1. Discovery and biological basis of
GREM1
GREM1 was initially isolated from v-mos transfected
rat fibroblasts by the BLAIR team in 1997 and named DRM
(down-regulated in v-mos-transfected cells) (1). Drm contains an open reading frame
(ORF) that encodes a 184-amino acid cysteine-rich protein with a
calculated relative molecular weight of 20,682. In 2000, the team
mapped the human homolog of DRM/Germlin, which maps to human
chromosome 15. It screened a human cDNA library to obtain a protein
size of 3.3 kb containing a 552 base region, and it is highly
conserved during biological evolution (2,3). DRM
is present on the outer surface of expressing cells and in the
endoplasmic reticulum and Golgi apparatus (4). Like many growth factor proteins, it
is a secreted protein (5). GREM1
antagonizes bone morphogenetic protein (BMP) signaling, regulates
BMP during development, and controls limb and kidney formation
(6,7). There is a single DRM-specific mRNA
expressed in different human tissues, including the brain, ovary,
intestine, and colon. GREM1 is expressed in normal cells and
tissues, including normal neurons, astrocytes, and fibroblasts.
GREM1 was earlier thought that expressed significantly in many
organs of the human body during embryonic development, such as
eyes, bones, lungs, kidneys, etc. With further research, GREM1
expression and distribution have also been found in many organs in
mature animals and adults. GREM1 expression was detected in normal
cells from several patients, but not in established tumor cell
lines from the same patient. These data suggest that the
downregulation of GREM1 is associated with tumor progression. GREM1
was detected both intracellularly and extracellularly, and it was
demonstrated to be a weakly essential protein.
2. Biological function of GREM1
McMahon group identified GREM1, the human homolog of
the rat developmental gene Drm/Gremlin, as a high glucose-triggered
gene in human mesangial cells (8).
GREM1 is an antagonist of Bone morphogenetic proteins (BMPs) and
belongs to the same family as Noggin, Chordin, and twisted
gastrulation-1 (Twsg1). GREM1 is able to graft BMP binding to its
cognate serine/threonine protein kinase receptor on lipid membranes
(9). During BMP signaling, BMPs
are processed by preprotein peptidases and then bind to type I and
II BMP receptors to generate mature dimers. The binding of BMP
homodimers to their cognate receptors results in the
phosphorylation of the type I receptor by the type II receptor in
the GS domain which is glycine and serine residue-rich region on
the type I receptor. Activated BMP receptors then phosphorylate
Smad1/5/8 protein, which dimerizes with Smad4 and accumulates in
the nucleus, mediating changes in BMP-regulated gene expression.
GREM1 plays an antagonistic role by regulating BMP dimer formation
during this process (10,11). In addition, Gremlin overexpression
may regulate both mesangial cell growth and transdifferentiation of
cultured tubular epithelial cells to a more fibroblast-like
phenotype (6). GREM1 can also
induce mesenteric cell proliferation and extracellular matrix
accumulation by enhancing the activation of the ERK1/2 pathway
(12). GREM1 was highly expressed
in proximal convoluted tubular epithelial cells (PTEC, HK-2)
exposed to aristolochic acid and down-regulated BMP-7 and
phosphorylated Smad1/5/8 levels, and restored BMP-7 signaling
activity and attenuated aristolochic acid-induced EMT-related
phenotypic changes after down-regulation of GREM1(13). In mouse renal fibroblast cells,
GREM1 was able to upregulate renal fibroblast and tubular
epithelial cell fibrosis by activating TGF-β. Endogenous GREM1
silencing was also able to downregulate TGF-β-mediated
extracellular matrix protein (ECM) production and
epithelial-mesenchymal transition (EMT) (14).
In addition to its antagonism with BMP, GREM1 has
also shown other new effects in recent years. Grem1 binds to fibrin
and binds to Sit proteins to regulate monocyte chemotaxis
negatively. GREM1 can promote angiogenesis by attaching to vascular
endothelial cytokine receptor (VEGFR2) as a regulatory factor that
regulates and promotes angiogenesis (15). This is due to the binding of Grem1
to heparin and heparan sulfate proteoglycans on the surface of
endothelial cells. Moreover, GREM1-mediated angiogenesis is also
mediated by AVB3 integrin binding and AVB3/VEGFR2 complex. The
identification of Grem1 as a new pro-angiogenic factor is of great
significance in the field of highly vascularized tumors and
endothelial cell biology. In the study of human umbilical cord
blood hematopoietic cells, GREM1 also plays a certain regulatory
role, participating in and regulating BMP2 and BMP4, which is
related to the generation of atherosclerotic plaques. In
GREM1-/- embryonic fibroblasts, the phosphorylation
level of ERK is reduced compared with wild-type, and the activation
of phosphorylated ERK1/2 is an effector downstream of GREM1.
3. Role of GREM1 in tumors
The discovery of GREM1 and cancer was first reported
by Topol in 2000. It was found that the expression of GREM1 was
higher in some normal cells but significantly lower in some cells
after transfection of oncogenes, such as fibroblasts. In further
gene sequencing studies, Topol showed that deletion of the GREM1
gene was associated with metastatic breast cancer and other
metastatic tumors. Suzuki M, by measuring the mRNA expression of
GREM1 found that GREM1 was expressed in some normal human cells,
still expression of GREM1 mRNA in the corresponding cells was
reduced or disappeared due to gene methylation after tumor
occurrence. These studies suggest that early investigators believe
that GREM1 is likely to inhibit tumor development and
progression.
Hong Namkoong and others are against this view
(16). The related research team
found that the mRNA expression of GREM1 was significantly increased
in many human malignant tumors compared with the corresponding
normal non-tumor tissues. His initial research was on cervical
cancer cells and later expanded to kidney, breast, and lung cancer,
all with strikingly similar results (16). Subsequently, a larger and larger
sample size study on the expression level of GREM1 mRNA in various
tumors confirmed identical views. This study involved 774 samples
of various solid tumors from human tissues, such as human lung
cancer, colon cancer, bladder cancer, skin basal cell carcinoma,
breast cancer, melanoma, uterine sarcoma, and 16 other kinds of
tumors (17). GREM1 is widely
expressed in the stroma of various tumors and can promote the
proliferation of tumor cells. It is one of the key factors in
tumorigenesis. Due to the different function of GREM1 in different
tumors, its expression level is also different.
Recent studies have written that GREM1 plays a
critical role in the occurrence and development of many tumors. As
reported by Davis et al (17), GREM1 mutation is an essential cause
of familial colon cancer, and its expression in colon cancer is
significantly increased. Karagiannis found that GREM1 could
regulate the progression of colorectal cancer (18). The possible mechanism is that GREM1
promotes EMT by activating TGFβ1/SMAD signaling pathway (19). Other studies have found that the
expression of GREM1 is closely related to the metastasis of
pancreatic cancer, which is one of the prognostic indicators of
pancreatic cancer (20). It is
believed that GREM1 is closely associated with angiogenesis. Its
association with many other malignancies is also under further
investigation.
Role of GREM1 in breast cancer
Breast cancer is a highly prevalent cancer in women
worldwide, and the incidence of breast cancer in women has
continued to rise slowly in recent years (0.5% per year). Breast
cancer is also a common type in cancer-related deaths (21). Based on the results of multiple
statistical analyses of breast cancer gene expression differences,
GREM1 expression is elevated in breast cancer patients. This
increase is associated with a decreased prognosis in breast cancer
patients (22,23). In estrogen receptor (ER)-negative
tumors, GREM1 knockout suppressed the proliferation of breast
cancer cells and the growth of xenogeneic breast tumors, whereas
overexpression enhanced the viability, growth, and invasiveness of
the cells. Estrogen-associated receptor alpha (ERRα) is an orphan
nuclear hormone receptor that increases GREM1 expression by
interacting with the GREM1 promoter. GREM1 activates EGFR, the
upstream regulator of ERRα, and also enhances the promoter of ESRRA
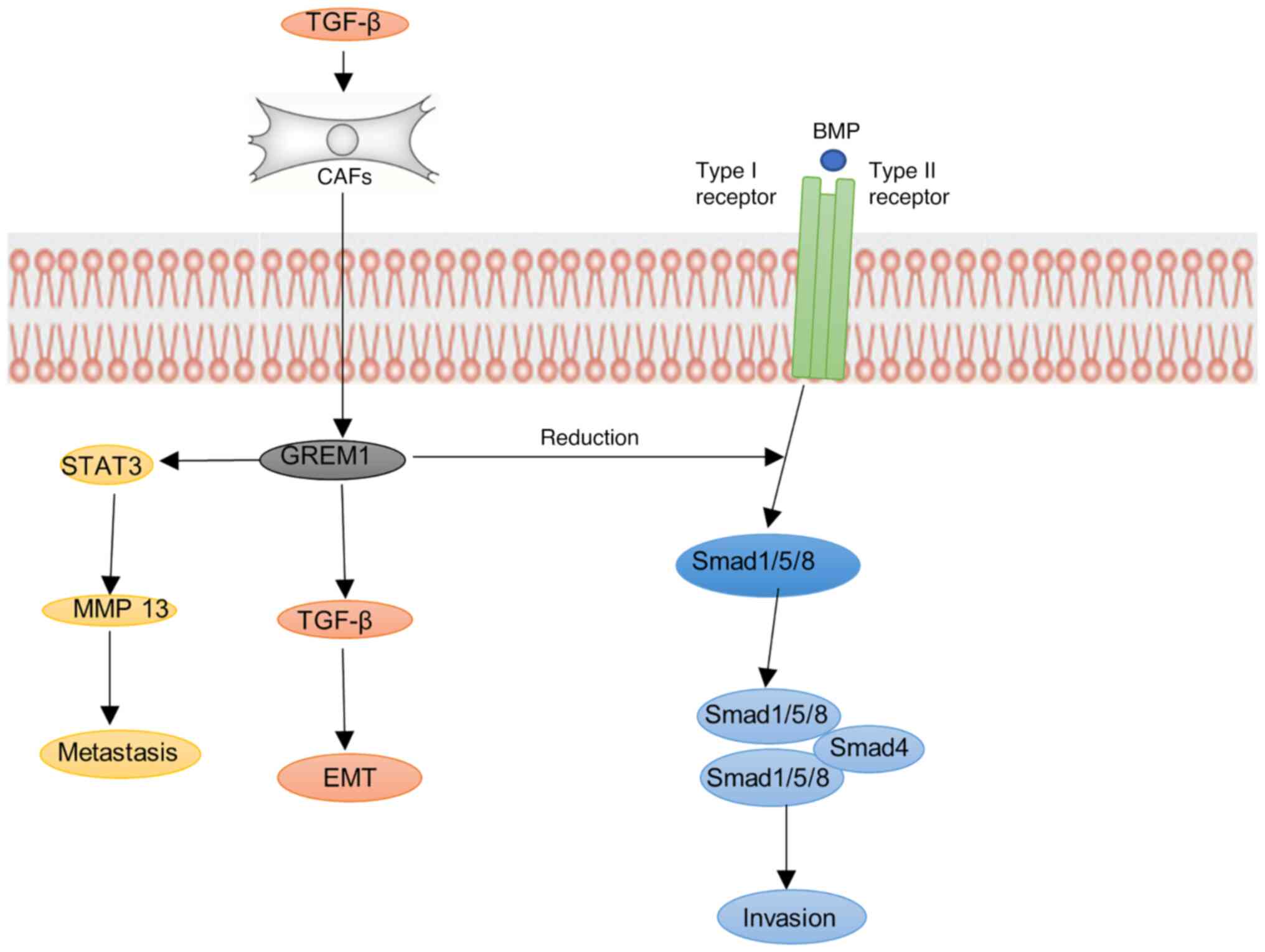
encoding ERRα, thus forming a positive feedback loop (24). In breast cancer-associated
fibroblasts (CAF), TGF-β and inflammatory cytokines can encourage
the increase of CAF-derived GREM1, thereby inhibiting the BMP/SMAD
pathway and increasing the mesenchymal phenotype, stemness and
invasion of tumors, which strongly promotes the fibrosis of CAF to
the internal and external exudation of breast cancer cells
(25). In recent years, the role
of the tumor immune microenvironment has emerged in breast cancer.
It has been shown that GREM1 is highly expressed in the
tumor-associated stroma of breast cancer (26). In molecular profiling of breast
cancer spread and metastasis, GREM1 was shown to be localized in
epithelial cells, and it is also a vital candidate gene associated
with breast cancer invasion (22).
During breast cancer metastasis, GREM1 promotes lung metastasis of
breast cancer cells by increasing matrix metalloproteinase13
(MMP13) expression by activating signal transducer and activator of
transcription3 (STAT3) (27). At
present, the molecular characteristics of clinical diagnosis of
breast cancer are mainly immunohistochemical markers such as ER,
PR, and HER2, proliferation markers such as Ki-67, genomic markers
such as PI3K, and immune markers such as PD-1(28). Based on the study of GREM1 in
breast cancer, it is likely to be a diagnostic marker for breast
cancer (Fig. 1).
Function of GREM1 in colorectal
cancer
Colorectal cancer is the second leading cause of
cancer-related death. In recent years, through the development of
surgery, radiotherapy and chemotherapy, the overall therapeutic
effect of colorectal cancer has also made significant progress. In
particular, the addition of fluorouracil and oxaliplatin to the
treatment regimen also greatly improved the overall survival rate
of patients with stage III colorectal cancer (29). Immune checkpoint inhibitor therapy,
such as pembrolizumab, has been approved as the preferred regimen.
There will also be many ICIs (approved or unapproved) immune
checkpoint-related inhibitors supported by data in the future
(30). Diabetes mellitus, and
hyperglycemia impact the incidence, prognosis and metastasis of
colorectal cancer (31). Increases
in GREM1 are associated with the development of type II diabetes
(32). In stage III colorectal
cancer, GREM1 overexpression is associated with poor prognosis
(33). BMP and TGF-β belong to the
TGFR2 receptor-associated Smad pathway of the same family and are
controlled by GREM1. GREM1 antagonism of BMP signaling can also
regulate the development of intestinal cancer and the growth of
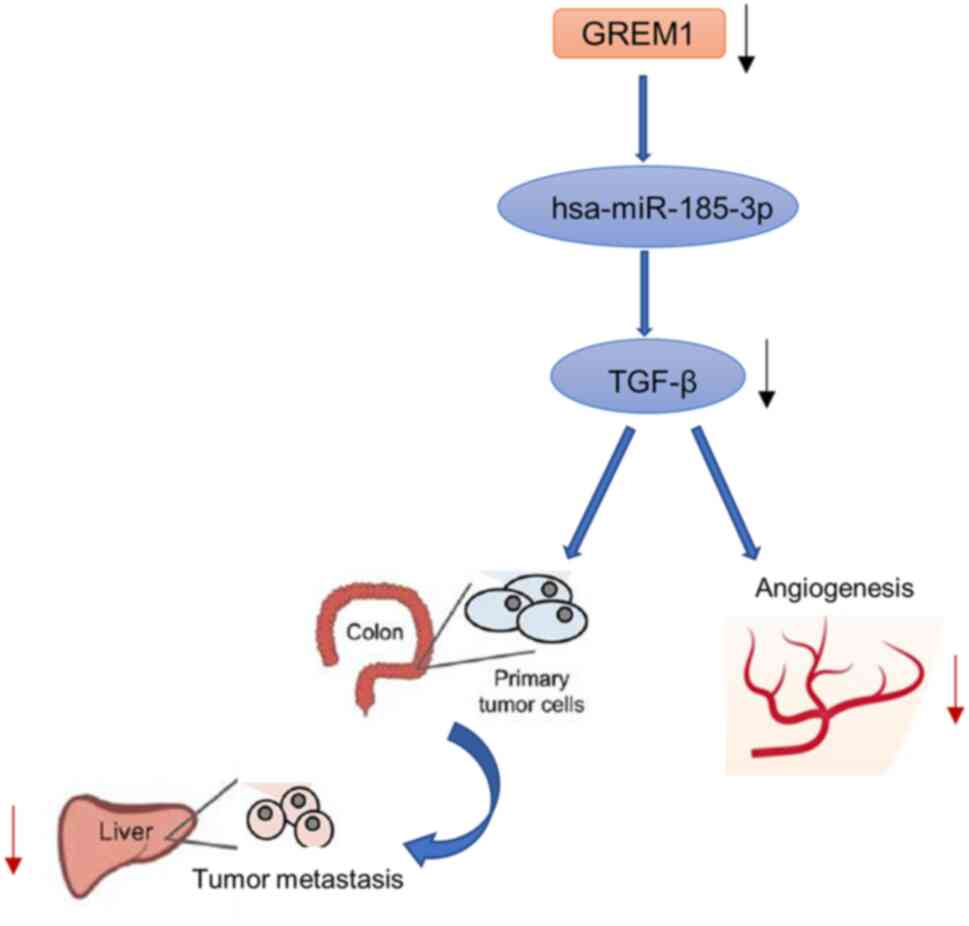
liver metastases (34). GREM1 is
an SNP (single nucleotide polymorphism) near the TGF-β gene
(35). It has been shown that
rs12915554, a low-frequency variant in the TGF-β-associated GREM1
3'UTR, increases GREM1 expression in a post-transcriptional manner
mediated by disrupting the hsa-miR-185-3p binding site, which may
be associated with a high incidence of colon cancer in the Chinese
population (36). In hereditary
mixed polyposis syndrome leading to colorectal cancer tumors, GREM1
is expressed in epithelial cells located in crypts. It is
associated with the development of the disease, and GREM1 can
enhance the increase in Lgr5-positive progenitor and stem cell
characteristics and form ectopic crypts, proliferate, and trigger
colorectal tumors (37). But in
patients with long-standing ulcerative colitis, low GREM1
expression was associated with an increased risk of
colitis-associated cancer (38)
(Fig. 2).
Role of GREM1 in pancreatic
cancer
Pancreatic cancer is highly malignant and its
incidence is increasing worldwide (39). And in this era, when most tumors
can prolong overall survival through various treatments, there is
still no effective treatment for pancreatic cancer that can extend
its overall survival, and patients are found late and inoperable.
With the gradual deepening of researchers' research on GREM1, its
application in pancreatic cancer has gradually increased. Lan et
al (40), showed in a recent
study that GREM1 is a critical regulator of heterogeneity in human
and mouse pancreatic cancer cells, and loss of GREM1 is
particularly significant for the transforming of pancreatic cancer
epithelial cells into mesenchymal cells. Loss or presentation of
GREM1 also plays an essential role in other earlier pancreatic
cancer-related studies. The expression of GREM1 is positively
correlated with high microvessel density in neuroendocrine tumors,
and the increase of high microvessel density is associated with a
good prognosis. At the same time, the loss of GREM1 is also
associated with a poor prognosis in pancreatic neuroendocrine
tumors (19). In terms of
mechanism, it has been shown that deletion of GREM1 can increase
the proliferation and migration of mouse embryonic fibroblasts by
activating the phosphorylation of Smad1/5/8 and decreasing
phosphorylated ERK by BMP4(41).
GREM1 silencing in many tumors is via promoter hypermethylation,
which is also associated with increased Fuhrman grade and decreased
overall survival in tumors and is associated with angiogenesis
(42). In addition, GREM1 is also
a soluble inhibitor of TGF-β, which also favors
epithelial-mesenchymal transition and distal metastasis (43,44).
Elevated GREM1, also increased the expression of
p21Cip1, and decreased the level of p42/44 MAPK, thus
playing a role in inhibiting tumorigenesis (45). However, in studies of chronic
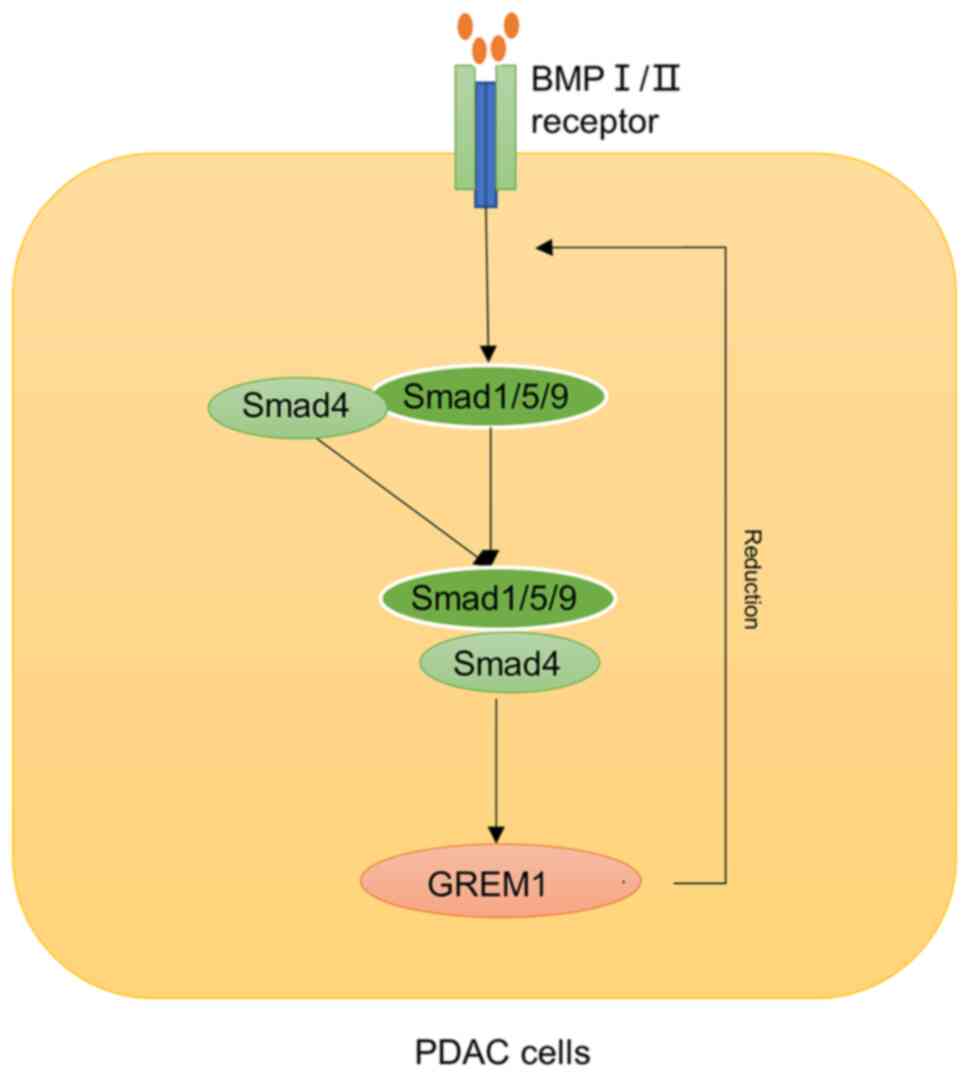
pancreatitis and pancreatic ductal adenocarcinoma (PDAC), it has
been shown that up-regulation of GREM1 in interstitial α-smooth
muscle actin (SMA)-positive fibroblasts in chronic pancreatitis may
promote the development of PDAC, and GREM1 may also play an
important role in macrophage phenotype through its endogenous
counter molecular macrophage migration inhibitory factor (MIF)
(46). GREM1 is also generally
overexpressed in cancer-associated stromal cells. In Sen Yang's
study, elevated GREM1 in serum was also associate with worse
patient prognosis, and loss of GREM1 increased metastasis in
tissues (47). In pancreatic
cancer, the expression level of GREM1 varies in different cell
populations, so whether the treatment regimen can be more
personalized for patients in clinical treatment provides a new idea
for future GREM1 in this type of cancer research and clinical
treatment (Fig. 3).
4. Discussion
GREM1 is an antagonist of BMP and can bind directly
to BMP to inhibit BMP ligand binding to the corresponding receptor
and participate in regulating embryonic kidney and other vital
organ development (33). GREM1
showed various tumor-promoting or tumor-inhibiting effects in
various cells, which were associated with different regulatory
effects of GREM1 on BMP4 and other mitogen-driven cell
proliferation. In different cancers, the expression and role of
GREM1 vary. In studies related to breast cancer and the immune
microenvironment, GREM1 expression is elevated in the relevant
stroma, and high GREM1 expression makes patients have a worse
prognosis and are more likely to metastasize. Increased GREM1
expression is also associated with the development of type II
diabetes, which is associated with poorer prognosis in colon
cancer. However, in pancreatic cancer studies, the expression level
of GREM1 is different from other cancers. Most studies have
confirmed that GREM1 is lowly expressed in pancreatic cancer,
especially in non-stromal cells. However, high expression of GREM1
has also been reported in mechanistic cells of pancreatic cancer,
in addition to increased serum GREM1 in patients with pancreatic
cancer. GREM1 is also an inhibitor of TGF-β, and loss of GREM1
relieves TGF-β regulation of tumors (34). Methylation of the CpG III region of
the GREM1 promoter, which leads to GREM1 silencing, is associated
with enhanced tumor malignancy and also increases active
angiogenesis, which is associated with decreased overall survival
in cancer patients (48). GREM1
binds VEGFR2 to initiate angiogenesis, whereas the knockdown of
GREM1 in MEF cells reduced phospho-ERK and enhanced fibrocyte cell
proliferation and migration by activating Smad1/5/8. GREM1
expression is generally elevated in different tumor
microenvironments and the tumor stroma. However, among diverse
marker cell populations and specific cancer types, loss of GREM1 is
more likely to cause cancers with worse prognoses and more severe
cancer metastasis. This suggests that GREM1 is essential for the
plasticity of tumor cells, and even different ways to maintain its
activity or related pathways activated can cause different final
directions of tumors. In many studies, GREM1 has undoubtedly been
considered by many researchers to be a target for the diagnosis and
treatment of emerging tumors with important clinical translational
significance. In the serum of patients with pancreatic cancer, the
combined diagnosis of GREM1 and CA199 may play an important role in
indicating the prognosis of patients with pancreatic cancer. In
studies of pancreatic cancer single-cell populations,
overexpression of GREM1 even reduced liver metastasis in pancreatic
cancer tumors. In addition, specific antibodies to GREM1 have also
progressed in mouse models of prostate cancer, which undoubtedly
provides new ideas and strong hope for future GREM1-related
research (48).
Acknowledgements
Not applicable.
Funding
Funding: This work was supported by a grant from the Shenyang
Science and Technology Plan Fund Project (grant no. 213463).
Availability of data and materials
Not applicable.
Authors' contributions
DaZ was mainly responsible for writing this
manuscript and collecting related documents. DoZ helped write the
section on breast cancer. NW helped write and modify the colon
cancer section. FC helped write the section on pancreatic cancer
and modified the syntax, and was involved in template drawing. MJ
helped revise the grammar and design of the article, and corrected
the preprint manuscript. ZZ designed the overall structure of the
paper, guided the revision of the paper and provided financial
support. Data authentication is not applicable. All authors have
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Authors' information
ZZ: Director of Department of Medical Oncology,
General Hospital of Northern Theater Command (Shenyang, China) and
engaged in clinical tumor treatment for a long time. DaZ:
Postdoctoral fellow of medical oncology, General Hospital of
Northern Theater Command (Shenyang, China) and specializes in basic
oncology research. DoZ: Postdoctoral fellow of medical oncology,
General Hospital of Northern Theater Command (Shenyang, China) and
specializes in basic oncology research. NW: Postgraduate medical
oncology, General Hospital of Northern Theater Command, Jinzhou
Medical University (Shenyang, China). FC: Postgraduate medical
oncology, Department of Oncology, General Hospital of Northern
Theater Command, China Medical University (Shenyang, China). MJ:
Doctor of medical Oncology, General Hospital of Northern Theater
Command (Shenyang, China).
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Topol LZ, Marx M, Laugier D, Bogdanova NN,
Boubnov NV, Clausen PA, Calothy G and Blair DG: Identification of
drm, a novel gene whose expression is suppressed in transformed
cells and which can inhibit growth of normal but not transformed
cells in culture. Mol Cell Biol. 17:4801–4810. 1997.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Pearce JJ, Penny G and Rossant J: A mouse
cerberus/Dan-related gene family. Dev Biol. 209:98–110.
1999.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Topol LZ, Modi WS, Koochekpour S and Blair
DG: DRM/GREMLIN (CKTSF1B1) maps to human chromosome 15 and is
highly expressed in adult and fetal brain. Cytogenet Genome Res.
89:79–84. 2000.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Topol LZ, Bardot B, Zhang Q, Resau J,
Huillard E, Marx M, Calothy G and Blair DG: Biosynthesis,
Post-translation modification, and functional characterization of
Drm/Gremlin. J Biol Chem. 275:8785–8793. 2000.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hsu DR, Economides AN, Wang X, Eimon PM
and Harland RM: The xenopus dorsalizing factor gremlin identifies a
novel family of secreted proteins that antagonize BMP Activities.
Mol Cell. 1:673–683. 1998.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Zúñiga A, Haramis AP, McMahon AP and
Zeller R: Signal relay by BMP antagonism controls the SHH/FGF4
feedback loop in vertebrate limb buds. Nature. 401:598–602.
1999.PubMed/NCBI View
Article : Google Scholar
|
|
7
|
Michos O, Panman L, Vintersten K, Beier K,
Zeller R and Zuniga A: Gremlin-mediated BMP antagonism induces the
epithelial-mesenchymal feedback signaling controlling metanephric
kidney and limb organogenesis. Development. 131:3401–3410.
2004.PubMed/NCBI View Article : Google Scholar
|
|
8
|
McMahon R, Murphy M, Clarkson M, Taal M,
Mackenzie HS, Godson C, Martin F and Brady HR: IHG-2, a mesangial
cell gene induced by high glucose, is human gremlin. Regulation by
extracellular glucose concentration, cyclic mechanical strain, and
transforming growth factor-beta1. J Biol Chem. 275:9901–9904.
2000.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Brazil DP, Church RH, Surae S, Godson C
and Martin F: BMP signalling: Agony and antagony in the family.
Trends Cell Biol. 25:249–264. 2015.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Kaplan FS, Xu M, Seemann P, Connor JM,
Glaser DL, Carroll L, Delai P, Fastnacht-Urban E, Forman SJ,
Gillessen-Kaesbach G, et al: Classic and atypical fibrodysplasia
ossificans progressiva (FOP) phenotypes are caused by mutations in
the bone morphogenetic protein (BMP) type I receptor ACVR1. Hum
Mutat. 30:379–390. 2009.PubMed/NCBI View Article : Google Scholar
|
|
11
|
López-Rovira T, Chalaux E, Massagué J,
Rosa JL and Ventura F: Direct binding of Smad1 and Smad4 to two
distinct motifs mediates bone morphogenetic protein-specific
transcriptional activation of Id1 gene. J Biol Chem. 277:3176–3185.
2002.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Huang H, Huang H, Li Y, Liu M, Shi Y, Chi
Y and Zhang T: Gremlin induces cell proliferation and extra
cellular matrix accumulation in mouse mesangial cells exposed to
high glucose via the ERK1/2 pathway. BMC Nephrol.
14(33)2013.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Li Y, Wang Z, Wang S, Zhao J, Zhang J and
Huang Y: Gremlin-mediated decrease in bone morphogenetic protein
signaling promotes aristolochic acid-induced
epithelial-to-mesenchymal transition (EMT) in HK-2 cells.
Toxicology. 297:68–75. 2012.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Rodrigues-Diez R, Lavoz C, Carvajal G,
Rayego-Mateos S, Rodrigues Diez RR, Ortiz A, Egido J, Mezzano S and
Ruiz-Ortega M: Gremlin is a downstream profibrotic mediator of
transforming growth factor-beta in cultured renal cells. Nephron
Exp Nephrol. 122:62–74. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Mezzano S, Droguett A, Lavoz C, Krall P,
Egido J and Ruiz-Ortega M: Gremlin and renal diseases: Ready to
jump the fence to clinical utility? Nephrol Dial Transplant.
33:735–741. 2018.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Namkoong H, Shin SM, Kim HK, Ha SA, Cho
GW, Hur SY, Kim TE and Kim JW: The bone morphogenetic protein
antagonist gremlin 1 is overexpressed in human cancers and
interacts with YWHAH protein. BMC Cancer. 6(74)2006.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Davis H, Irshad S, Bansal M, Rafferty H,
Boitsova T, Bardella C, Jaeger E, Lewis A, Freeman-Mills L, Giner
FC, et al: Aberrant epithelial GREM1 expression initiates colonic
tumorigenesis from cells outside the stem cell niche. Nat Med.
21:62–70. 2015.PubMed/NCBI View
Article : Google Scholar
|
|
18
|
Karagiannis GS, Treacy A, Messenger D,
Grin A, Kirsch R, Riddell RH and Diamandis EP: Expression patterns
of bone morphogenetic protein antagonists in colorectal cancer
desmoplastic invasion fronts. Mol Oncol. 8:1240–1252.
2014.PubMed/NCBI View Article : Google Scholar
|
|
19
|
O'Reilly S: Gremlin: A complex molecule
regulating wound healing and fibrosis. Cell Mol Life Sci.
78:7917–7923. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Chen MH, Yeh YC, Shyr YM, Jan YH, Chao Y,
Li CP, Wang SE, Tzeng CH, Chang PM, Liu CY, et al: Expression of
gremlin 1 correlates with increased angiogenesis and
progression-free survival in patients with pancreatic
neuroendocrine tumors. J Gastroenterol. 48:101–108. 2013.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Siegel RL, Miller KD, Fuchs HE and Jemal
A: Cancer statistics, 2022. CA Cancer J Clin. 72:7–33.
2022.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Schuetz CS, Bonin M, Clare SE, Nieselt K,
Sotlar K, Walter M, Fehm T, Solomayer E, Riess O, Wallwiener D, et
al: Progression-specific genes identified by expression profiling
of matched ductal carcinomas in situ and invasive breast tumors,
combining laser capture microdissection and oligonucleotide
microarray analysis. Cancer Res. 66:5278–5286. 2006.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tevaarwerk AJ, Gray RJ, Schneider BP,
Smith ML, Wagner LI, Fetting JH, Davidson N, Goldstein LJ, Miller
KD and Sparano JA: Survival in patients with metastatic recurrent
breast cancer after adjuvant chemotherapy: Little evidence of
improvement over the past 30 years. Cancer. 119:1140–1148.
2013.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Park SA, Sung NJ, Choi BJ, Kim W, Kim SH
and Surh YJ: Gremlin-1 augments the oestrogen-related receptor α
signalling through EGFR activation: Implications for the
progression of breast cancer. Br J Cancer. 123:988–999.
2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Ren J, Smid M, Iaria J, Salvatori DCF, van
Dam H, Zhu HJ, Martens JWM and Ten Dijke P: Cancer-associated
fibroblast-derived Gremlin 1 promotes breast cancer progression.
Breast Cancer Res. 21(109)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ma XJ, Dahiya S, Richardson E, Erlander M
and Sgroi DC: Gene expression profiling of the tumor
microenvironment during breast cancer progression. Breast Cancer
Res. 11(R7)2009.PubMed/NCBI View
Article : Google Scholar
|
|
27
|
Sung NJ, Kim NH, Surh YJ and Park SA:
Gremlin-1 promotes metastasis of breast cancer cells by activating
STAT3-MMP13 signaling pathway. Int J Mol Sci.
21(9227)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Loibl S, Poortmans P, Morrow M, Denkert C
and Curigliano G: Breast cancer. Lancet. 397:1750–1769.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Wolpin BM, Meyerhardt JA, Mamon HJ and
Mayer RJ: Adjuvant treatment of colorectal cancer. CA Cancer J
Clin. 57:168–185. 2007.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Boukouris AE, Theochari M, Stefanou D,
Papalambros A, Felekouras E, Gogas H and Ziogas DC: Latest evidence
on immune checkpoint inhibitors in metastatic colorectal cancer: A
2022 update. Crit Rev Oncol Hematol. 173(103663)2022.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Cheng HC, Chang TK, Su WC, Tsai HL and
Wang JY: Narrative review of the influence of diabetes mellitus and
hyperglycemia on colorectal cancer risk and oncological outcomes.
Transl Oncol. 14(101089)2021.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Ng MC, Shriner D, Chen BH, Li J, Chen WM,
Guo X, Liu J, Bielinski SJ, Yanek LR, Nalls MA, et al:
Meta-analysis of genome-wide association studies in African
Americans provides insights into the genetic architecture of type 2
diabetes. PLoS Genet. 10(e1004517)2014.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Saxena R, Elbers CC, Guo Y, Peter I, Gaunt
TR, Mega JL, Lanktree MB, Tare A, Castillo BA, Li YR, et al:
Large-scale gene-centric meta-analysis across 39 studies identifies
type 2 diabetes loci. Am J Hum Genet. 90:410–425. 2012.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Kobayashi H, Gieniec KA, Wright JA, Wang
T, Asai N, Mizutani Y, Lida T, Ando R, Suzuki N, Lannagan TRM, et
al: The balance of stromal BMP signaling mediated by GREM1 and ISLR
drives colorectal carcinogenesis. Gastroenterology.
160:1224–1239.e30. 2021.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Bellam N and Pasche B: TGF-beta signaling
alterations and colon cancer. Cancer Treat Res. 155:85–103.
2010.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Li J, Liu H, Zou L, Ke J, Zhang Y, Zhu Y,
Yang Y, Gong Y, Tian J, Zou D, et al: A functional variant in GREM1
confers risk for colorectal cancer by disrupting a hsa-miR-185-3p
binding site. Oncotarget. 8:61318–61326. 2017.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Dutton LR, Hoare OP, McCorry AMB, Redmond
KL, Adam NE, Canamara S, Bingham V, Mullan PB, Lawler M, Dunne PD
and Brazil DP: Fibroblast-derived Gremlin1 localises to epithelial
cells at the base of the intestinal crypt. Oncotarget.
10:4630–4639. 2019.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Low END, Mokhtar NM, Wong Z and Raja Ali
RA: Colonic mucosal transcriptomic changes in patients with
long-duration ulcerative colitis revealed colitis-associated cancer
pathways. J Crohns Colitis. 13:755–763. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Park W, Chawla A and O'Reilly EM:
Pancreatic cancer: A review. JAMA. 326:851–862. 2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Lan L, Evan T, Li H, Hussain A, Ruiz EJ,
Zaw Thin M, Ferreira RMM, Ps H, Riising EM, Zen Y, et al: GREM1 is
required to maintain cellular heterogeneity in pancreatic cancer.
Nature. 607:163–168. 2022.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Curran SP, Hickey FB, Watson A, Godson C
and Brazil DP: Deletion of Gremlin1 increases cell proliferation
and migration responses in mouse embryonic fibroblasts. Cell
Signal. 24:889–898. 2012.PubMed/NCBI View Article : Google Scholar
|
|
42
|
van Vlodrop IJ, Baldewijns MM, Smits KM,
Schouten LJ, van Neste L, van Criekinge W, van Poppel H, Lerut E,
Schuebel KE, Ahuja N, et al: Prognostic significance of Gremlin1
(GREM1) promoter CpG island hypermethylation in clear cell renal
cell carcinoma. Am J Pathol. 176:575–584. 2010.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Miao H, Wang N, Shi LX, Wang Z and Song
WB: Overexpression of mircoRNA-137 inhibits cervical cancer cell
invasion, migration and epithelial-mesenchymal transition by
suppressing the TGF-β/smad pathway via binding to GREM1. Cancer
Cell Int. 19(147)2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Hong SB, Oh H, Valera VA, Stull J, Ngo DT,
Baba M, Merino MJ, Linehan WM and Schmidt LS: Tumor suppressor FLCN
inhibits tumorigenesis of a FLCN-null renal cancer cell line and
regulates expression of key molecules in TGF-beta signaling. Mol
Cancer. 9(160)2010.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Chen B, Athanasiou M, Gu Q and Blair DG:
Drm/Gremlin transcriptionally activates p21(Cip1) via a novel
mechanism and inhibits neoplastic transformation. Biochem Biophys
Res Commun. 295:1135–1141. 2002.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Davis JM, Cheng B, Drake MM, Yu Q, Yang B,
Li J, Liu C, Younes M, Zhao X, Bailey JM, et al: Pancreatic stromal
Gremlin 1 expression during pancreatic tumorigenesis. Genes Dis.
9:108–115. 2020.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Yang S, Zhang Y, Hua Y, Cui M, Wang M, Gao
J, Liu Q and Liao Q: GREM1 is a novel serum diagnostic marker and
potential therapeutic target for pancreatic ductal adenocarcinoma.
Front Oncol. 12(968610)2022.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Cheng C, Wang J, Xu P, Zhang K, Xin Z,
Zhao H, Ji Z, Zhang M, Wang D, He Y, et al: Gremlin1 is a
therapeutically targetable FGFR1 ligand that regulates lineage
plasticity and castration resistance in prostate cancer. Nat
Cancer. 3:565–580. 2022.PubMed/NCBI View Article : Google Scholar
|