects during the
antitumor immune responses in patients with CRC (11,35),
preventing their use for accurate prediction of survival.
Therefore, a comprehensive immunoprofile nomogram for predicting
survival was created using the independent prognostic variables
(Fig. 3A). The C-index for the
predicted OS was 0.861 (95% CI: 0.796-0.925) in the internal
validation and 0.759 (95% CI: 0.714-0.804) in the external
validation cohort. The calibration plot for the probability of
5-year OS between the actual observed survival and the prediction
by the nomogram was in good accordance in the primary cohort
(Fig. 3B) as well as in the
validation cohort (Fig. 3C).
To facilitate clinical application, a recent study
by our group proposed modifying a nomogram into a simple
immunoprofile system based on a nomogram for patients with
esophageal cancer (24). In the
present study, an immunoprofile for patients with CRC was
developed. In the immunoprofile system, 0, 1, 2 and 3 points
corresponded to stages I, II, III and IV, respectively. The points
for CD8-poor and CD33+MDSC ≥1% were between stage II and
III in the nomogram; thus, 1.5 points was assigned in the
immunoprofile. Next, each patient was assigned an immunoprofile
index by adding the values of CD8 and CD33. Patients with different
immunoprofile indices had variant clinical outcomes: Patients with
a high immunoprofile index had unfavorable OS; patients with the
lowest immunoprofile index had the highest OS; other patients had
intermediate clinical outcomes (Fig.
S4D).
Comparison of the predictive accuracy for OS
between the immunoprofile and TNM staging systems. The ability
of the immunoprofile to identify differences in patients at the
same TNM stage was further investigated. As presented in Fig. 4A, all patients (n=249) were divided
into two risk subgroups based on their immunoprofile for
CD8+TIL and CD33+MDSCs, regardless of TNM
stage, using a value of 1.5 as the cutoff. During the observation
period, 86.7% of patients with an immunoprofile index <1.5 were
still alive and only 43.7% of cases were still alive in the high
immunoprofile index (≥1.5) group. The immunoprofile system was able
to separate patients at the same tumor stage into different risk
subgroups (Fig. 4B-E),
particularly for TNM stages II (P<0.0001) and III (P=0.0002),
where significant separation was noted in these patients.
For the patients at stage II/III (n=216), the
predictive accuracy for OS between the immunoprofile and TNM stage
system was compared by ROC curve analysis. The area under the ROC
curve (AUC) of the immunoprofile and TNM staging system was 0.702
(95% CI: 0.636-0.762) and 0.613 (95% CI: 0.0544-0.0678),
respectively (Fig. 4F). The
Z-statistic value of the pairwise comparison of ROC curves was
1.995, exhibiting a statistically significant difference (P=0.046).
These results demonstrated that the immunoprofile provided a more
accurate prognosis for patients with stage II/III CRC compared with
the TNM staging system.
Discussion
Cancer has traditionally been defined with a
cellular-centered vision, but this is being replaced by a holistic
vision that includes the microenvironment (36). The immune microenvironment has an
important role in the evolution of tumors (37), and the immune components within the
microenvironment have a crucial impact on the clinical outcomes for
tumor patients (8). CRC is a
chronic mucosal inflammatory-related malignancy (2). Pagès et al (13) first established an immunoscore in
CRC that included only CD3/CD8 adaptive lymphocytes and did not
consider immunosuppressive markers. In the present study, to
improve the application of immune parameters in the prognosis
assessment of patients, for the first time, the density and
location of 11 immunological markers in situ was
simultaneously analyzed and an immune-related prognostic nomogram
for patients with CRC was constructed. To facilitate routine
clinical usage and discriminate the nomogram system from the
previous immunoscore algorithm, the system of the present study was
modified and defined as an ‘immunoprofile’. The present results
indicated that the immunoprofile system had a good predictive
performance. The AUC for predicting OS in patients with stage
II/III was 0.702, significantly better than that for the TNM II/III
stage (AUC=0.613) (Z-value=1.995, P=0.046).
The present immunoprofile model utilized
tumor-infiltrating CD8+TIL and CD33+MDSCs,
which are significantly associated with clinical outcomes for CRC.
CD8+T lymphocytes are the most important immune effector
and have a central role in eliminating TCs (38). The prognostic role of
CD8+TIL as the most robust immune biomarker has been
reported in various cancer types (8,17).
The results of the present study are consistent with those of
previous reports (11,39) in terms of high CD8+TIL
infiltration being associated with improved patient survival and
being an independent prognostic factor according to multivariate
analyses. Furthermore, the distribution of CD8+TILs in
the human tumor regions was not random, which corroborated reports
that infiltration to specific regions varies according to the
different tumor types (6,40,41).
In the present study, it was observed that the increased density of
CD8-TILs in the parenchyma was more essential for the
prognosis of CRC. Compared with CD8+TILs infiltrating
the mesenchyme, CD8+TILs in the parenchyma exerted a
greater antitumor immune response. Correlation analysis suggested
that infiltration of CD8+TIL was significantly
correlated with the expression of PD-L1, PD-1, TIM3, OX40 and ICOS.
The PD-1/PD-L1 pathway has an important role in inhibiting the
immune response and controls the induction and maintenance of
immune tolerance in the tumor microenvironment (42). TIM3, another inhibitive immune
checkpoint molecule, is expressed in CD8+T cells and is
thought to be involved in T-cell differentiation and activation,
and its persistence may be associated with an exhaustion status
(43). OX40 is a co-stimulatory
molecule, which, if in contact with the OX40 ligand expressed by
antigen-presenting cells, promotes the proliferation of
CD4+ and CD8+T cells and the survival of
antigen-specific memory T cells (44). Evidence suggests that OX40
expression improves the prognostic significance of
CD8+T-cell infiltration in CRC (45). ICOS is another T-cell
co-stimulatory molecule. ICOS is expressed on activated T cells,
memory T cells and regulatory T cells. Similarly, ICOS binds to its
ligand (ICOSL), which regulates T-cell proliferation and survival,
as well as stimulates the production of cytokines (46). Thus, these T-cell inhibitory and
activating receptors modulate the balance between immune tolerance
and immune response in the CRC tumor (29). However, these co-inhibitors and
co-stimulators are expressed simultaneously or at different times
on CD8+T cells, resulting in different phenotypes
(47). The relationship between
these phenotypes and the prognosis of patients with CRC remains to
be fully elucidated and requires to be further investigated. In
addition, there is growing evidence that CD4+T cells
have a vital role in anti-tumor immunity, specifically designed to
activate the CD8+ cytotoxic T-lymphocyte response
(48). The present correlation
analysis indicated that the infiltration of CD8+TIL was
associated with increased CD4+TIL, suggesting that
CD4+TIL have an adjunct role in the efficacy of
anti-tumor CD8+TIL responses in the CRC
microenvironment. Furthermore, correlation analysis also revealed
that the infiltration of CD8+TIL was associated with
increased Foxp3+Tregs, which may also demonstrate the
negative feedback mechanism of the immune system. However, this
hypothesis requires to be further analyzed and verified by Digital
Spatial Profiling (49).
MDSCs represent a heterogeneous population of
pathologically activated immature myeloid cells that have potent
immune suppressive activity in peripheral lymphoid organs and tumor
in situ, and the abnormal accumulation of these cells is an
important mechanism for tumor immune evasion (22,50).
Previous studies have indicated that the proportion of circulating
CD33+/CD11b+ HLA-DR-MDSCs in patients with
CRC was higher than that in healthy individuals and the percentage
of MDSCs in the peripheral blood of patients with CRC was closely
correlated with clinical cancer stage and distant metastasis
(35,51). Furthermore, recent evidence has
revealed that the expansion of circulating granulocytic MDSCs was
associated with poor prognosis in patients with metastatic CRC
treated with FOLFOX-Bevacizumab chemotherapy (52). Although tumor tissue was included
in certain studies (35,51), the significance was limited by low
sample numbers. The present study first analyzed the relationship
between the infiltration of CD33+MDSCs in situ
and the risk of death in patients with CRC. It was revealed that a
high density of CD33+MDSCs in the tumor in situ
was associated with poor outcome compared with a low density of
CD33+MDSCs. Multivariate analyses suggested that
CD33+MDSCs were an independent prognostic factor in CRC.
Furthermore, it was observed that tumor-infiltrating
CD33+MDSCs were closely correlated with the infiltration
of CD4+TILs (P<0.001), suggesting that MDSCs affect
antitumor immunity by CD4+T cells in CRC in situ
(53); however, further
investigation is required to confirm their immunosuppressive
function.
In the tumor microenvironment, different ICs lead
to different TME statuses and affect the prognosis of patients.
Based on immune effector CD8+TIL and immunosuppressive
CD33+MDSC, the present immunoprofile system demonstrated
good predictive performance for patients with stage II/III CRC.
However, the present study also has several limitations. First, the
sample size of stage I/IV patients was too small, limiting the
performance evaluation of the immunoprofile system, which thus
requires further research. In addition, the immune factors used in
the present study were quite limited. With the development of
cancer and immune microenvironment research, there is no doubt that
continuous improvement of the immunoprofile will be implemented,
incorporating more prognostic parameters and improving the
predictive model. Furthermore, the prognostic value of
tumor-infiltrating Foxp3+Tregs in CRC remains
controversial. Certain studies have indicated that
Foxp3+Tregs are associated with favorable prognosis in
CRC, whereas the results of the present study did not indicate such
positive prognostic value. Therefore, future investigations should
distinguish Treg-cell subpopulations (21) or define a suppressive index of Treg
cells (40) in CRC, and to explore
the underlying mechanisms governing the roles of these cells.
In conclusion, in the present study, a
nomogram-based immunoprofile system based on CD8+TILs
and CD33+MDSCs was established in patients with CRC. The
results demonstrated that the immunoprofile provides accurate
prognosis prediction and is an important supplement to the TNM
staging system for patients with stage II/III CRC.
Supplementary Material
Heatmap displaying the hierarchical
clustering analysis of tumor-infiltrating immune cells and six
co-signaling molecules or the immunomodulatory factor IDO1 using
immunohistochemical staining in the primary cohort. Each colored
square in the figure indicates the Spearman correlation r-value
between the two markers. Red color denotes a strong positive
correlation (r=0.9, P<0.00001); yellow, no correlation (r=0);
and green, negative correlation (r= 0.9). P, parenchyma; M,
mesenchyme; TC, tumor cell; IC, immune cell; Foxp3, forkhead box
p3; PD1, programmed cell death 1; PD-L1, PD1 ligand 1; TIM3, T-cell
immunoglobulin mucin family member 3; LAG3, lymphocyte-activating
3; OX40, tumor necrosis factor receptor superfamily, member 4;
ICOS, inducible T-cell costimulator; IDO1, indoleamine
2,3-dioxygenase 1.
Overall survival of patients
stratified by different CD8+TIL infiltration status in
both tumor regions was analyzed in combination. CD8+TIL
lowlow represents ≤1% CD8+T cells infiltrating in the
parenchyma with ≤5% CD8+T cells infiltrating in the
mesenchyme. CD8+TIL highhigh indicates >1%
CD8+T cells infiltrating in parenchyma with >5%
CD8+T cells infiltrating in mesenchyme.
CD8+TIL heterogeneous is another combination
(lowhigh/highlow) of CD8+T cell infiltration status in
both tumor regions. TIL, tumor-infiltrating lymphocytes.
Survival analysis of immune markers in
the primary cohort. Survival curves comparing overall survival of
patients stratified by different (A) CD4H+TIL
infiltration status and Foxp3+T-regulatory cell
infiltration status, (B) PD1+TIL infiltrating status, as
well as (C) PD-L1/LAG3/TIM3/OX40/ICOS/IDO1 expression status. TIL,
tumor-infiltrating lymphocytes; Foxp3, forkhead box p3; PD1,
programmed cell death 1; PD-L1, PD1 ligand 1; TIM3, T-cell
immunoglobulin mucin family member 3; LAG3, lymphocyte-activating
3; OX40, tumor necrosis factor receptor superfamily, member 4;
ICOS, inducible T-cell costimulator; IDO1, indoleamine
2,3-dioxygenase 1.
Survival curves comparing overall
survival of patients stratified by different (A) TNM stage, (B) T
stage, (C) N stage in the primary cohort. (D) Survival curves
grouped by different immunoprofile in all patients with colorectal
cancer (n=249).
Primary antibodies used for
IHC/multiplex IHC.
Kaplan-Meier survival analyses for the
primary cohort (n=96) according to the minimum P-value
cutoffs.
Acknowledgements
The authors would like to acknowledge Dr Lai Song
from Beijing DCTY Bio-information Technology Co. (Beijing, China)
for excellent technical assistance in data analysis.
Funding
This study was supported by the National Natural Science
Foundation of China (grant no. 81672274) and the Clinical and
Scientific Research Support Fund of the General Hospital of the
Chinese PLA (grant no. 2017-FC-TSYS-3022).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author upon
reasonable request.
Authors' contributions
LXW, SCJ, QW and JQH contributed to the conception
and design of the study. LXW, NJC and JFL collected the data. LXW,
NJC, ZZ and LJZ performed IHC and mIHC staining. LXW, LLW and YC
performed the statistical analysis and interpretation. LXW was a
major contributor in writing the manuscript. All authors
contributed to critical revision of the final manuscript and
approved the manuscript for submission. LXW and SCJ confirm the
authenticity of the raw data.
Ethics approval and consent to
participate
In this study, the Chinese PLA General Hospital was
authorized to use the cohort of Qingyang People's Hospital. The
study was approved by the Ethics Committee of the Chinese PLA
General Hospital (Beijing, China; approval no. S2019-228-02) and
written informed consent was provided by all of the patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wittekind C, Compton CC, Greene FL and
Sobin LH: TNM residual tumor classification revisited. Cancer.
94:2511–2516. 2002.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014.PubMed/NCBI View Article : Google Scholar
|
|
3
|
André T, Boni C, Mounedji-Boudiaf L,
Navarro M, Tabernero J, Hickish T, Topham C, Zaninelli M, Clingan
P, Bridgewater J, et al: Multicenter International Study of
Oxaliplatin/5-Fluorouracil/Leucovorin in the Adjuvant Treatment of
Colon Cancer (MOSAIC) Investigators: Oxaliplatin, fluorouracil, and
leucovorin as adjuvant treatment for colon cancer. N Engl J Med.
350:2343–2351. 2004.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Sobrero A, Lonardi S, Rosati G, Di
Bartolomeo M, Ronzoni M, Pella N, Scartozzi M, Banzi M, Zampino MG,
Pasini F, et al: FOLFOX or CAPOX in Stage II to III Colon Cancer:
Efficacy Results of the Italian Three or Six Colon Adjuvant Trial.
J Clin Oncol. 36:1478–1485. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gill S, Meyerhardt JA, Arun M and Veenstra
CM: Translating IDEA to Practice and Beyond: Managing Stage II and
III Colon Cancer. Am Soc Clin Oncol Educ Book. 39:226–235.
2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Galon J, Costes A, Sanchez-Cabo F,
Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M,
Berger A, Wind P, et al: Type, density, and location of immune
cells within human colorectal tumors predict clinical outcome.
Science. 313:1960–1964. 2006.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Broussard EK and Disis ML: TNM staging in
colorectal cancer: T is for T cell and M is for memory. J Clin
Oncol. 29:601–603. 2011.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Fridman WH, Pagès F, Sautès-Fridman C and
Galon J: The immune contexture in human tumours: Impact on clinical
outcome. Nat Rev Cancer. 12:298–306. 2012.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Galon J, Angell HK, Bedognetti D and
Marincola FM: The continuum of cancer immunosurveillance:
Prognostic, predictive, and mechanistic signatures. Immunity.
39:11–26. 2013.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Mei Z, Liu Y, Liu C, Cui A, Liang Z, Wang
G, Peng H, Cui L and Li C: Tumour-infiltrating inflammation and
prognosis in colorectal cancer: Systematic review and
meta-analysis. Br J Cancer. 110:1595–1605. 2014.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Mlecnik B, Tosolini M, Kirilovsky A,
Berger A, Bindea G, Meatchi T, Bruneval P, Trajanoski Z, Fridman
WH, Pagès F, et al: Histopathologic-based prognostic factors of
colorectal cancers are associated with the state of the local
immune reaction. J Clin Oncol. 29:610–618. 2011.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Van den Eynde M, Mlecnik B, Bindea G,
Fredriksen T, Church SE, Lafontaine L, Haicheur N, Marliot F,
Angelova M, Vasaturo A, et al: The Link between the Multiverse of
Immune Microenvironments in Metastases and the Survival of
Colorectal Cancer Patients. Cancer Cell. 34:1012–1026.e3.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Pagès F, Mlecnik B, Marliot F, Bindea G,
Ou F-S, Bifulco C, Lugli A, Zlobec I, Rau TT, Berger MD, et al:
International validation of the consensus Immunoscore for the
classification of colon cancer: A prognostic and accuracy study.
Lancet. 391:2128–2139. 2018.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kerkar SP and Restifo NP: Cellular
constituents of immune escape within the tumor microenvironment.
Cancer Res. 72:3125–3130. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chen L: Co-inhibitory molecules of the
B7-CD28 family in the control of T-cell immunity. Nat Rev Immunol.
4:336–347. 2004.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Mahoney KM, Rennert PD and Freeman GJ:
Combination cancer immunotherapy and new immunomodulatory targets.
Nat Rev Drug Discov. 14:561–584. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Bethmann D, Feng Z and Fox BA:
Immunoprofiling as a predictor of patient's response to cancer
therapy-promises and challenges. Curr Opin Immunol. 45:60–72.
2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Salama P, Phillips M, Grieu F, Morris M,
Zeps N, Joseph D, Platell C and Iacopetta B: Tumor-infiltrating
FOXP3+ T regulatory cells show strong prognostic
significance in colorectal cancer. J Clin Oncol. 27:186–192.
2009.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Sinicrope FA, Rego RL, Ansell SM, Knutson
KL, Foster NR and Sargent DJ: Intraepithelial effector
(CD3+)/regulatory (FoxP3+) T-cell ratio
predicts a clinical outcome of human colon carcinoma.
Gastroenterology. 137:1270–1279. 2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
deLeeuw RJ, Kost SE, Kakal JA and Nelson
BH: The prognostic value of FoxP3+ tumor-infiltrating
lymphocytes in cancer: A critical review of the literature. Clin
Cancer Res. 18:3022–3029. 2012.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Saito T, Nishikawa H, Wada H, Nagano Y,
Sugiyama D, Atarashi K, Maeda Y, Hamaguchi M, Ohkura N, Sato E, et
al: Two FOXP3(+)CD4(+) T cell subpopulations distinctly control the
prognosis of colorectal cancers. Nat Med. 22:679–684.
2016.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Gabrilovich DI: Myeloid-Derived Suppressor
Cells. Cancer Immunol Res. 5:3–8. 2017.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Shipp C, Speigl L, Janssen N, Martens A
and Pawelec G: A clinical and biological perspective of human
myeloid-derived suppressor cells in cancer. Cell Mol Life Sci.
73:4043–4061. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Duan J, Xie Y, Qu L, Wang L, Zhou S, Wang
Y, Fan Z, Yang S and Jiao S: A nomogram-based immunoprofile
predicts overall survival for previously untreated patients with
esophageal squamous cell carcinoma after esophagectomy. J
Immunother Cancer. 6(100)2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Li F, Zhao Y, Wei L, Li S and Liu J:
Tumor-infiltrating Treg, MDSC, and IDO expression associated with
outcomes of neoadjuvant chemotherapy of breast cancer. Cancer Biol
Ther. 19:695–705. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Chen L and Han X: Anti-PD-1/PD-L1 therapy
of human cancer: Past, present, and future. J Clin Invest.
125:3384–3391. 2015.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Gong J, Chehrazi-Raffle A, Reddi S and
Salgia R: Development of PD-1 and PD-L1 inhibitors as a form of
cancer immunotherapy: A comprehensive review of registration trials
and future considerations. J Immunother Cancer. 6(8)2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ganesh K, Stadler ZK, Cercek A, Mendelsohn
RB, Shia J, Segal NH and Diaz LA Jr: Immunotherapy in colorectal
cancer: Rationale, challenges and potential. Nat Rev Gastroenterol
Hepatol. 16:361–375. 2019.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Marin-Acevedo JA, Dholaria B, Soyano AE,
Knutson KL, Chumsri S and Lou Y: Next generation of immune
checkpoint therapy in cancer: New developments and challenges. J
Hematol Oncol. 11(39)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhai L, Ladomersky E, Lenzen A, Nguyen B,
Patel R, Lauing KL, Wu M and Wainwright DA: IDO1 in cancer: A
Gemini of immune checkpoints. Cell Mol Immunol. 15:447–457.
2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Wang Y, Li J, Xia Y, Gong R, Wang K, Yan
Z, Wan X, Liu G, Wu D, Shi L, et al: Prognostic nomogram for
intrahepatic cholangiocarcinoma after partial hepatectomy. J Clin
Oncol. 31:1188–1195. 2013.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Fong Y: Textbook Outcome Nomograms as
Multivariate Clinical Tools for Building Cancer Treatment Pathways
and Prognosticating Outcomes. JAMA Surg.
154(e190572)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Camp RL, Dolled-Filhart M and Rimm DL: RL
C. X-tile: A new bio-informatics tool for biomarker assessment and
outcome-based cut-point optimization. Clin Cancer Res.
10:7252–7259. 2004.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Stack EC, Wang C, Roman KA and Hoyt CC:
Multiplexed immunohistochemistry, imaging, and quantitation: A
review, with an assessment of Tyramide signal amplification,
multispectral imaging and multiplex analysis. Methods. 70:46–58.
2014.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Zhang B, Wang Z, Wu L, Zhang M, Li W, Ding
J, Zhu J, Wei H and Zhao K: Circulating and tumor-infiltrating
myeloid-derived suppressor cells in patients with colorectal
carcinoma. PLoS One. 8(e57114)2013.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Chen DS and Mellman I: Elements of cancer
immunity and the cancer-immune set point. Nature. 541:321–330.
2017.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Golstein P and Griffiths GM: An early
history of T cell-mediated cytotoxicity. Nat Rev Immunol.
18:527–535. 2018.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Pagès F, Kirilovsky A, Mlecnik B, Asslaber
M, Tosolini M, Bindea G, Lagorce C, Wind P, Marliot F, Bruneval P,
et al: In situ cytotoxic and memory T cells predict outcome in
patients with early-stage colorectal cancer. J Clin Oncol.
27:5944–5951. 2009.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Feng Z, Bethmann D, Kappler M,
Ballesteros-Merino C, Eckert A, Bell RB, Cheng A, Bui T, Leidner R,
Urba WJ, et al: Multiparametric immune profiling in HPV-
oral squamous cell cancer. JCI Insight. 2(2)2017.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Stromnes IM, Hulbert A, Pierce RH,
Greenberg PD and Hingorani SR: T-cell Localization, Activation, and
Clonal Expansion in Human Pancreatic Ductal Adenocarcinoma. Cancer
Immunol Res. 5:978–991. 2017.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Han Y, Liu D and Li L: PD-1/PD-L1 pathway:
Current researches in cancer. Am J Cancer Res. 10:727–742.
2020.PubMed/NCBI
|
|
43
|
Solinas C, De Silva P, Bron D,
Willard-Gallo K and Sangiolo D: Significance of TIM3 expression in
cancer: From biology to the clinic. Semin Oncol. 46:372–379.
2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Webb GJ, Hirschfield GM and Lane PJ: OX40,
OX40L and Autoimmunity: A Comprehensive Review. Clin Rev Allergy
Immunol. 50:312–332. 2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Weixler B, Cremonesi E, Sorge R, Muraro
MG, Delko T, Nebiker CA, Däster S, Governa V, Amicarella F, Soysal
SD, et al: OX40 expression enhances the prognostic significance of
CD8 positive lymphocyte infiltration in colorectal cancer.
Oncotarget. 6:37588–37599. 2015.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Xiao Z, Mayer AT, Nobashi TW and Gambhir
SS: ICOS Is an Indicator of T-cell-Mediated Response to Cancer
Immunotherapy. Cancer Res. 80:3023–3032. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Nakano M, Ito M, Tanaka R, Yamaguchi K,
Ariyama H, Mitsugi K, Yoshihiro T, Ohmura H, Tsuruta N, Hanamura F,
et al: PD-1+ TIM-3+ T cells in malignant
ascites predict prognosis of gastrointestinal cancer. Cancer Sci.
109:2986–2992. 2018.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Tay RE, Richardson EK and Toh HC:
Revisiting the role of CD4+ T cells in cancer
immunotherapy-new insights into old paradigms. Cancer Gene Ther.
28:5–17. 2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Merritt CR, Ong GT, Church SE, Barker K,
Danaher P, Geiss G, Hoang M, Jung J, Liang Y, McKay-Fleisch J, et
al: Multiplex digital spatial profiling of proteins and RNA in
fixed tissue. Nat Biotechnol. 38:586–599. 2020.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Kumar V, Patel S, Tcyganov E and
Gabrilovich DI: The Nature of Myeloid-Derived Suppressor Cells in
the Tumor Microenvironment. Trends Immunol. 37:208–220.
2016.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sun HL, Zhou X, Xue YF, Wang K, Shen YF,
Mao JJ, Guo HF and Miao ZN: Increased frequency and clinical
significance of myeloid-derived suppressor cells in human
colorectal carcinoma. World J Gastroenterol. 18:3303–3309.
2012.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Limagne E, Euvrard R, Thibaudin M, Rébé C,
Derangère V, Chevriaux A, Boidot R, Végran F, Bonnefoy N, Vincent
J, et al: Accumulation of MDSC and Th17 Cells in Patients with
Metastatic Colorectal Cancer Predicts the Efficacy of a
FOLFOX-Bevacizumab Drug Treatment Regimen. Cancer Res.
76:5241–5252. 2016.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Nagaraj S, Nelson A, Youn JI, Cheng P,
Quiceno D and Gabrilovich DI: Antigen-specific CD4(+) T cells
regulate function of myeloid-derived suppressor cells in cancer via
retrograde MHC class II signaling. Cancer Res. 72:928–938.
2012.PubMed/NCBI View Article : Google Scholar
|