Introduction
Thyroid cancer (TC) has become a global concern due
to its increasing incidence rate and was ranked 9th among all
cancers in 2020, with ~586,000 cases globally and 3 times higher
incidence in women compared with men (1). Due to more stringent diagnostic
practices, TC incidence has begun to decline in women (2,3). TC
generally derives from epithelial follicular cells, also known as C
cells, and ~90% of TC cases are well-differentiated (WD). DTC is
further classified as papillary (PTC) or follicular (FTC),
depending on histopathological criteria (4). DTCs frequently have genetic
alterations in the gladiators, molecules associated with the
mitogen activated protein kinase (MAPK) signaling pathway,
including RET/PTC rearrangement and point mutations in
RAS and BRAF genes, leading to activation of the MAPK
pathway (5).
BRAF serine-threonine kinase belongs to the family
of RAF proteins. BRAF mutations are oncogenic driver
mutations associated with solid tumors, including thyroid
carcinoma. Among all BRAF mutations identified,
BRAFV600E accounts for >90% (5). Missense mutation results in T>A
transversion at nucleotide position 1799 (c.T1799A), leading to
substitution of valine (V) into glutamic acid (E) at codon 600,
which disrupts interactions between the activation loop and ATP
binding site and allows formation of new interactions that keep the
protein in a catalytically active conformation, resulting in
continuous phosphorylation of MEK (6,7). The
prevalence of BRAFV600E in TC is more
heterogenous in Asian populations, spanning 28.2-90.0% (8). BRAF mutations are highly
prevalent in papillary carcinoma with classical histology and in
the tall cell variant, but are rare in the follicular variant
(9). In many studies, the presence
of BRAF mutation has been associated with aggressive tumor
characteristics, such as extrathyroidal extension, advanced tumor
stage at presentation, tumor recurrence and lymph node (LN) or
distant metastasis (9,10). Mutations in RAS and
BRAF genes rarely overlap in the same tumor and are mutually
exclusive (5,11).
Mammalian cells contain three functional
potooncogenes of the RAS family known as KRAS
(Kristan), HRAS (Harvey) and NRAS
(Neuroblastoma) (12).
These genes encode small GTPases, which are primary participants in
the transmission of growth signals from cell membrane receptors to
the nucleus (13). Gain of
function mutations in the RAS gene family result in
continuous stimulation of cell growth and proliferation, even in
the absence of extracellular signals (14), resulting in tumorigenesis. Point
mutations in the RAS gene (exon 1; codons 12 and 13)
increase its affinity for GTP or inactivate its autocatalytic
GTPase function (exon 2; codon 61) (15) thereby, permanently activating the
MAPK and P13K-AKT pathways. Point mutations of RAS occur
variably in all types of thyroid follicular cell-derived tumors. In
FTC, RAS mutations are found in 40-50% of tumors (16) and may also correlate with tumor
dedifferentiation and less favorable prognosis (17). In PTC, RAS mutations are
relatively infrequent and occur in ~10% of tumors (18). Papillary carcinoma with RAS
mutations almost always exhibit follicular variant histology; this
mutation also correlates with significantly less prominent nuclear
features of papillary carcinoma, more frequent encapsulation and
lower rate of LN metastasis (19).
Studies have reported an association between RAS mutations
and more aggressive behavior of PTC and higher frequency of distant
metastasis (20,21).
Besides the mutation hotspots of KRAS,
HRAS and NRAS, inherited single nucleotide
polymorphism (SNP) in exon 1 of HRAS T81C (rs12628)
is associated with risk of different types of human cancer. HRAS
T81C homozygous variant genotype (CC) has been associated with
bladder cancer, chronic myeloid leukemia and TC (22,23).
This SNP, located at codon 27 of exon 1, does not disturb the p21
protein structure and function as codons CAC and CAT both encode
histidine (His27His). This variation instead disturbs
expression of HRAS by inducing overexpression (24) and may be associated with additional
polymorphic loci inside regulatory sections of HRAS. Earlier
studies also investigated the role of HRAS T81C SNP in TC
and associated it with aneuploidy in follicular tumors of thyroid
(22,24).
The present study hypothesized that mutations in
hotspot regions of RAS (KRAS, HRAS,
NRAS); BRAF and presence of HRAS T81C
variation may modulate susceptibility to TC. To verify this
hypothesis, these mutations and polymorphisms in TC were assessed
to ascertian their association and functional role in thyroid
carcinogenesis of Pakistani population.
Materials and methods
Study design
The present investigation was a prospective cohort
study conducted by the Department of Biological Sciences,
International Islamic University, Islamabad; Department of
Biochemistry, Pakistan Institute of Medical Sciences (PIMS)
Islamabad and Combined Military Hospital (CMH) Muzaffarabad,
Pakistan. Ethical approval was obtained from Ethical Review Board
of PIMS. All participants enrolled in the study provided written
informed consent allowing the use of their tissue and blood
samples. Patients with any genetic disorder, other type of cancer
or receiving chemotherapy were excluded from the study.
Study subjects and sample collection
for analysis of BRAF and RAS mutations
Thyroid tumor and adjacent normal tissue (n=72;
distance, 5-10 mm) samples were obtained from histologically
confirmed patients with DTC who underwent total or
hemi-thyroidectomy in the Department of General Surgery, PIMS and
CMH, Pakistan between 2016 and 2018. Tissue samples were collected
in sterile vials and immediately stored at -80˚C until further
processing.
Study subjects and sample collection
for HRAS T81C genotyping
A total of 180 peripheral blood samples from
patients with DTC were collected from Department of General
Surgery, PIMS and CMH, Pakistan, between 2017 and 2019. In
addition, 220 blood samples were collected from ethnicity-matched
healthy controls free from any type of malignancy, who visited
hospitals for routine checkup. A total of ~3 ml each blood was
collected in EDTA-coated vials from patients with DTC and healthy
controls and stored at -80˚C until further processing.
DNA extraction
DNA from fresh tumor and adjacent normal tissue was
isolated using PureLink Genomic DNA Mini kit (Invitrogen; Thermo
Fisher Scientific, Inc.). A total of 5 formalin-fixed, paraffin
embedded (FFPE) tissue samples were retrieved from Department
Pathology, PIMS and CMH, Pakistan, which had not been immediately
collected following resection of thyroid gland. DNA was isolated
from FFPE tissue using QIAamp DNA FFPE Tissue kit (Qiagen GmbH).
Furthermore, DNA was isolated from blood samples of cases and
controls using QIAamp DNA Blood Mini kit (Qiagen GmbH).
Quantification of isolated DNA was performed using NanoDrop
Microvolume Spectrophotometer (Thermo Fisher Scientific, Inc.).
PCR
The primer sequences used to amplify exon 15 of
BRAF; exon 1 and 2 of each KRAS, HRAS and
NRAS gene along with their annealing temperatures and
amplicon size are given in Table
I. Each PCR reaction was executed in a final volume of 50 µl
containing 2 each forward and reverse primers (20 pM/µl), 3 genomic
DNA, 18 sterile water and 25 µl GoTaq 2X Green Master mix (Promega
Corporation). PCR reaction was performed using the following
thermocycling conditions: Initial denaturation for 5 min at 95˚C,
followed by denaturation of template DNA for 35 sec at 94˚C,
annealing for 35 sec and primer extension for 35 sec at 72˚C.
Denaturation, annealing and primer extension steps were repeated
for 35 cycles. Final extension was performed for 5 min at 72˚C as
previously described (22,25). The PCR product was run on 2%
agarose gel and analysed using AlphaImager™ Gel Imaging System
(ProteinSimple). Double distilled water (ddH2O) was used
as a negative control.
 | Table IPrimer sequences, annealing
temperature and product size of exons for PCR amplification. |
Table I
Primer sequences, annealing
temperature and product size of exons for PCR amplification.
| Gene | Exon | Primer Sequence,
5'→3' | Annealing
temperature, ˚C | Product size,
bp |
|---|
| BRAF | 15 | F:
TCATAATGCTTGCTCTGATAGGA | | |
| | | R:
GGCCAAAAATTTAATCAGTGGA | 58 | 224 |
| NRAS | 1 | F:
AGTACTGTAGATGTGGCTCGCC | | |
| | | R:
CCTCACCTCTATGGTGGGATC | 60 | 185 |
| NRAS | 2 | F:
CCCCTTACCCTCCACAC | | |
| | | R:
AGGTTAATATCCGCAAATGAC | 55 | 196 |
| HRAS | 1 | F:
CAGGAGACCCTGTAGGAGGA | | |
| | | R:
GGCACCTGGACGGCGGCGCTAG | 60 | 186 |
| HRAS | 2 | F:
TCCTGCAGGATTCCTACCGG | | |
| | | R:
GGTTCACCTGTACTGGTGGA | 55 | 194 |
| KRAS | 1 | F:
GTACTGGTGGAGTATTTGAT | | |
| | | R:
TGAAAATGGTCAGAGAAACC | 55 | 285 |
| KRAS | 2 | F:
CCTTCTCAGGATTCCTACAG | | |
| | | R:
TTATTTATGGCAAATACACAAATA | 55 | 1585 |
Di-deoxy Sanger sequencing
Gene JET PCR Purification kit (Thermo Fisher
Scientific, Inc.; cat. no. K0702) was used to purify PCR products
according to the manufacturer's instructions. The purified PCR
products of different DNA samples were subjected to Sanger
sequencing using SeqStudio Genetic Analyzer (Applied Biosystems;
Thermo Fisher Scientific, Inc.).
PCR-restriction fragment length
polymorphism (RFLP)
For HRAS T81C genotyping, exon 1 of
HRAS was amplified by PCR as previously described (15,22),
yielding a 186 bp product (Table
I). For RFLP, PCR product was subjected to digestion with
DraIII (Thermo Fisher Scientific, Inc.) restriction enzyme
at 37˚C for 16 h. The homozygous variant genotype (CC) was cut into
fragments of 128 and 58 bp; homozygote wild genotype (TT) yielded a
single fragment of 186 bp while heterozygous variant (TC) yielded
186, 128 and 58 bp fragments. Digestion products were subjected to
3% agarose gel electrophoresis and documented using AlphaImager™
Gel Imaging System (ProteinSimple).
Statistical analysis
Data are presented as the mean ± SD of three
independent repeats. χ2 test was used to compare cases
and controls in terms of categorical variables, such as age, sex,
histological type, thyroid stimulating hormone (TSH) levels,
residence and smoking status using multiple logistic regression
analysis. A goodness of fit test was applied to assess whether
polymorphisms between cases and controls were present in
Hardy-Weinberg equilibrium. Estimation of the relative risk and
degree of association between genotytpes and risk factors of TC
were determined by calculation of the odds ratio (OR) with 95%
confidence interval (CI). P≤0.05 was considered to indicate a
statistically significant difference. All statistical analysis was
performed using SPSS V. 23.0. software (IBM Corp.).
Results
Characteristics of patients with TC
for tissue analysis
For mutation analysis of BRAF, HRAS,
KRAS and NRAS genes, a total of 72 DTC and adjacent
normal tissue samples were taken. Table II shows the frequency distribution
of selected socio-demographic and clinicopathological
characteristics of DTC cases for mutational analysis. Among DTC
cases, 30.6% (22 of 72) were male and 69.4% (50 of 72) were female.
A total of 54 of 72 (75%) patients were <55 years of age and 18
of 72 (25%) were ≥55 years of age. The number of non-smokers and
smokers were 65 (90%) and 7 (10%) respectively. Furthermore, TSH
levels were normal and elevated in 58.4% (42 of 72) and 41.6% (30
of 72) of cases, respectively. The normal reference range for TSH
was taken as 0.35-6.0 µIU/ml. History of benign thyroid disease
(BTD; including thyroid adenoma, goitre and thyrotoxicosis) was
found in 80% of patients. WDTC was present in 94.0% (68 of 72) of
patients. Vascular/capsular invasion and lymph node metastasis was
positive in 43.1 and 55.8% of patients, respectively. Other
clinicopathological details of TC cases are shown in Table II.
 | Table IIFrequency distribution of selected
socio-demographic and clinicopathological characteristics of DTC
cases for mutational analysis. |
Table II
Frequency distribution of selected
socio-demographic and clinicopathological characteristics of DTC
cases for mutational analysis.
| Characteristic | Cases, n=72
(%) |
|---|
| Sex | |
|
Male | 22.0 (30.6) |
|
Female | 50.0 (69.4) |
| Age, years | |
|
<55 | 54.0 (75.0) |
|
≥55 | 18.0 (25.0) |
| Smoking status | |
|
Non-smoker | 65.0 (90.0) |
|
Smoker | 7.0 (10.0) |
| TSH levels | |
|
Normal | 42.0 (58.4) |
|
Elevated | 30.0 (41.6) |
| BTD | |
|
Absent | 58.0 (80.5) |
|
Present | 14.0 (19.5) |
| Histological
type | |
|
CPTC | 45.0 (62.5) |
|
FPTC | 27.0 (37.5) |
| Grade | |
|
WD | 68.0 (94.0) |
|
PD | 4.0 (6.0) |
| Tumor focality | |
|
Unifocal | 40.0 (55.6) |
|
Multifocal | 32.0 (44.4) |
| Stage, <55
years | |
|
I | 21.0 (39.0) |
|
II | 33.0 (61.0) |
| Stage, ≥55
years | |
|
I+II | 7.0 (39.0) |
|
≥III | 11.0 (61.0) |
| V/C invasion | |
|
Absent | 31.0 (43.1) |
|
Present | 41.0 (56.9) |
| LN metastasis | |
|
Absent | 33.0 (45.8) |
|
Present | 39.0 (54.2) |
Mutational analysis of BRAF and RAS
genes
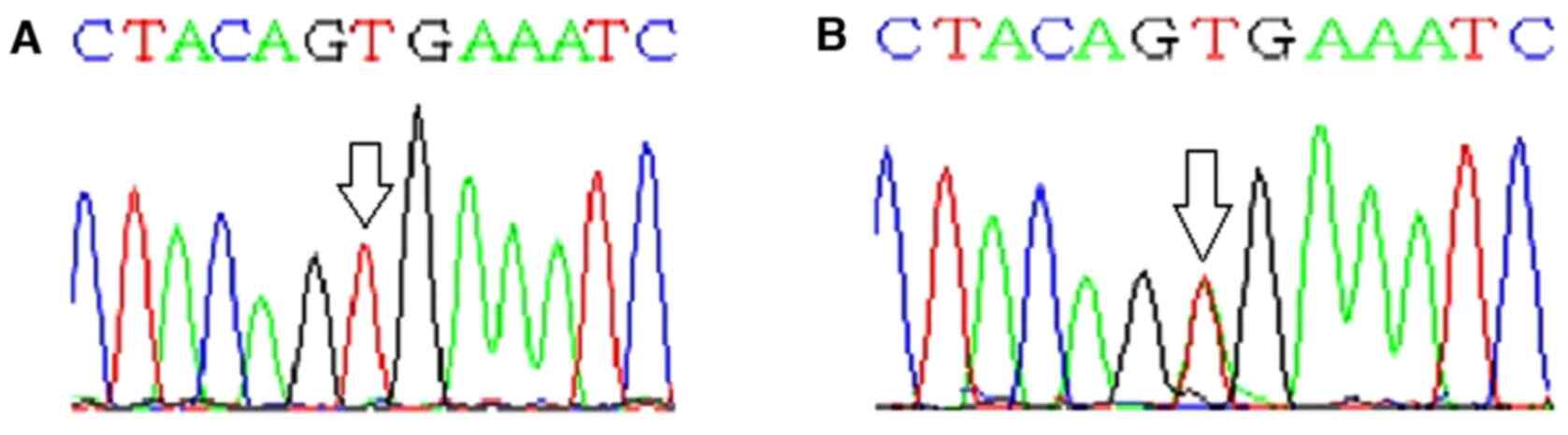
Exon 15 of BRAF gene was screened for
presence of hotspot mutations in DTC tumor and adjacent normal
tissue. T to A transversion was noted at nucleotide position 1,799
(c.T1799A) in 28% (20 of 72) of patients with DTC, resulting in
substitution of V into E at codon 600. BRAF mutation was not
found in adjacent normal tissue samples. Partial electropherograms
showing T to A tranversion in BRAFV600E mutation
are depicted in Fig. 1.
BRAFV600E mutation was not found in any patients
with follicular variant of PTC (FPTC). BRAFV600E
mutation was confined to classical variant of PTC (CPTC)
(P=0.0001). A higher frequency of BRAFV600E
mutation (41%) was significantly associated with higher tumour
focality and LN metastasis (P=0.03 and 0.005; Table III). Table III shows the association between
BRAFV600E mutation status with demographic and
clinicopathological features of patients with DTC.
 | Table IIIAssociation of
BRAFV600E mutation status with demographic and
clinicopathological features of patients with DTC. |
Table III
Association of
BRAFV600E mutation status with demographic and
clinicopathological features of patients with DTC.
| |
BRAFV600E mutation | |
|---|
| Characteristic | Cases, n=72
(%) | Positive, n=20
(27.7%) | Negative, n=52
(72.3%) | P-value |
| Sex | | | | |
|
Male | 22.0 (30.6) | 9.0 (41.0) | 13.0 (59.0) | 0.1000 |
|
Female | 50.0 (69.4) | 11.0 (22.0) | 39.0 (78.0) | |
| Age, years | | | | |
|
<55 | 54.0 (75.0) | 14.0 (26.0) | 40.0 (74.0) | 0.7000 |
|
≥55 | 18.0 (25.0) | 6.0 (33.3) | 12.0 (66.7) | |
| Smoking status | | | | |
|
Non-smoker | 65.0 (90.0) | 18.0 (24.6) | 47.0 (75.4) | 0.6000 |
|
Smoker | 7.0 (10.0) | 2.0 (85.7) | 5.0 (14.3) | |
| TSH levels | | | | |
|
Normal | 42.0 (58.4) | 10.0 (23.8) | 32.0 (76.2) | 0.4000 |
|
Elevated | 30.0 (41.6) | 10.0 (33.3) | 20.0 (66.7) | |
| BTD | | | | |
|
Absent | 58.0 (80.5) | 15.0 (25.8) | 43.0 (74.2) | 0.3000 |
|
Present | 14.0 (19.5) | 5.0 (35.7) | 9.0 (64.3) | |
| Histological
type | | | | |
|
CPTC | 45.0 (62.5) | 20.0 (44.4) | 25.0 (55.6) | 0.0001a |
|
FPTC | 27.0 (37.5) | 0.0 (0.0) | 27.0 (100.0) | |
| Grade | | | | |
|
WD | 68.0 (94.0) | 18.0 (26.4) | 50.0 (73.6) | 0.5000 |
|
PD | 4.0 (6.0) | 2.0 (50.0) | 2.0 (50.0) | |
| Tumor focality | | | | |
|
Unifocal | 40.0 (55.6) | 7.0 (17.5) | 33.0 (82.5) | 0.0300a |
|
Multifocal | 32.0 (44.4) | 13.0 (41.0) | 19.0 (59.0) | |
| Stage, <55
years | | | | |
|
I | 21.0 (39.0) | 8.0 (38.0) | 13.0 (62.0) | 0.1000 |
|
II | 33.0 (61.0) | 6.0 (18.0) | 27.0 (81.0) | |
| Stage, ≥55
years | | | | |
|
I+II | 7.0 (39.0) | 2.0 (28.5) | 5.0 (71.5) | 0.5000 |
|
≥III | 11.0 (61.0) | 4.0 (36.4) | 7.0 (63.6) | |
| V/C Invasion | | | | |
|
Absent | 31.0 (43.1) | 6.0 (19.0) | 25.0 (81.0) | 0.1000 |
|
Present | 41.0 (56.9) | 14.0 (34.1) | 27.0 (65.8) | |
| LN metastasis | | | | 0.0050a |
|
Absent | 33.0 (45.8) | 4.0 (15.2) | 29.0 (84.8) | |
|
Present | 39.0 (54.2) | 16.0 (41.0) | 23.0 (59.0) | |
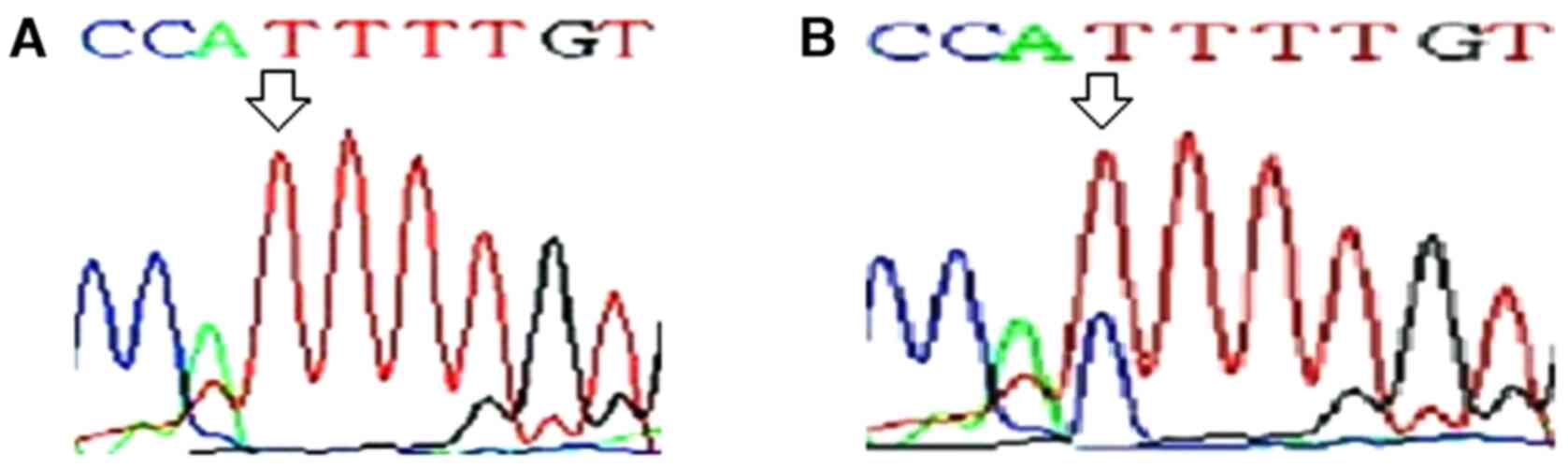
Exons 1 and 2 of KRAS, NRAS and
HRAS genes were screened for presence of hotspot mutations
in codons 12, 13 and 61 of a RAS gene family. No mutation
was found in any of these codons in DTC tumor or adjacent normal
tissue samples. Following DNA sequencing of HRAS, a frequent
substitution of T to C was found in exon 1 at codon 27 (cDNA
position 81), which was present in wobble base position (Fig. 2). HRAS T81C mutation was
found in 21% (15 of 72) of DTC tumor tissue. Therefore, this
mutation was investigated in blood samples of DTC cases and
controls as a potential genetic polymorphism.
Characteristics of patients with TC
for blood analysis
A total of 180 patients with DTC, along with 220
healthy controls, were selected for the study of HRAS T81C
polymorphism. Out of 180 DTC cases, 72.8% (131 of 180) were <55
years of age and 27.2% (49 of 180) were ≥55 years of age; 37.7% (68
of 180) were male and 62.3% (112 of 180) were female. The
proportion of non-smokers was 68.8% (124 of 180) and that of
smokers was 31.2% (56 of 180). The cases and controls were matched
with respect to sex, age, dwelling and smoking status (P=0.18;
0.91; 0.46 and 0.83). Socio-demographic and clinicopathological
characteristics of DTC cases and controls are listed in Table IV.
 | Table IVFrequency distribution of
socio-demographic and clinicopathological characteristics of DTC
cases and controls for HRAS T81C genotyping |
Table IV
Frequency distribution of
socio-demographic and clinicopathological characteristics of DTC
cases and controls for HRAS T81C genotyping
| Characteristic | Cases, n=180
(%) | Controls, n=220
(%) | χ2 | P-value |
|---|
| Sex | | | 1.87 | 0.18 |
|
Male | 68.0 (37.7) | 98.0 (44.5) | | |
|
Female | 112.0 (62.3) | 122.0 (55.5) | | |
| Age, years | | | | |
|
<55 | 131.0 (72.8) | 158.0 (71.8) | 0.05 | 0.91 |
|
≥55 | 49.0 (27.2) | 62.0 (28.2) | | |
| Dwelling | | | | |
|
Rural | 120.0 (66.6) | 138.0 (62.7) | 0.67 | 0.46 |
|
Urban | 60.0 (33.4) | 82.0 (37.2) | | |
| Smoking status | | | | |
|
Non-smoker | 124.0 (68.8) | 149.0 (67.7) | 0.06 | 0.83 |
|
Smoker | 56.0 (31.2) | 71.0 (32.3) | | |
| BTD | | | | |
|
Absent | 75.0 (41.6) | | | |
|
Present | 105.0 (58.3) | | | |
| Histological
type | | | | |
|
PTC | 151.0 (83.9) | | | |
|
FTC | 29.0 (16.1) | | | |
| TSH levels | | | | |
|
Normal | 52.0 (28.9) | | | |
|
Elevated | 128.0 (71.1) | | | |
| Grade | | | | |
|
WD | 176.0 (97.8) | | | |
|
PD | 4.0 (2.2) | | | |
| Tumor focality | | | | |
|
Unifocal | 96.0 (53.3) | | | |
|
Multifocal | 84.0 (46.7) | | | |
| Stage, <55
years | | | | |
|
I | 65.0 (36.1) | | | |
|
II | 66.0 (36.7) | | | |
| Stage, ≥55
years | | | | |
|
I+II | 26.0 (14.4) | | | |
|
≥III | 23.0 (12.8) | | | |
| V/C invasion | | | | |
|
Absent | 93.0 (51.7) | | | |
|
Present | 87.0 (48.3) | | | |
| LN metastasis | | | | |
|
Absent | 110.0 (61.1) | | | |
|
Present | 70.0 (38.9) | | | |
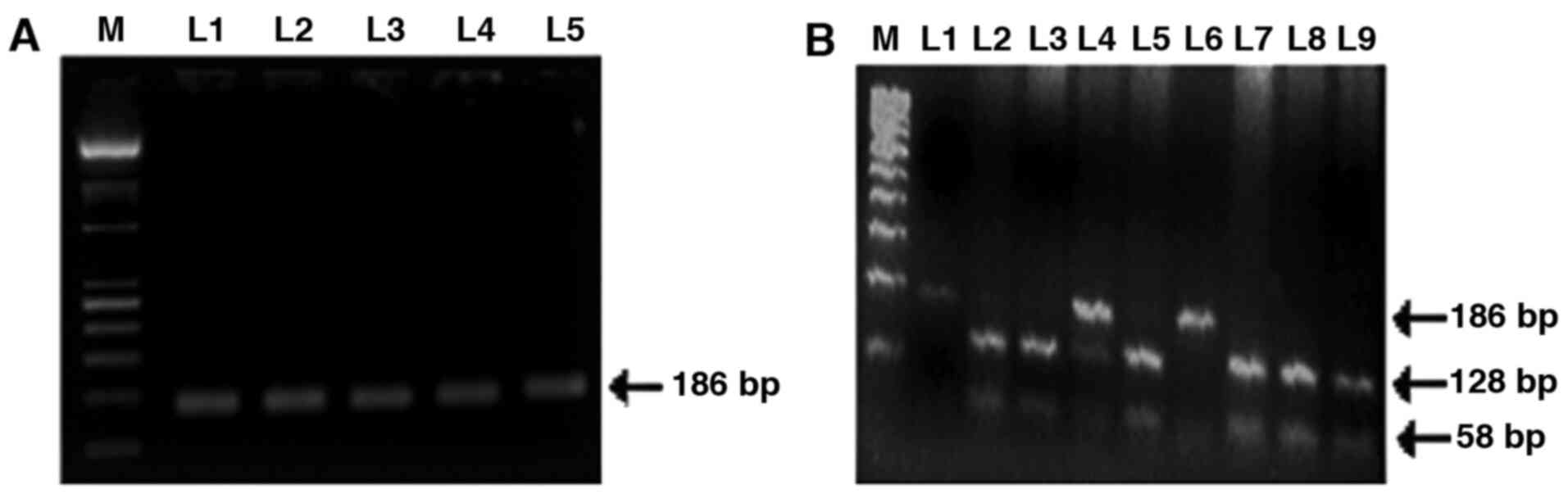
Analysis of HRAS T81C SNP
PCR amplified product of HRAS exon 1 and
fragment digestion of PCR product by DraIII restriction
enzyme is shown in Fig. 3.
Frequency distribution of HRAS T81C genotypes TT, TC and CC
among cases were 37.7, 46.1 and 16.1%, respectively, in patients,
compared with 54.5, 34.1 and 11.4%, respectively, in controls.
Furthermore, the allele frequency of T and C among cases was 60.8
and 39.2%, respectively, and 71.5 and 28.5%, respectively, in
controls. The difference in genotypic and allele frequency between
cases and controls was statistically significant (P≤0.05; Table V). As the frequency of homozygous
mutant (CC) genotype was low, the combined variant (TC+ CC)
genotype was compared with homozygous wild genotype (TT) between
cases and controls to investigate the increased cancer risk
associated with variant genotypes. Overall frequency of TC + CC was
significantly greater in cases compared with controls (62.2 vs.
45.4%) with OR of 2.0 (95% CI, 1.3-2.9; P=0.0009; Table V).
 | Table VDistribution of HRAS T81C
genotypes and its allele frequency in DTC cases and controls for
HRAS T81C genotyping. |
Table V
Distribution of HRAS T81C
genotypes and its allele frequency in DTC cases and controls for
HRAS T81C genotyping.
| Type | DTC cases, n=180
(%) | Controls, n=220
(%) | OR (95% CI) | P-value |
|---|
| Genotype | | | | |
|
TT | 68.0 (37.7) | 120.0 (54.5) | 1.00 (Ref) | |
|
TC | 83.0 (46.1) | 75.0 (34.1) | 1.90
(1.30-3.00) | 0.0020a |
|
CC | 29.0 (16.1) | 25.0 (11.4) | 2.05
(1.10-3.80) | 0.0300a |
|
TC + CC | 112.0 (62.2) | 100.0 (45.4) | 2.00
(1.30-2.90) | 0.0009a |
| Allele | | | | |
|
T | 219.0 (60.8) | 315.0 (71.5) | 1.00 (Ref) | |
|
C | 141.0 (39.2) | 125.0 (28.5) | 1.60
(1.20-2.20) | 0.0010a |
Genotype frequencies of TT and TC + CC were compared
between DTC cases and controls with respect to different
socio-demographic and clinicopathological parameters (Table VI). The results indicated
significantly higher frequency of variant genotype (TC + CC) in
male patients with DTC compared with males in the control group
(61.7 vs. 40.8%; P=0.01). Age was a strong risk factor for DTC as
the difference in the frequency of TC + CC genotype between cases
and controls ≥55 years of age was significant (59.27 vs. 19.3%;
P=0.00002; OR=6.0). Similarly, patients with DTC living in rural
areas had significantly higher frequency of variant genotype (TC +
CC) compared with controls (62.57 vs. 42.1%; P=0.001). Furthermore,
combined TC and CC genotype was significantly greater in non-smoker
DTC patientcompared with non-smoker control group (61.37 vs. 44.9%;
P=0.007). TC and CC were frequently observed in patients with DTC
without history of BTD compared with patients with BTD (76.07 vs.
52.3%; P=0.002). A high frequency of variant genotype (TC + CC) was
found in patients with DTC with multifocal disease (70.27 vs.
55.2%; P=0.04) and LN metastasis (84.37 vs. 48.2%; P=0.00001)
compared with patients with unifocal disease and without LN
metastasis. Association of rare variants (TC + CC) with other
socio-demographic and clinicopathological parameters of DTC cases
and controls is shown in Table
VI.
 | Table VIAssociation of HRAS T81C
genotypes with socio-demographic and clinicopathological parameters
of DTC cases and controls for HRAS T81C genotyping. |
Table VI
Association of HRAS T81C
genotypes with socio-demographic and clinicopathological parameters
of DTC cases and controls for HRAS T81C genotyping.
| | Cases | Controls | |
|---|
| Parameter | n=180.0 (%) | TT, n=68.0
(37.7%) | TC + CC, n=112.0
(62.3%) | n=220.0 (%) | TT, n=120.00
(54.6%) | TC + CC, n=100.0
(45.4%) | OR (95% CI) 2.00
(1.30-2.90) | P-value
0.00090 |
| Sex |
|
Male | 68.0 (37.7) | 26.0 (38.2) 42.0
(37.5) | 42.0 (61.7) | 98.0 (44.5) | 58.0 (59.2) | 40.0 (40.8) | 2.30
(1.20-4.40) |
0.01000a |
|
Female | 112.0 (62.3) | | 70.0 (62.5) | 122.0 (55.5) | 62.0 (50.8) | 60.0 (49.2) | 1.70
(1.00-2.90) | 0.06000 |
| Age, years | | | | | | | | |
|
<55 | 131.0 (72.8) | 48.0 (36.6) | 83.0 (63.4) | 158.0 (71.8) | 70.0 (44.3) | 88.0 (55.7) | 1.40
(0.80-2.20) | 0.20000 |
|
≥55 | 409.0 (27.2) | 20.0 (40.8) | 29.0 (59.2) | 62.0 (28.2) | 50.0 (80.6) | 12.0 (19.3) | 6.00
(2.60-14.10) |
0.00002a |
| Dwelling | | | | | | | | |
|
Rural | 120.0 (66.6) | 45.0 (37.5) | 75.0 (62.5) | 138.0 (62.7) | 80.0 (57.9) | 58.0 (42.1) | 2.30
(1.40-3.80) |
0.00100a |
|
Urban | 60.0 (33.4) | 23.0 (38.3) | 37.0 (61.6) | 82.0 (37.2) | 40.0 (48.8) | 42.0 (51.2) | 1.40
(0.70-2.70) | 0.30000 |
| Smoking status | | | | | | | | |
|
Non-smoker | 124.0 (68.8) | 48.0 (38.7) | 76.0 (61.3) | 149.00 (67.7) | 82.0 (55.1) | 67.0 (44.9) | 1.90
(1.20-3.10) |
0.00700a |
|
Smoker | 56.0 (31.2) | 20.0 (35.7) | 36.0 (64.3) | 71.00 (32.3) | 38.00 (53.5) | 33.0 (46.5) | 2.10
(1.00-4.20) | 0.05000 |
| BTD | | | | | | | | |
|
No | 75.0 (41.6) | 18.0 (24.0) | 57.0 (76.0) | | | | 2.90
(1.50-5.50) |
0.00200a |
|
Yes | 105.0 (58.3) | 50.0 (47.6) | 55.0 (52.3) | | | | | |
| Histological
type | | | | | | | | |
|
PTC | 151.0 (83.9) | 60.0 (45.7) | 91.0 (60.3) | | | | 0.60
(0.20-1.40) | 0.29000 |
|
FTC | 29.0 (16.1) | 8.0 (27.6) | 21.0 (72.4) | | | | | |
| TSH levels | | | | | | | | |
|
Normal | 52.0 (28.8) | 17.0 (32.7) | 35.0 (67.3) | | | | 1.40
(0.70-2.70) | 0.40000 |
|
High | 128.0 (71.1) | 51.0 (39.8) | 77.0 (60.1) | | | | | |
| Tumor grade | | | | | | | | |
|
WD | 176.0(97.7) | 66.0 (37.5) | 110.0 (62.5) | | | | 1.7.0
(0.20-12.10) | 100.000 |
|
PD | 4.0 (2.2) | 2.0 (50.0) | 2.0 (50.0) | | | | | |
| Tumor focality | | | | | | | | |
|
Unifocal | 96.0 (53.3) | 43.0 (44.8) | 53.0 (55.2) | | | | 0.50
(0.30-0.97) |
0.04000a |
|
Multifocal | 84.0 (46.7) | 25.0 (29.8) | 59.0 (70.2) | | | | | |
| Stage <55
years | | | | | | | | |
|
I | 65.0 (36.1) | 27.0 (41.5) | 38.0 (58.5) | | | | | |
|
II | 66.0 (36.7) | 21.0 (31.8) | 45.0 (68.1) | | | | 0.60
(0.30-1.30) | 0.30000 |
| Stage ≥55
years | | | | | | | | |
|
I+II | 26.0 (14.4) | 16.0 (61.5) | 10.0 (38.4) | | | | 0.10
(0.03-0.50) | 0.03000 |
|
≥III | 23.0 (12.8) | 4.0 (17.4) | 19.0 (82.6) | | | | | |
| V/C invasion | | | | | | | | |
|
No | 93.0 (51.7) | 43.0 (46.2) | 50.0 (53.7) | | | | 0.50
(0.30-0.90) | 0.02000 |
|
Yes | 87.0 (48.3) | 25.0 (28.7) | 62.0 (71.2) | | | | | |
| LN metastasis | | | | | | | | |
|
No | 110.0 (61.0) | 57.0 (51.8) | 53.0 (48.2) | | | | 0.20
(0.10-0.40) |
0.00001a |
|
Yes | 70.0 (38.9) | 11.0 (15.7) | 59.0 (84.3) | | | | | |
Genetic association study of HRAS T81C
polymorphism
Various inheritance models were applied to asses the
inhertence pattern of polymorphism. A significantly higher
frequency of of variant genotype (TC+CC) was observed in DTC cases
as compared with controls (62.27 vs. 45.4; P=0.0009) indicating the
ominant mode of inheritance. Table
VII depicts the results of the association study for HRAS
T81C SNP.
 | Table VIIGenetic association study of HRAS
T81C polymorphism. |
Table VII
Genetic association study of HRAS
T81C polymorphism.
| Model | Genotype | Cases | Controls | OR (95% CI) | P-value |
|---|
| Co-dominant | T/T | 68.0 (37.8) | 120.0 (54.5) | 1.0 (Ref) | |
| | T/C | 83.0 (46.1) | 75.0 (34.1) | 1.9 (1.3-3.0) | 0.0020 |
| | C/C | 29.0 (16.1) | 25.0 (11.4) | 2.0 (1.1-3.8) | 0.0300 |
| Dominant | T/T | 68.0 (37.8) | 120.0 (54.5) | 1.0 (Ref) | |
| | T/C + C/C | 112.0 (62.2) | 100.0 (45.4) | 2.0 (1.3-3.0) | 0.0009 |
| Recessive | T/T + T/C | 151.0 (83.9) | 195.0 (88.6) | 1.0 (Ref) | |
| | C/C | 29.0 (16.1) | 25.0 (11.4) | 1.5 (0.8-2.7) | 0.2000 |
| Over-dominant | T/T + C/C | 97.0 (53.9) | 145.0 (65.9) | 1.0 (Ref) | |
| | T/C | 83.0 (46.1) | 75.0 (34.1) | 1.6 (1.1-2.5) | 0.0200 |
| Additive | T/T | 68.0 (37.8) | 120.0 (54.5) | 1.0 (Ref) | |
| | C/C | 29.0 (16.1) | 25.0 (11.4) | 2.0 (1.1-3.8) | 0.0300 |
Patient follow-up
The patients were followed until the end of
radioiodine therapy (data not shown). In patients with DTC lacking
BRAFV600E mutation, low doses of I-131 (2.5-3
mCi) were given and patients responded well with high uptake. In
addition, patients with DTC with BRAFV600E
mutation exhibited decreased uptake of I-131 at low doses;
therefore high doses of I-131 were given (75-80 mCi) for proper
uptake and subsequent response to radio-iodine therapy.
Discussion
The MAP kinase pathway serves as a signal transducer
between the extracellular environment and the nucleus (26). Extracellular signals, such as
hormones and growth factors, interact with RET to activate small
G-proteins of the RAS family, which activate and recruit RAF
protein to the cell membrane where it is activated (27). Active BRAF signals via MEK to
activate ERK, which activates downstream transcription factors to
induce cell differentiation, proliferation, growth and apoptosis
(28).
Triggering kinase activity makes BRAF a potent
activator of MEK. BRAFV600E mutation increases
the kinase activity of BRAF by nearly 700-fold, thereby stimulating
constitutive activation of MEK/ERK signaling in tumor cells in the
absence of extracellular stimuli, allowing the cell to become
self-sufficient in growth signals within this pathway (28). Here, BRAFV600E
mutation was found in 28% of patients with DTC.
BRAFV600E mutation has been detected in 40-70% of
malignant melanoma, 45-55% of DTC and 10% of colorectal cancer
(29). In addition the
BRAFV600E mutation has also been identified in
ovarian, breast and lung cancer (9,30,31).
Clinically, FPTC metastasizes to cervical lymph nodes less
frequently than CPTC but has a similar survival rate (29,32).
In the present study, the BRAFV600E mutation was
present in 44.4% of CPTC cases but absent in FPTC cases. The
prevalence of BRAFV600E mutation in CPTC is
50-60% (9,10,33).
Studies have shown that FPTC molecular profile may be different
from that of CPTC (34,35). Additionally, it has been reported
that FPTC has lower BRAF but higher RAS mutation rate
compared with CPTC (36). The
present study demonstrated an association between
BRAFV600E mutation and multifocality and LN
metastasis in DTC. Accumulating data have shown that
BRAFV600E mutation is associated with
unfavourable clinicopathological characteristics, such as
extrathyroidal extension, LN metastasis, recurrence and advanced
disease stage in DTC (37,38). BRAFV600E is
associated with silencing of multiple thyroid-specific
iodine-metabolizing genes such as sodium/iodide symporter and
apical iodide transporter, responsible for transportation of
inorganic iodine into thyroid cells (39,40).
Consequently, tumors harbouring the mutation are, to an extent,
resistant to radio iodine abalation used for the treatment of TC,
which may explain the more aggressive phenotype exhibited by DTC
harboring BRAFV600E mutation (41).
RAS is the most commonly mutated gene family
in TC that contributes to cancer initiation and progression via
inhibition of GTP hydrolysis by diminishing GTPase activity
(42). Gain of function mutations
in the hotspot regions of the RAS gene family affect codons
12 and 13 in exon 1 and codon 61 in exon 2. No mutation in any of
the RAS genes was found in the present study, which supports
previous studies indicating that mutations in BRAF and
RAS generally occur in a mutually exclusive manner (42,43).
Mutual exclusiveness suggests that MAP kinase pathway is controlled
at different levels to regulate TC pathogenesis. Certain studies
observed an increase in RAS mutation in dietary
iodine-deficient countries, such as eastern Hungary and Japan
(44,16) whereas, Vuong et al (45) reported no difference in frequency
of RAS mutations between iodine-rich and -deficient
countries.
To the best of our knowledge, the present study is
the first multicentric study from Pakistan exploring the utility of
HRAS T81C SNP analysis in TC risk, which may help in future
treatment modalities. HRAS T81C SNP was found to be a strong
risk factor for TC (22,24). The HRAS T81C variant (TC +
CC) and heterozygous genotype (TC) were found in 62.2 and 46.1% of
patients with TC compared with 45.4 and 34.1% of controls,
respectively; these rates are higher in comparison with other
ethnic groups (21,46). The reason for the higher frequency
of variant genotype in DTC cases compared withcontrols may be
attributed to geographical differences and relatively small sample
size (21). In addition, HRAS
T81C was significantly associated with the risk of DTC in the
present study (P=0.0009). Earlier studies have also reported
significant association of HRAS T81C SNP with risk of
gastric, colon and bladder cancer (47,48),
although, the frequency of variant genotype reported by those
studies was relatively less compared with the present study
Following stratification of HRAS T81C
genotypes with clinicopathological risk factors in patients with
DTC, TC and CC variants have been significantly associated with
higher age, which is in line with previous studies that identified
higher age as a key risk factor for tumorogenesis in relation to
HRAS T81C gene polymorphism (48,49).
However, an earlier study demonstrated no significant association
of age with risk of thyroid cancer in relation to HRAS T81C
genotypes (22). Males and rural
dwellers with DTC exhibited greater frequency of TC + CC compared
with control males and rural dwellers, which differs from earlier
studies (22,47). The present results demonstrated
that variant genotype (TC + CC) was inversely associated to smoking
status. Ciggarete smoking may lower endogenous TSH levels in the
body and hence lower the risk of TC (50). These results were different to
earlier studies, in which no association of TSH level with smoking
was found (51,52). Patients with DTC with no history of
BTD had greater frequency of TC + CC genotype. Khan et al
(22) found no association between
HRAS T81C genotype and history of BTD. As BTD is a risk
factor and molecular crosstalk occurs during the initiation and
progression of cancer, there may be other molecular changes
responsible for the development of BTD, which may serve a role in
the development of cancer phenotype. Although previous history of
BTD was recorded, the present study included histologically
confirmed patients with DTC rather than patients with any BTD.
Investigation of MAP kinase pathway aberrations in Pakistani
individuals with BTD will improve understanding of the
etiopathogenesis of TC in this region. In the present study,
HRAS T81C variant genotype was associated with multifocality
and LN metastasis (P≤0.05). An earlier studies demonstrated no
significant association of variant genotype with multifocality and
LN metastasis (53). The present
study did not observe any association between histological type,
tumor grade, TSH levels, V/C invasion and tumor stage with HRAS
T81C TC + CC genotype. However, Krishna et al (54) showed high expression of HRAS
protein in WDTC and higher stage. A previous study has reportedno
association between HRAS T81C variants and histological
types of TC (55).
Although the mechanism underlying the role of
HRAS T81C SNP in cancer initiation is not completely known,
this SNP may not be involved in delaying GTP-bound activated state
(56) and alteration of the
RAS protein structure (24), but rather affect cancer
susceptibility via linkage with other polymorphic sites in
functional intron regions of HRAS (57,58).
HRAS T81C exon 1 may be linked to rs112587690 SNP in intron
1 and L-myc rs3134613 SNP in the development of cutaneous melanoma
and colorectal cancer, respectively (57,58).
The polymorphism may also be linked to a candidate region with
variable tandem repeat present downstream of exon 4, exhibiting
possible transcriptional enhancer activity (59). Furthermore, reports have shown the
association of HRAS T81C SNP with hexanucleotide region
present 80 bp upstream of 5' exon 1 (55,60,61).
Additionally, the HRAS T81C SNP is also associated with
polymorphic intron D2 (dopamine) region of HRAS that may
serve as a regulator of Intron D Exon inclusion (24). HRAS T81C polymorphism
follows a dominant mode of inheritance, which assumes that carriers
of wild genotypes are associated with lower cancer risk compared
with heterozygous and rare genotypes (62).
As the sample size of the present study was modest,
further studies with larger sample size and follow-up of patients
are required to authenticate the association to better distinguish
racial and ethnic differences affecting the pathogenesis and
severity of DTC.
In summary, BRAFV600E mutation
may be implicated in the pathogenesis of DTC in a mutual exclusive
manner with RAS mutations in Kashmiri population.
BRAFV600E mutation was confined to CPTC variant
and was significantly associated with multifocality and LN
metastasis, suggesting that BRAFV600E mutation
may be useful for evaluation of prognosis of patients with DTC.
These results indicated that BRAF may be a promising target
for pharmacological intervention in DTC. HRAS T81C varant
genotype was increased in DTC with dominant pattern of inheritence.
HRAS T81C variant genotype increased risk of DTC with no
history of smoking, males, higher age, multifocality and LN
metatasis. Further analysis of other genetic markers and long-term
clinical follow-up may improve understanding of DTC.
Acknowledgements
Not applicable.
Funding
Funding: The present study was funded by International Islamic
University, Islamabad (grant no. NRPU 8038).
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
FAR, GHB and MSK conceptualized the study,
collected data and wrote the manuscript. GHB and MSK analyzed the
data and reviewed and edited the manuscript. FAR and ST performed
the experiments. ST supervized the study and provided resources.
FAR and ST visualized the data. MSK and GHB confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Review Board of PIMS (approval no.F.1-1/2015/ERB/SZABMU). All
samples were collected with written informed consent from patients
and proper ethical procedures were followed.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sung H, Ferlay J, Siegel RL, Laversanne M,
Soerjomataram I, Jemal A and Bray F: Global Cancer Statistics 2020:
GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36
Cancers in 185 Countries. CA Cancer J Clin. 71:209–249.
2021.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Morris LG, Tuttle RM and Davies L:
Changing trends in the incidence of thyroid cancer in the United
States. JAMA Otolaryngol Head Neck Surg. 142:709–711.
2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Nikiforov YE, Seethala RR, Tallini G,
Baloch ZW, Basolo F, Thompson LD, Barletta JA, Wenig BM, Al Ghuzlan
A, Kakudo K, et al: Nomenclature revision for encapsulated
follicular variant of papillary thyroid carcinoma: A paradigm shift
to reduce overtreatment of indolent tumors. JAMA Oncol.
2:1023–1029. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Laha D, Nilubol N and Boufraqech M: New
Therapies for Advanced Thyroid Cancer. Front Endocrinol (Lausanne).
11(82)2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Nikiforov YE: Thyroid carcinoma: Molecular
pathways and therapeutic targets. Mod Pathol. 21 (Suppl 2):S37–S43.
2008.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Dvorak K, Aggeler B, Palting J, McKelvie
P, Ruszkiewicz A and Waring P: Immunohistochemistry with the
anti-BRAF V600E (VE1) antibody: Impact of pre-analytical conditions
and concordance with DNA sequencing in colorectal and papillary
thyroid carcinoma. Pathology. 46:509–517. 2014.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Wan PT, Garnett MJ, Roe SM, Lee S,
Niculescu-Duvaz D, Good VM, Jones CM, Marshall CJ, Springer CJ,
Barford D, et al: Cancer Genome Project: Mechanism of activation of
the RAF-ERK signaling pathway by oncogenic mutations of B-RAF.
Cell. 116:855–867. 2004.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Oh HS, Kwon H, Park S, Kim M, Jeon MJ, Kim
TY, Shong YK, Kim WB, Choi J, Kim WG, et al: Comparison of
Immunohistochemistry and Direct Sanger Sequencing for Detection of
the BRAF(V600E) Mutation in Thyroid Neoplasm. Endocrinol Metab
(Seoul). 33:62–69. 2018.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xing M: BRAF mutation in thyroid cancer.
Endocr Relat Cancer. 12:245–262. 2005.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kim TY, Kim WB, Rhee YS, Song JY, Kim JM,
Gong G, Lee S, Kim SY, Kim SC, Hong SJ, et al: The BRAF mutation is
useful for prediction of clinical recurrence in low-risk patients
with conventional papillary thyroid carcinoma. Clin Endocrinol
(Oxf). 65:364–368. 2006.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kondo T, Ezzat S and Asa SL: Pathogenetic
mechanisms in thyroid follicular-cell neoplasia. Nat Rev Cancer.
6:292–306. 2006.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Mo SP, Coulson JM and Prior IA: RAS
variant signalling. Biochem Soc Trans. 46:1325–1332.
2018.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Takai Y, Sasaki T and Matozaki T: Small
GTP-binding proteins. Physiol Rev. 81:153–208. 2001.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Castellano E and Santos E: Functional
specificity of ras isoforms: So similar but so different. Genes
Cancer. 2:216–231. 2011.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chakladar J, Li WT, Bouvet M, Chang EY,
Wang-Rodriguez J and Ongkeko WM: Papillary thyroid carcinoma
variants are characterized by co-dysregulation of immune and cancer
associated genes. Cancers (Basel). 11(1179)2019.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Motoi N, Sakamoto A, Yamochi T, Horiuchi
H, Motoi T and Machinami R: Role of ras mutation in the progression
of thyroid carcinoma of follicular epithelial origin. Pathol Res
Pract. 196:1–7. 2000.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Garcia-Rostan G, Zhao H, Camp RL, Pollan
M, Herrero A, Pardo J, Wu R, Carcangiu ML, Costa J and Tallini G:
ras mutations are associated with aggressive tumor phenotypes and
poor prognosis in thyroid cancer. J Clin Oncol. 21:3226–3235.
2003.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ezzat S, Zheng L, Kolenda J, Safarian A,
Freeman JL and Asa SL: Prevalence of activating ras mutations in
morphologically characterized thyroid nodules. Thyroid. 6:409–416.
1996.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zhu Z, Gandhi M, Nikiforova MN, Fischer AH
and Nikiforov YE: Molecular profile and clinical-pathologic
features of the follicular variant of papillary thyroid carcinoma.
An unusually high prevalence of ras mutations. Am J Clin Pathol.
120:71–77. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hara H, Fulton N, Yashiro T, Ito K,
DeGroot LJ and Kaplan EL: N-ras mutation: An independent prognostic
factor for aggressiveness of papillary thyroid carcinoma. Surgery.
116:1010–1016. 1994.PubMed/NCBI
|
|
21
|
Howell GM, Hodak SP and Yip L: RAS
mutations in thyroid cancer. Oncologist. 18:926–932.
2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Khan MS, Pandith AA, Ul Hussain M, Iqbal
M, Khan NP, Wani KA, Masoodi SR and Mudassar S: Lack of mutational
events of RAS genes in sporadic thyroid cancer but high risk
associated with HRAS T81C single nucleotide polymorphism
(case-control study). Tumour Biol. 34:521–529. 2013.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Wang Y, Gao W, Guo Y and Peng C: The role
of HRAS rs12628 polymorphism in cancer risks: Evidence from a
meta-analysis of 19 case-control studies. Int J Clin Exp Med.
10:2386–2396. 2017.
|
|
24
|
Castro P, Soares P, Gusmão L, Seruca R and
Sobrinho-Simões M: H-RAS 81 polymorphism is significantly
associated with aneuploidy in follicular tumors of the thyroid.
Oncogene. 25:4620–4627. 2006.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Khan MS, Pandith AA, Azad N, Hussain MU,
Masoodi SR, Wani KA, Andrabi KI and Mudassar S: Impact of molecular
alterations of BRAF in the pathogenesis of thyroid cancer.
Mutagenesis. 29:131–137. 2014.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Morrison DK: MAP kinase pathways. Cold
Spring Harb Perspect Biol. 4(a011254)2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Garnett MJ and Marais R: Guilty as
charged: B-RAF is a human oncogene. Cancer Cell. 6:313–319.
2004.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Cantwell-Dorris ER, O'Leary JJ and Sheils
OM: BRAFV600E: Implications for carcinogenesis and molecular
therapy. Mol Cancer Ther. 10:385–394. 2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Pakneshan S, Salajegheh A, Smith RA and
Lam AK: Clinicopathological relevance of BRAF mutations in human
cancer. Pathology. 45:346–356. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Davies H, Bignell GR, Cox C, Stephens P,
Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W,
et al: Mutations of the BRAF gene in human cancer. Nature.
417:949–954. 2002.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Sahin IH and Klostergaard J: BRAF
mutations as actionable targets: A paradigm shift in the management
of colorectal cancer and novel avenues. JCO Oncol Pract: Jun 2,
2021 (Epub ahead of print). doi: 10.1200/OP.21.00160.
|
|
32
|
Kebebew E and Clark OH: Differentiated
thyroid cancer: ‘complete’ rational approach. World J Surg.
24:942–951. 2000.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Kim J, Giuliano AE, Turner RR, Gaffney RE,
Umetani N, Kitago M, Elashoff D and Hoon DS: Lymphatic mapping
establishes the role of BRAF gene mutation in papillary thyroid
carcinoma. Ann Surg. 244:799–804. 2006.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Lupi C, Giannini R, Ugolini C, Proietti A,
Berti P, Minuto M, Materazzi G, Elisei R, Santoro M, Miccoli P, et
al: Association of BRAF V600E mutation with poor
clinicopathological outcomes in 500 consecutive cases of papillary
thyroid carcinoma. J Clin Endocrinol Metab. 92:4085–4090.
2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Abdullah MI, Junit SM, Ng KL, Jayapalan
JJ, Karikalan B and Hashim OH: Papillary Thyroid Cancer: Genetic
Alterations and Molecular Biomarker Investigations. Int J Med Sci.
16:450–460. 2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Adeniran AJ, Zhu Z, Gandhi M, Steward DL,
Fidler JP, Giordano TJ, Biddinger PW and Nikiforov YE: Correlation
between genetic alterations and microscopic features, clinical
manifestations, and prognostic characteristics of thyroid papillary
carcinomas. Am J Surg Pathol. 30:216–222. 2006.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Xing M, Westra WH, Tufano RP, Cohen Y,
Rosenbaum E, Rhoden KJ, Carson KA, Vasko V, Larin A, Tallini G, et
al: BRAF mutation predicts a poorer clinical prognosis for
papillary thyroid cancer. J Clin Endocrinol Metab. 90:6373–6379.
2005.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kebebew E, Weng J, Bauer J, Ranvier G,
Clark OH, Duh QY, Shibru D, Bastian B and Griffin A: The prevalence
and prognostic value of BRAF mutation in thyroid cancer. Ann Surg.
246:466–471. 2007.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Liu D, Hu S, Hou P, Jiang D, Condouris S
and Xing M: Suppression of BRAF/MEK/MAP kinase pathway restores
expression of iodide-metabolizing genes in thyroid cells expressing
the V600E BRAF mutant. Clin Cancer Res. 13:1341–1349.
2007.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Makboul R, Mostafa NM, El-Deek HEM,
Aboulhagag NA, Shehata MR and Abdelhafez YG: Do BRAFV600E mutation
and sodium-iodide symporter expression affect the response to
radioactive iodine therapy in patients with papillary thyroid
carcinoma? Nucl Med Commun. 41:416–425. 2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Durante C, Puxeddu E, Ferretti E, Morisi
R, Moretti S, Bruno R, Barbi F, Avenia N, Scipioni A, Verrienti A,
et al: BRAF mutations in papillary thyroid carcinomas inhibit genes
involved in iodine metabolism. J Clin Endocrinol Metab.
92:2840–2843. 2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li ZN, Zhao L, Yu LF and Wei MJ: BRAF and
KRAS mutations in metastatic colorectal cancer: Future perspectives
for personalized therapy. Gastroenterol Rep (Oxf). 8:192–205.
2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Oikonomou E, Koustas E, Goulielmaki M and
Pintzas A: BRAF vs. RAS oncogenes: Are mutations of the same
pathway equal? Differential signalling and therapeutic
implications. Oncotarget. 5:11752–11777. 2014.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Shi YF, Zou MJ, Schmidt H, Juhasz F,
Stensky V, Robb D and Farid NR: High rates of ras codon 61 mutation
in thyroid tumors in an iodide-deficient area. Cancer Res.
51:2690–2693. 1991.PubMed/NCBI
|
|
45
|
Vuong HG, Kondo T, Oishi N, Nakazawa T,
Mochizuki K, Inoue T, Tahara I, Kasai K, Hirokawa M, Tran TM, et
al: Genetic alterations of differentiated thyroid carcinoma in
iodine-rich and iodine-deficient countries. Cancer Med.
5:1883–1889. 2016.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Jung CK, Kim Y, Jeon S, Jo K, Lee S and
Bae JS: Clinical utility of EZH1 mutations in the diagnosis of
follicular-patterned thyroid tumors. Hum Pathol. 81:9–17.
2018.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Zhang Y, Jin M, Liu B, Ma X, Yao K, Li Q
and Chen K: Association between H-RAS T81C genetic polymorphism and
gastrointestinal cancer risk: A population based case-control study
in China. BMC Cancer. 8(256)2008.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Pandith AA, Shah ZA, Khan NP, Baba KM,
Wani MS and Siddiqi MA: HRAS T81C polymorphism modulates risk of
urinary bladder cancer and predicts advanced tumors in ethnic
Kashmiri population. Urol Oncol. 31:487–492. 2013.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Rostami M, Kalaei Z, Pourhoseingholi MA
and Kadivar M: Study on association between H-ras gene polymorphism
and gastric adenocarcinoma risk. Gastroenterol Hepatol Bed Bench.
6:146–151. 2013.PubMed/NCBI
|
|
50
|
Cho A, Chang Y, Ahn J, Shin H and Ryu S:
Cigarette smoking and thyroid cancer risk: A cohort study. Br J
Cancer. 119:638–645. 2018.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Jee SH, Samet JM, Ohrr H, Kim JH and Kim
IS: Smoking and cancer risk in Korean men and women. Cancer Causes
Control. 15:341–348. 2004.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Navarro Silvera SA, Miller AB and Rohan
TE: Risk factors for thyroid cancer: A prospective cohort study.
Int J Cancer. 116:433–438. 2005.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Dou R, Zhang L, Lu T, Liu D, Mei F, Huang
J and Qian L: Identification of a novel HRAS variant and its
association with papillary thyroid carcinoma. Oncol Lett.
15:4511–4516. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Krishna A, Singh S, Singh V, Kumar V,
Singh US and Sankhwar SN: Does Harvey-Ras gene expression lead to
oral squamous cell carcinoma? A clinicopathological aspect. J Oral
Maxillofac Pathol. 22:65–72. 2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Suarez HG, du Villard JA, Severino M,
Caillou B, Schlumberger M, Tubiana M, Parmentier C and Monier R:
Presence of mutations in all three ras genes in human thyroid
tumors. Oncogene. 5:565–570. 1990.PubMed/NCBI
|
|
56
|
Lowy DR and Willumsen BM: Function and
regulation of ras. Annu Rev Biochem. 62:851–891. 1993.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Tomei S, Adams S, Uccellini L, Bedognetti
D, De Giorgi V, Erdenebileg N, Ascierto ML, Reinboth J, Liu Q,
Bevilacqua G, et al: Association between HRAS rs12628 and
rs112587690 polymorphisms with the risk of melanoma in the North
American population. Med Oncol. 29:3456–3461. 2012.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Ni Q, Zhang YJ, Zhang SC, Jin MJ, Ma XY,
Yao KY, Li QL and Chen K: Association between H-ras and L-myc gene
polymorphisms and susceptibility to colorectal cancer. Zhonghua
Zhong Liu Za Zhi. 34:15–20. 2012.PubMed/NCBI(In Chinese).
|
|
59
|
Trepicchio WL and Krontiris TG: Members of
the rel/NF-kappa B family of transcriptional regulatory proteins
bind the HRAS1 minisatellite DNA sequence. Nucleic Acids Res.
20:2427–2434. 1992.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Kotsinas A, Gorgoulis VG, Zacharatos P,
Mariatos G, Kokotas S, Liloglou T, Ikonomopoulos J, Zoumpourlis V,
Kyroudi A, Field JK, et al: Additional characterization of a
hexanucleotide polymorphic site in the first intron of human H-ras
gene: Comparative study of its alterations in non-small cell lung
carcinomas and sporadic invasive breast carcinomas. Cancer Genet
Cytogenet. 126:147–154. 2001.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Sol-Church K, Stabley DL, Nicholson L,
Gonzalez IL and Gripp KW: Paternal bias in parental origin of HRAS
mutations in Costello syndrome. Hum Mutat. 27:736–741.
2006.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Wang Q: Cancer predisposition genes:
Molecular mechanisms and clinical impact on personalized cancer
care: Examples of Lynch and HBOC syndromes. Acta Pharmacol Sin.
37:143–149. 2016.PubMed/NCBI View Article : Google Scholar
|