Introduction
Lung cancer is the most common malignancy in men and
the second most common among women (1). It remains one of the leading causes
of cancer mortality worldwide (2)
and is classified broadly into small-cell lung cancer (SCLC) and
non-small cell lung cancer (NSCLC), the latter being significantly
more common and further subdivided into various histological types
(2). Numerous advances in lung
cancer treatment have been made in recent years, perhaps the most
important being molecular testing for NSCLC to establish targeted
therapy towards driver mutations (3,4).
However, the frequency of these mutations appears to vary
significantly in different geographic regions and ethnicities
(4). For example, EGFR, PTEN,
ALK, ROS1 and RET mutations are predominant in East Asia
and are also more commonly identified in females and non-smokers
(5,6), while KRAS, TP53, BRAF
non-V600E, STK11 and JAK2/3 mutations are more common
in smokers (7). Programmed
death-ligand 1 (PD-L1), a transmembrane protein involved in
immunosuppression, is also expressed in several malignancies,
including NSCLC. It binds to programmed cell death 1 (PD-1),
inhibiting effector T cells and protects malignant cells from the
immune system, hence blocking this phenomenon. In addition, it
assists in the death of malignant cells and has become an essential
entity of lung cancer treatment. Therefore, mutation testing of
PD-L1 expression, has shown promising results in targeted therapy
(8). As with other molecular
targets, PD-L1 expression has a significant geographic and
epidemiologic variation with higher prevalence in East Asia
(9) and in females (9).
Data on the prevalence of molecular drivers of NSCLC
in the Middle East is scarce, with only one cross-sectional study
reporting the EGFR mutation pattern in the region (10). Therefore, in the present study, the
histological patterns and molecular drivers of NSCLC in the United
Arab Emirates (UAE), were reviewed.
Materials and methods
The electronic health records (EHR) of all patients
diagnosed with lung cancer between April 2015 and September 2022
were retrospectively analysed at the Respiratory Institute,
Cleveland Clinic Abu Dhabi (Abu Dhabi, UAE) tertiary care hospital.
Approval (REC approval number A-2018-006) for the study was
obtained from The Cleveland Clinic Abu Dhabi Research Ethics
Committee (Abu Dhabi, UAE) and patient consent was waived due to
the retrospective nature of the present study.
Study population
EHRs were searched to locate all patients diagnosed
with lung cancer. Patients met inclusion criteria if they were
adults and had a new diagnosis of lung cancer. While the patients
with SCLC and those without pathology results were excluded from
the final analysis. The data were collected between May 2019 and
December 2022.
Study variables
Recorded data points included demographics (age,
sex, ethnicity, smoking status, height and weight) and tumour
characteristics (focality, type, histological grade,
visceral/lymphatic invasion, clear margins, TNM classification,
staging, tumour mutations and PD-L1 expression).
Data analysis
Quantitative variables are expressed as the mean and
standard deviation for normally distributed data and the median and
interquartile range for all other data. Categorical variables are
expressed as numbers and percentages. Statistical comparisons
between continuous characteristics were carried out using an
unpaired t-test while comparison for categorical variables was
performed thorough chi-squared test, and P<0.05 was considered
to indicate a statistically significant difference. The data
analysis was conducted using MS Excel 2019 (Microsoft Corp.).
Logistical regression model was created to study the relationship
between EGFR and other categorical variables using R-studio version
23.12.0 (Posit Software).
Results
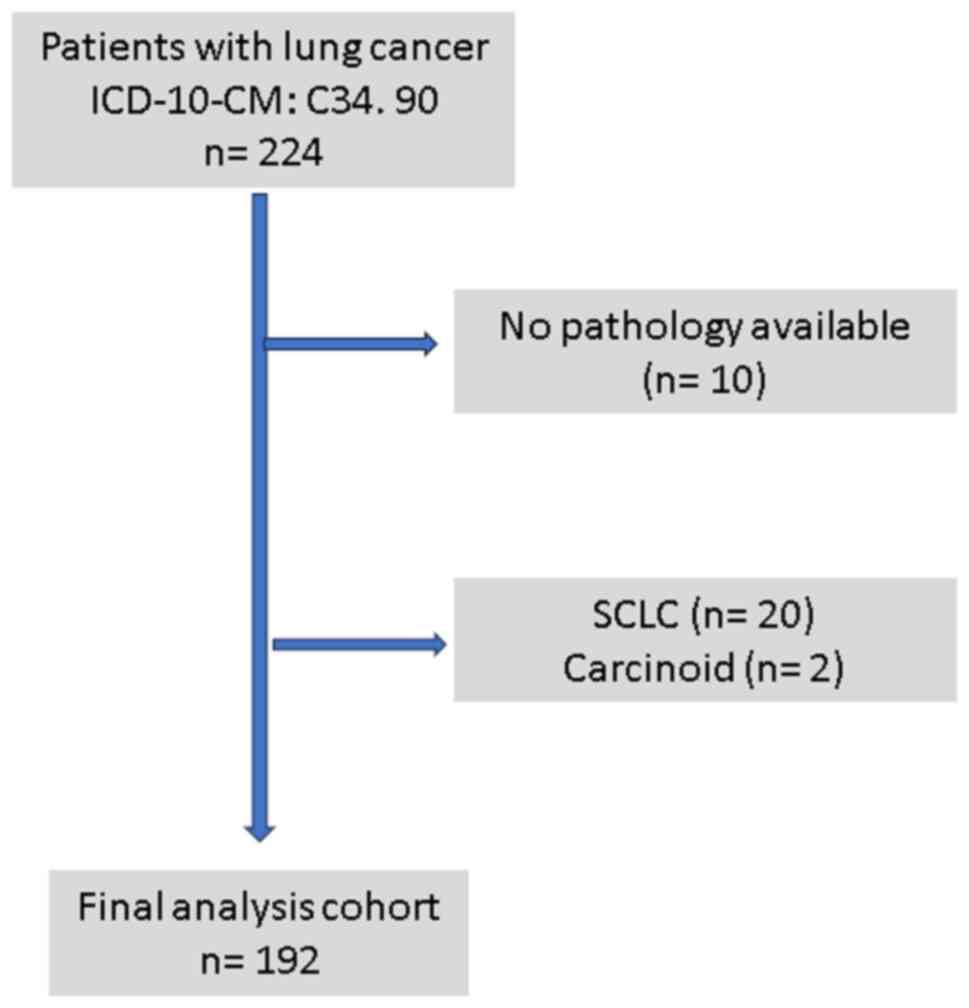
A total of 224 patients were noted to have a
diagnosis of lung cancer on a search of EHR; 32 were excluded
(Fig. 1) due to incomplete records
or diagnosis other than NSCLC, and hence, 192 patients (138 males
and 54 females) were included in the final analysis. The mean age
of patients was 66.3 years (std deviation, ±12.52), and the mean
BMI was 26.5 (std deviation, ±5.56). A total of 155 patients (81%)
were either current or ex-smokers, while 19% had never smoked. The
baseline characteristics and results are shown in Table I. Adenocarcinoma was the most
common type of lung cancer and accounted for 134 patients (70%),
followed by squamous cell cancer in 47 (24%) patients, while large
cell lung cancer was noted in only 5 (3%) patients.
 | Table IBaseline characteristics and results
for lung cancer cohort. |
Table I
Baseline characteristics and results
for lung cancer cohort.
| Variables | n | % | Mean | Std dev |
|---|
| Baseline
characteristics | | | | |
|
Age | 192 | | 66.3 | 12.52 |
|
BMI | 192 | | 26.5 | 5.56 |
|
Male | 138 | 72 | | |
|
Female | 54 | 28 | | |
|
Current or
ex-smoker | 155 | 81 | | |
| Tumour type | | | | |
|
Adenocarcinoma | 134 | 70 | | |
|
Squamous
cell carcinoma | 47 | 24 | | |
|
Large cell
carcinoma | 5 | 3 | | |
|
Other | 6 | 3 | | |
| Tumour
characteristics | | | | |
|
Visceral
pleural invasion | 12 | 6.3 | | |
|
Lymphovascular
invasion | 13 | 6.8 | | |
|
Positive
margins on surgical biopsy | 8 | 6.3 | | |
|
Cavitation
on CT | 13 | 6.9 | | |
|
Ground glass
area present on CT | 32 | 16.8 | | |
|
Endobronchial
involvement | 40 | 21.3 | | |
| Molecular
markers | | | | |
|
EGFR | 29 | 15 | | |
|
BRAF | 3 | 1.6 | | |
|
ALK1 | 8 | 4.2 | | |
|
ROS1 | 2 | 1 | | |
|
MET | 1 | 0.5 | | |
|
KRAS | 16 | 8.3 | | |
|
RET | 0 | 0 | | |
|
PD-L1 | 56 | 24.7 | | |
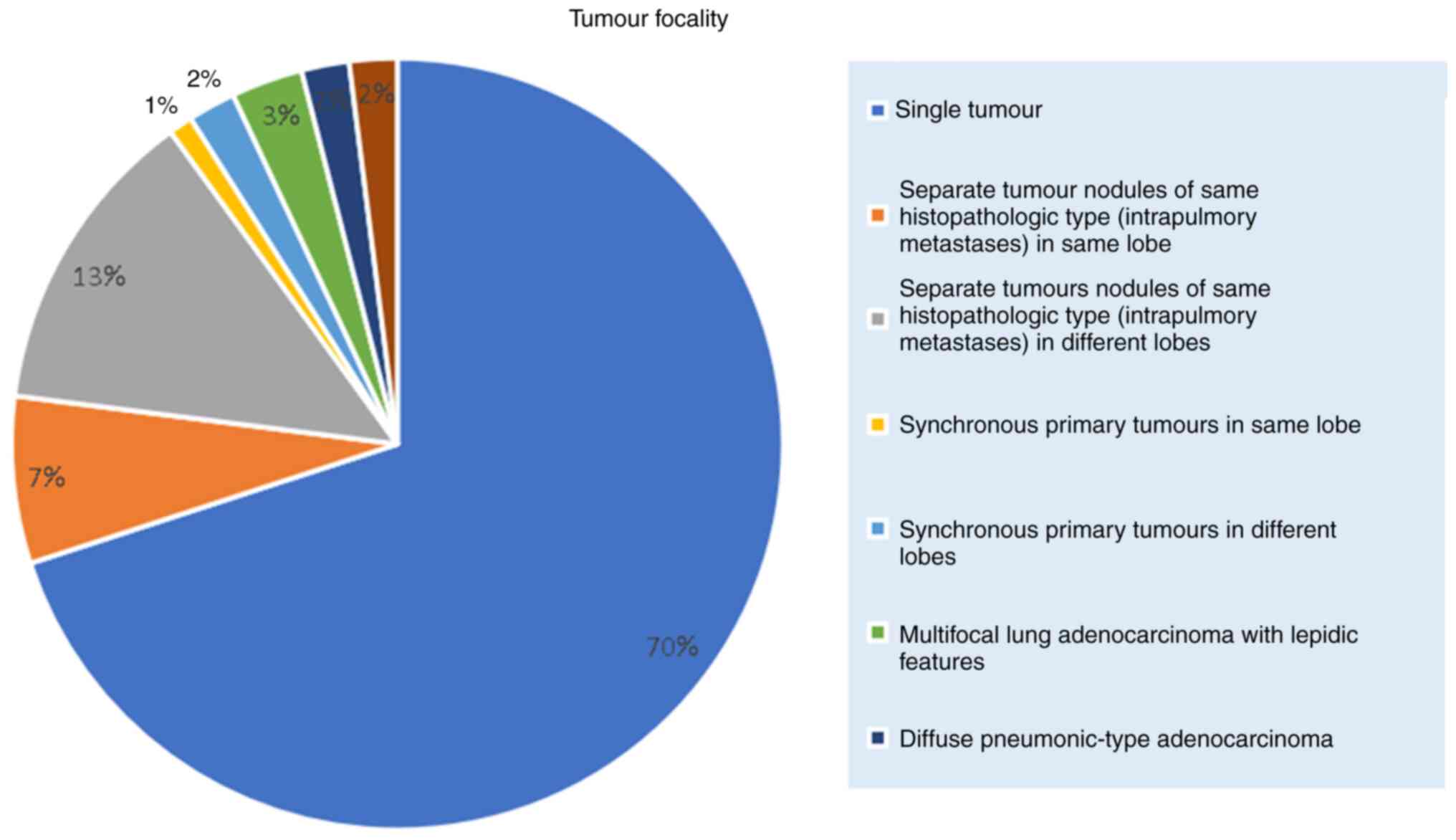
In terms of tumour focality, 138 patients (71.8%)
had a single tumour, and 24 patients (12.5%) had separate tumour
nodules of the same histopathologic type. The tumour focality
breakdown is shown in Fig. 2.
Diagnosis in most of the patients was performed using bronchoscopy
(108 patients); 45 were diagnosed by CT-guided biopsy and 39
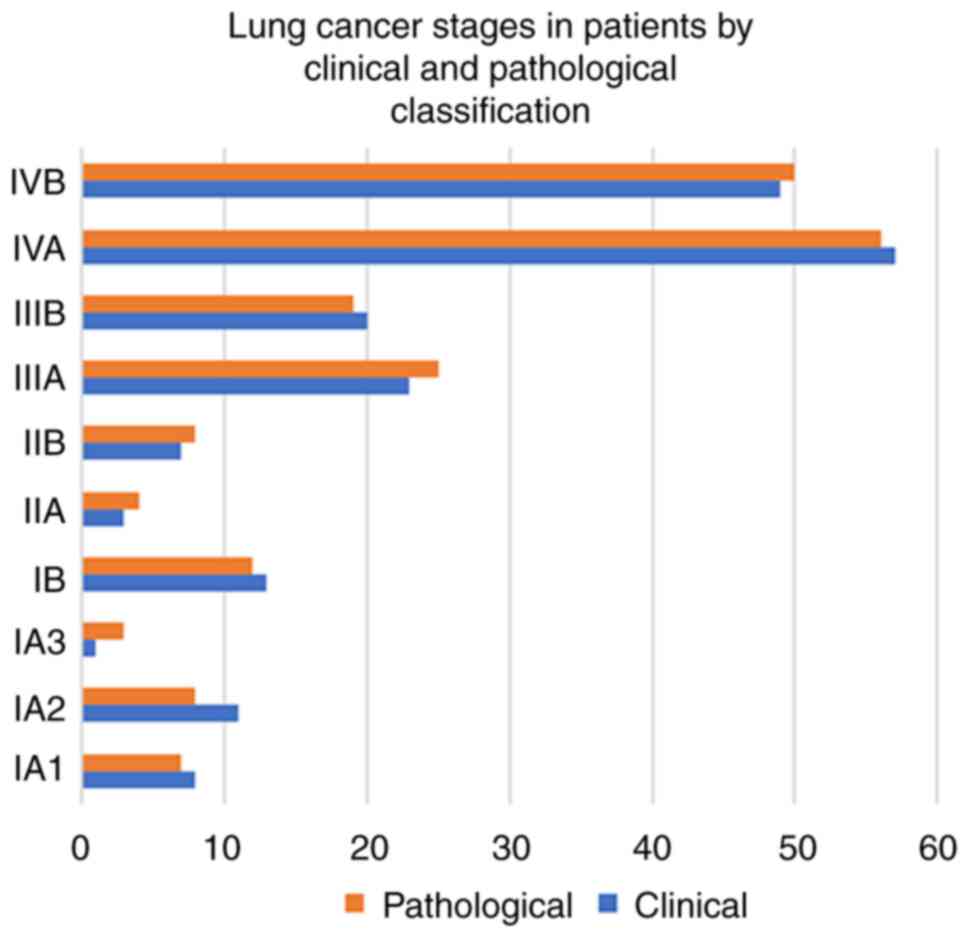
patients by surgery. A total of 106 (55%) patients had stage IV
cancer (stage IVA, 53% and stage IVB, 47%), followed by 43 (22%)
with stage III and 33 (18%) with stage I, and 10 (5%) patients had
stage II cancer. There was a slight reduction in the number of
patients in stage I, with a slight increase in stage II and III on
pathological staging, while the number of patients with stage IV
remained the same (Fig. 3).
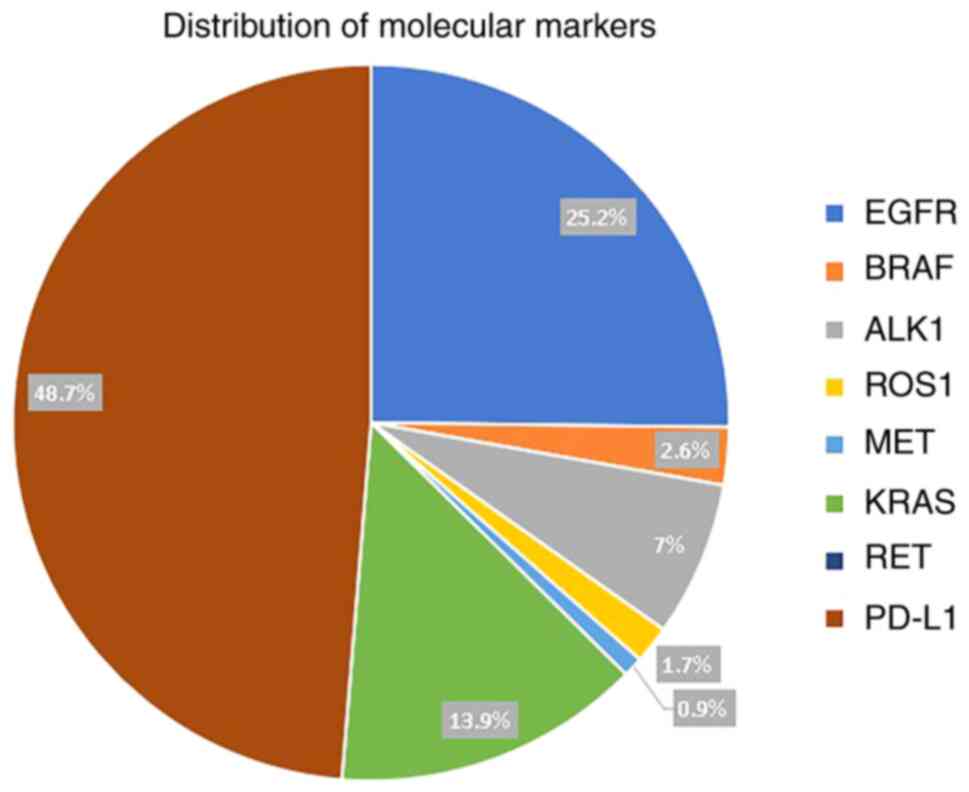
PD-L1 expression was present in 56 (24.7%) patients
and was expressed in >50% of the tumour cells in 27 (12%)
patients. Of the seven mutations tested, EGFR mutations were the
most common and were detected in 29 (15%) patients, followed by
KRAS in 16 (8.3%) patients, activin A receptor like type 1 (ALK1)
in 8 (4.2%) patients, while BRAF, ROS proto-oncogene 1, receptor
tyrosine kinase (ROS1) and MET proto-oncogene, receptor tyrosine
kinase (MET) were present in 3 (1.6%), 2 (1%) and 1 (0.5%),
respectively (Table I). Ret
proto-oncogene (RET) mutation was not detected in any patients.
Distribution of molecular markers is shown in Fig. 4. A total of 22 out of 29 patients
with EGFR mutation were non-smokers, which was statistically
significant (χ2=22.2, dof 1; P<0.005) (data not
shown). Conversely, only 27 out of 56 PD-L1-positive patients were
non-smokers (χ2=0.21, dof 1; P=0.64). No difference was
observed with regard to the age of PD-L1-positive and -negative
patients (mean age, 66.7 and 64.6, respectively; P=0.479) (data not
shown).
There was no significant difference between
EGFR-positive or negative patients in terms of age (mean age, 67.2
and 64.6, respectively; P=0.168) (data not shown). Logistical
regression also showed a significantly reduced odds ratio for male
sex and smoking but and increased odds ratio for adenocarcinomas.
The results obtained regarding the relationship between, age and
tumour stage were not significant. The results of logistical
regression are detailed in Table
II.
 | Table IIResults of logistical regression,
detailing the relationship between EGFR positivity and dependant
variables. |
Table II
Results of logistical regression,
detailing the relationship between EGFR positivity and dependant
variables.
| Characteristic | OR | 95% CI | P-value |
|---|
| Age | 0.98 | 0.95, 1.02 | 0.4 |
| Sex M | 0.25 | 0.1, 0.64 | <0.005 |
| Smoker | 0.24 | 0.09, 0.61 | <0.005 |
| Stage 3,4 | 1.07 | 0.05,7.45 | 0.4 |
| Adenocarcinoma | 0.08 | 0, 0.41 | 0.016 |
Discussion
The present study investigated a cohort of 192 cases
of NSCLC at a tertiary care hospital in the UAE. The population of
the present study comprised 138 males and 54 females, with an
average age of 66.3 years. Notably, the mean age of the population
of the present study was older than previously published studies in
the Gulf and Asian region (10-12).
A significant majority of the patients, 81%, had a history of
smoking, while 19% had never smoked. The prevalence of
adenocarcinoma was notably high, accounting for 70% of the cases,
followed by squamous cell carcinoma (24%) and large cell lung
cancer (3%). The prevalence of smoking and the type of NSCLC was
similar to another reported study in the region (10). Tumour focality, stage at diagnosis
and modality of diagnosis in the present study was similar to
another study from the region (10). No correlation between the tumour
focality and the cancer stage was established, which appears
counterintuitive. However, the data is heavily skewed towards stage
III and IV cancer with very few patients having early-stage
disease. Similarly, most of the patients had only a single tumour
or satellite nodules in the same lobe. This discrepancy is one of
the limitations of the present study and larger datasets would be
more appropriate to show such a relationship.
Molecular characteristics
EGFR mutations were detected in 15% of the
population of the present study. Significant heterogenicity exists
in the frequency of EGFR mutations in different ethnicities, with
East Asian populations showing greater mutations in comparison to
European populations (13). The
EGFR gene is located on the short arm of chromosome 7. It is
200-kb long and contains 28 exons encoding for the EGFR protein
that contains 1,210 amino acids (14). Mutation in this gene is usually
associated with adenocarcinoma, Asian ethnicity and light-smoking
but not with sex. There are several variant subtypes of EGFR
mutations most of which are located on exons 18-21 with exon 19
mutations being the most common. Exon 19 deletions are more
commonly associated with the male sex while exon 21 deletions
appear to be prevalent in females (15).
In the population of the present study, the mutation
frequency was lower than in the East Asian study but higher than
the reported frequencies in the European study (14,15).
The results obtained in the present study were similar to a recent
systematic review, which revealed an overall prevalence of 17% for
EGFR mutations in the Middle East and North African region;
however, they reported significant variations within countries
ranging from 11-30% (16). In the
present study, EGFR mutations were significantly more frequent in
non-smokers, with 22 out of 29 patients with EGFR positive
mutations falling into this category. These findings are well
reported in a number of studies (16-18);
in particular, the meta-analysis by Ren et al (18) which indicated that the odds ratio
for EGFR mutation in non-smokers was 4.8 compared with smokers.
PD-L1
The present study revealed that PD-L1 expression was
present in 24.7% of the patients. The prevalence of PD-L1
expression in the present study was similar to data published in
the only other study examining PD-L1 expression from our region
(19); however, both studies are
corroborated by Dietel et al (20) in their large multicentre study
investigating PD-L1 expression in 18 different countries. PD-L1
positivity did not show a significant association with smoking
status, as 27 out of 69 PD-L1-positive patients were
non-smokers.
Other mutations
KRAS mutations were present in 8.3% of patients,
followed by ALK1 in 4.2%. Other mutations, such as BRAF, ROS1 and
MET, were observed in smaller proportions, while no RET mutations
were detected. Τhe findings of the present study offer important
insights into the epidemiological, clinical, and molecular
characteristics of NSCLC in the UAE. The data also revealed
presentation with advanced stage, which underlines the need for
early detection and intervention strategies. The molecular analysis
provided valuable information about the prevalence of genetic
mutations in patients with NSCLC. EGFR mutations were notably
frequent and were strongly associated with non-smoking status.
PD-L1 expression, on the other hand, did not show a significant
association with smoking. Major limitations of the present study
included the retrospective design. Furthermore, the molecular
testing was at the discretion of the multidisciplinary tumour
board. In conclusion, the present study contributes to the
understanding of NSCLC by providing a comprehensive overview of
patient demographics, tumour characteristics, and molecular
profiles in the UAE. Further research is warranted to explore the
clinical implications of these findings and can serve as a guide
for future research, clinical decision-making, and treatment
approaches for patients with NSCLC in the UAE.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
IS conceptualized and designed the manuscript. SI,
NK, MU, ZZ, SS and AW collected the data. IS, NK, MU analyzed the
data. NK, MU, ZZ, SS and AW performed the critical review of the
manuscript. IS and SI drafted the manuscript. SI and AW confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved (REC approval number
A-2018-006) by The Cleveland Clinic Abu Dhabi Research Ethics
Committee (Abu Dhabi, UAE) and patient consent was waived due to
the retrospective nature of the present study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Mattiuzzi C and Lippi G: Current cancer
epidemiology. J Epidemiol Glob Health. 9:217–22. 2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Bach PB: Smoking as a factor in causing
lung cancer. JAMA. 301:539–541. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Gregg JP, Li T and Yoneda KY: Molecular
testing strategies in non-small cell lung cancer: Optimizing the
diagnostic journey. Transl Lung Cancer Res. 8:286–301.
2019.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Lu F, Li S, Dong B, Zhang S, Lv C and Yang
Y: Identification of lung adenocarcinoma mutation status based on
histologic subtype: Retrospective analysis of 269 patients. Thorac
Cancer. 7:17–23. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Yang H, Jiang P, Liu D, Wang HQ, Deng Q,
Niu X, Lu L, Dai H, Wang H and Yang W: Matrix metalloproteinase 11
is a potential therapeutic target in lung adenocarcinoma. Mol Ther
Oncolytics. 14:82–93. 2019.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Chen YJ, Roumeliotis TI, Chang YH, Chen
CT, Han CL, Lin MH, Chen HW, Chang GC, Chang YL, Wu CT, et al:
Proteogenomics of non-smoking lung cancer in East Asia delineates
molecular signatures of pathogenesis and progression. Cell.
182:226–244.e17. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Paver E, O'Toole S, Cheng XM, Mahar A and
Cooper WA: Updates in the molecular pathology of non-small cell
lung cancer. Semin Diagn Pathol. 38:54–61. 2021.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Yu H, Boyle TA, Zhou C, Rimm DL and Hirsch
FR: PD-L1 expression in lung cancer. J Thorac Oncol. 11:964–975.
2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gu Y, Tang YY, Wan JX, Zou JY, Lu CG, Zhu
HS, Sheng SY, Wang YF, Liu HC, Yang J and Hong H: Sex difference in
the expression of PD-1 of non-small cell lung cancer. Front
Immunol. 13(1026214)2022.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Jazieh AR, Jaafar H, Jaloudi M, Mustafa
RS, Rasul K, Zekri J, Bamefleh H and Gasmelseed A: Patterns of
epidermal growth factor receptor mutation in non-small-cell lung
cancers in the Gulf region. Mol Clin Oncol. 3:1371–1374.
2015.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Fakhruddin N, Mahfouz R, Farhat F, Tfayli
A, Abdelkhalik R, Jabbour M, Yehia L, Mahfoud Z and Zaatari G:
Epidermal growth factor receptor and KRAS mutations in lung
adenocarcinoma: A retrospective study of the Lebanese population.
Oncol Rep. 32:2223–2229. 2014.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Shi Y, Au JSK, Thongprasert S, Srinivasan
S, Tsai CM, Khoa MT, Heeroma K, Itoh Y, Cornelio G and Yang PC: A
prospective, molecular epidemiology study of EGFR mutations in
Asian patients with advanced non-small-cell lung cancer of
adenocarcinoma histology (PIONEER). J Thorac Oncol. 9:154–162.
2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Melosky B, Kambartel K, Häntschel M,
Bennetts M, Nickens DJ, Brinkmann J, Kayser A, Moran M and Cappuzzo
F: Worldwide prevalence of epidermal growth factor receptor
mutations in non-small cell lung cancer: A meta-analysis. Mol Diagn
Ther. 26:7–18. 2022.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Jurišić V, Obradovic J, Pavlović S and
Djordjevic N: Epidermal growth factor receptor gene in
non-small-cell lung cancer: The importance of promoter polymorphism
investigation. Anal Cell Pathol (Amst).
2018(6192187)2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Tanaka T, Matsuoka M, Sutani A, Gemma A,
Maemondo M, Inoue A, Okinaga S, Nagashima M, Oizumi S, Uematsu K,
et al: Frequency of and variables associated with the EGFR mutation
and its subtypes. Int J Cancer. 126:651–655. 2010.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Boustany Y, Laraqui A, El Rhaffouli H,
Bajjou T, El Mchichi B, El Anaz H, Amine IL, Chahdi H, Oukabli M,
Souhi H, et al: Prevalence and patterns of EGFR mutations in
non-small cell lung cancer in the Middle East and North Africa.
Cancer Control. 29(10732748221129464)2022.
|
|
17
|
Tseng CH, Chiang CJ, Tseng JS, Yang TY,
Hsu KH, Chen KC, Wang CL, Chen CY, Yen SH, Tsai CM, et al: EGFR
mutation, smoking, and gender in advanced lung adenocarcinoma.
Oncotarget. 8:98384–98393. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Ren JH, He WS, Yan GL, Jin M, Yang KY and
Wu G: EGFR mutations in non-small-cell lung cancer among smokers
and non-smokers: A meta-analysis. Environ Mol Mutagen. 53:78–82.
2012.PubMed/NCBI View
Article : Google Scholar
|
|
19
|
Jazieh AR, Bounedjar A, Bamefleh H,
Alfayea T, Almaghraby HQ, Belarabi A, Ouahioune W, Derbouz Z,
Alkaiyat M, Alkattan K, et al: Expression of immune response
markers in arab patients with lung cancer. JCO Glob Oncol.
6:1218–1224. 2020.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Dietel M, Savelov N, Salanova R, Micke P,
Bigras G, Hida T, Antunez J, Guldhammer Skov B, Hutarew G, Sua LF,
et al: Real-world prevalence of programmed death ligand 1
expression in locally advanced or metastatic non-small-cell lung
cancer: The global, multicenter EXPRESS study. Lung Cancer.
134:174–179. 2019.PubMed/NCBI View Article : Google Scholar
|