Introduction
The heart is the first organ to form in vertebrate
embryogenesis (1). In vertebrates,
the survival of the developing embryo depends on the heart and the
circulatory systems, therefore cardiac abnormalities not only cause
miscarriage and stillbirth, but also seriously affect the quality
of life after birth (2). The heart
is extremely sensitive to embryonic environmental changes during
development. Certain physical, chemical and biological contaminants
are known to contribute to cardiovascular disease (3,4). The
current study shows that exposure to persistent organic pollutants,
such as polychlorinated biphenyls (PCBs), is significantly
correlated with heart disease (5,6). In
China, PCB contamination has become a serious environmental
problem. For example, in Jiangsu Province the concentration of PCBs
in some surface water has exceeded the standard (20 ng/l) for the
‘Environmental Quality Standard for Surface Water’ (7). Total PCB concentration in human
adipose tissue ranged from 4.1 to 125 ng/g lipid (mean 32.8 ng/g
lipid), which significantly exceeds the mean total World Health
Organization (WHO) toxicity equivalent (TEQ) values for PCBs in
human adipose tissue, which is 16.2 pg/g lipid (8). Studies have also demonstrated that
PCBs may pass through the placenta to the fetus in mothers who have
been exposed before and during pregnancy (9,10).
The AHR belongs to the basic helix-loop-helix
Per-Arnt-Sim family of transcriptional regulators. Several studies
have shown that members of this family play key roles in a broad
range of biological functions and that the biochemical and
toxicological effects of PCBs act through the AHR pathway (11,12).
However, little is known about the underlying effects of the AHR
during the differentiation of embryonic carcinoma cell line P19
into cardiomyocytes.
In this study, we constructed two short hairpin
(sh)RNA plasmid vectors against AHR that were capable of
persistently generating small interfering (si)RNA in cells. We
transfected these vectors into the P19 cells to determine the
effects of AHR gene silencing on their differentiation.
Furthermore, we examined the expression levels of four critical
genes (ARNT, CYP1A1, GSK3β and β-catenin), and
determined which are components of the AHR and Wnt signaling
pathways using quantitative polymerase chain reaction (qPCR).
Materials and methods
Cell culture
P19 cells were obtained from the American Type
Culture Collection (ATCC, Manassas, VA, USA). The cells were
cultured in α-minimal essential medium (α-MEM; Gibco-BRL, Grand
Island, NY, USA) containing 10% fetal bovine serum (FBS;
Gibco-BRL), 100 U/ml penicillin and 100 μg/ml streptomycin in
bacteriological dishes in an atmosphere of 5% CO2 in air
at 37°C. Embryoid bodies (EB) were transferred to 10-cm bacterial
dishes that contained 15 ml α-MEM supplemented with 1% DMSO (Sigma,
St. Louis, MO, USA), 10% FBS, 100 U/ml penicillin and 100 μg/ml
streptomycin to induce cardiac differentiation, and then cultured
for 96 h. The EBs were then collected and transferred to 6-cm
bacterial dishes supplemented with α-MEM containing 10% FBS for an
additional 6 days. Cell morphological changes during the growth and
differentiation of P19 were observed under an inverted microscope
(Nikon, Tokyo, Japan).
Construction of shRNA expression vector
for AHR
Two target DNA fragments were designed and
constructed for AHR based on shRNA design, enzyme insertion sites
in the pGPU6-GFP-Neo expression vector and the AHR exons (GenBank
accession number: NM_013464.4) as cited in GenBank. The sequences
of the primers used were: shRNA1 sense:
5′-CAGAGCGTATATGAGCTCATCCATA-3′; and antisense:
5′-GTCTCGCATATACTCGAGTAGGTAT-3′; shRNA2 sense:
5′-CCTCCACAGGCAGCAGTCTATTATA-3′; and antisense:
5′-GGAGGTGTCCGTCGTCAGATAATAT-3′. Another unrelated sequence was
used as the control. No homologous sequence was found by BLAST
analysis. Loop-stem structure was a non-homologous base
(TTCAAGAGA), which was non-complementary to AHR. Enzyme insertion
sites for BbsI and BamHI were constructed into the
ends of the oligonucleotide fragments, and the specificity of
constructed oligonucleotides strands was analyzed by the BLAST
program. The primer sequences were as follows: shRNA1 sense:
5′-CACCGCAGAGCGTATATGAGCTCATCCATATTCAAG
AGATATGGATGAGCTCATATACGCTCTGTTTTTTG-3′; and antisense:
5′-GATCCAAAAAACAGAGCGTATATGAGCTCATCCATATCTCTTGATATGGATGAGCTCATATACGCTCTGC-3′;
shRNA2 sense:
5′-CACCGCCTCCACAGGCAGCAGTCTATTATATTCAAGAGATATAATAGACTGCTGCCTGTGGAGGTTTTTTG-3′;
and antisense:
5′-GATCCAAAAAACCTCCACAGGCAGCAGTCTATTATATCTCTTGAATATAATAGACTGCTGCCTGTGGAGGC-3′.
Sense and antisense oligonucleotides were annealed to generate a
double-stranded oligonucleotide, and the annealed shRNA
oligonucleotide fragment template was inserted into the
pGPU6-GFP-Neo vector using T4 DNA ligase. The recombinant plasmid
was then transformed into competent bacillus Escherichia
coli, and the bacteria were cultured overnight in LB medium
that contained kanamycin. Recombinant plasmids were extracted,
purified and cut using restriction enzymes BbsI,
BamHI and PstI in order to identify the correct
fragments.
Plasmid transfection
P19 cells were digested with trypsin and seeded into
6-well plates. When the density of the cells on the slide reached
80–90% confluence they were transfected in four groups: the first
group was the blank control, the second was the negative control,
the third was the shRNA1 group and the fourth was the shRNA2 group.
Transfection was performed in accordance with the manufacturer’s
instructions. Briefly, P19 cells were seeded into 6-well plates at
a density of 2.5×105 cells/well and cultured for 24 h.
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) was mixed with
Opti-MEM I medium, shRNA expression vectors were added to the
solution and the cells were incubated at room temperature for 20–25
min. The transfection mixture was added to each well with 600 μl
FBS-free α-MEM medium. The cells were incubated at 37°C for 4–6 h,
and the medium was changed. α-MEM medium (1.5 ml) containing 10%
FBS was added and the cells were incubated for another 24 h.
RNA and real-time qPCR
Total RNA was extracted from cells using TRIzol
reagent according to the manufacturer’s instructions (Invitrogen).
Total RNA (1 μg) was transcribed to cDNA using M-MLV reverse
transcriptase. Real-time qPCR reactions were carried out using an
MX3000 real-time instrument (Stratagene, Cedar Creek, TX, USA).
Each PCR amplification was performed in triplicate, using the
following conditions: samples were incubated at 95°C for 3 min for
initial denaturation, and then subjected to 40 PCR cycles. Each PCR
cycle consisted of 95°C for 30 sec and 62°C for 40 sec.
Additionally, normalization to the housekeeping gene glyceraldehyde
3-phosphate dehydrogenase (GAPDH) was performed. The primer
sequences used are listed in Table
I.
 | Table IPrimers used for real-time polymerase
chain reaction (PCR). |
Table I
Primers used for real-time polymerase
chain reaction (PCR).
| Gene name | Sequences
(5′-3′) | Product length
(bp) |
|---|
| AHR | F:
ATGGAGAGGTGCTTCAGGTGCCG
R: ATGGAGGGTGGCTGAAGTGGAGT | 185 |
| GATA4 | F:
CCTGCGGCCTCTACATGA
R: AGGGTCTCACCAGCAGGA | 136 |
| Nkx2.5 | F:
CCTGCGGCCTCTACATGA
R: AGGGTCTCACCAGCAGGA | 222 |
| ARNT | F:
GACAGACCACAGGACAGTTCC
R: AGCATGGACAGCATTTCTTGAA | 172 |
| CYP1A1 | F:
GGTTAACCATGACCGGGAACT
R: TGCCCAAACCAAAGAGAGTGA | 122 |
| GSK3β | F:
TGGCAGCAAGGTAACCACAG
R: CGGTTCTTAAATCGCTTGTCCTG | 189 |
| β-catenin | F:
ATGGAGCCGGACAGAAAAGC
R: CTTGCCACTCAGGGAAGGA | 108 |
| GAPDH | F:
TTCACCACCATGGAGAAGGC
R: GGCATGGACTGTGGTCATGA | 237 |
Statistical analysis
Values are shown as the mean ± standard deviation
(SD). Statistical analyses were performed using one-way analysis of
variance (ANOVA) and t-tests or Student’s-tests with a correction
for multiple comparisons. P<0.05 was considered to indicate a
statistically significant result.
Results
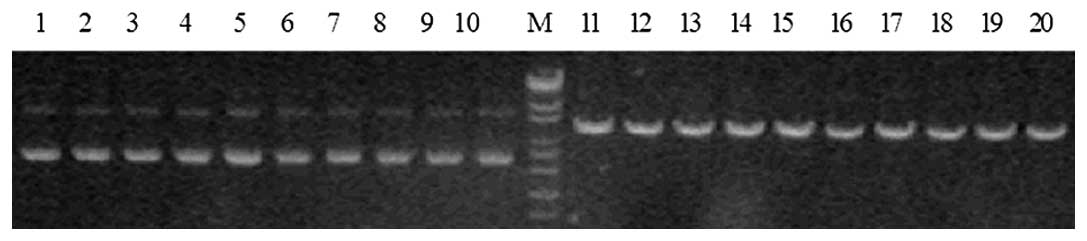
Identification of enzyme digestion
AHR-targeted shRNA expression vectors were cut using
PstI. A DNA band of 5,180 bp was able to be digested, which
indicated that the target gene segment AHR had been inserted into
pGPU6-GFP-Neo vector (Fig. 1).
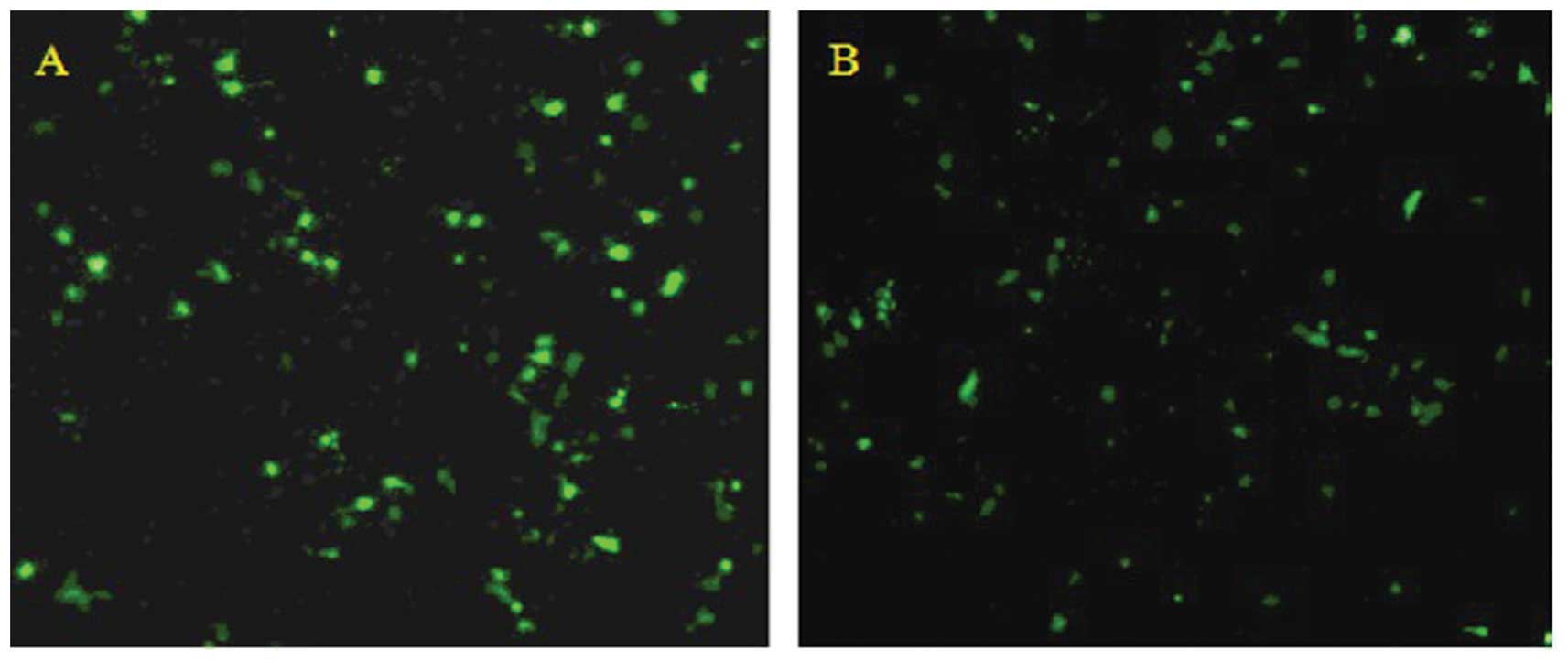
Observation of transfection results
Transfection was carried out using Lipofectamine
2000 (Invitrogen) according to the manufacturer’s instructions.
After 48 h, green fluorescence was observed in transfected cells
under the fluorescence microscope, and the transfection rate was
found to be approximately 50% (Fig.
2).
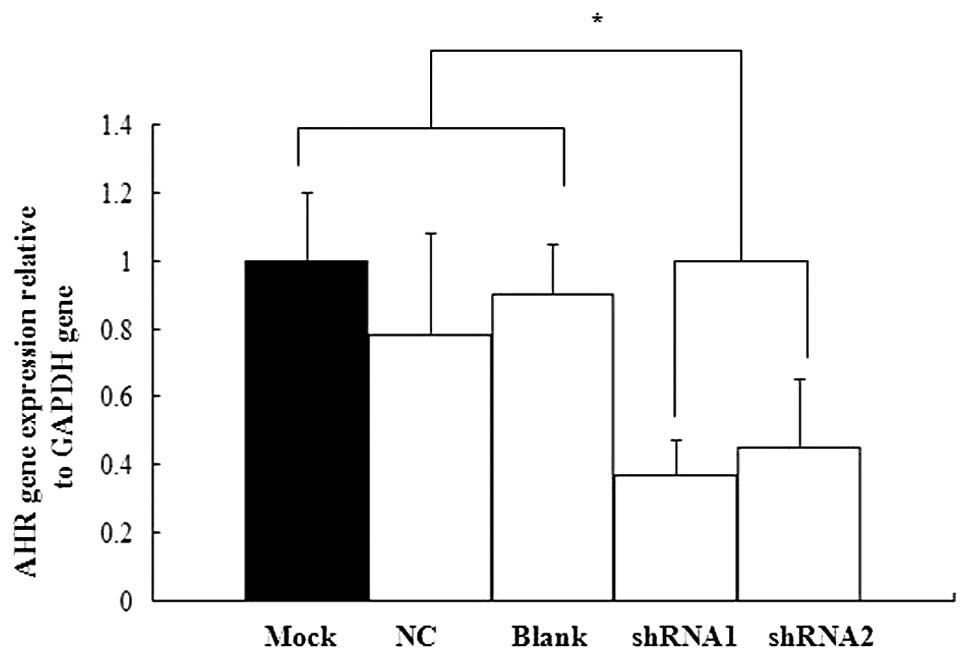
shRNA targeting AHR inhibited AHR mRNA
expression in P19 cells
We used real-time qPCR to confirm the efficiency of
shRNA silencing on AHR expression. No statistically significant
difference was found in AHR mRNA expression between the mock
treatment group and the blank control group. Moreover, the two
shRNA groups showed varying degrees of inhibitory effect (Fig. 3). Therefore, the AHR-targeted shRNA
expression vectors were selected for subsequent experiments.
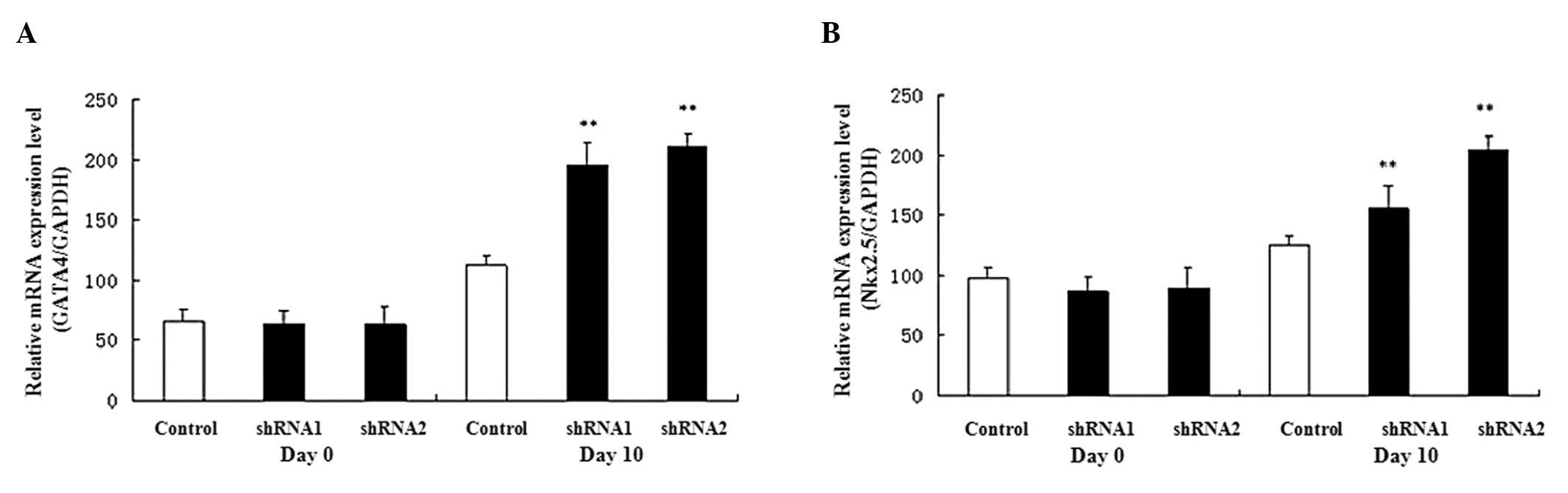
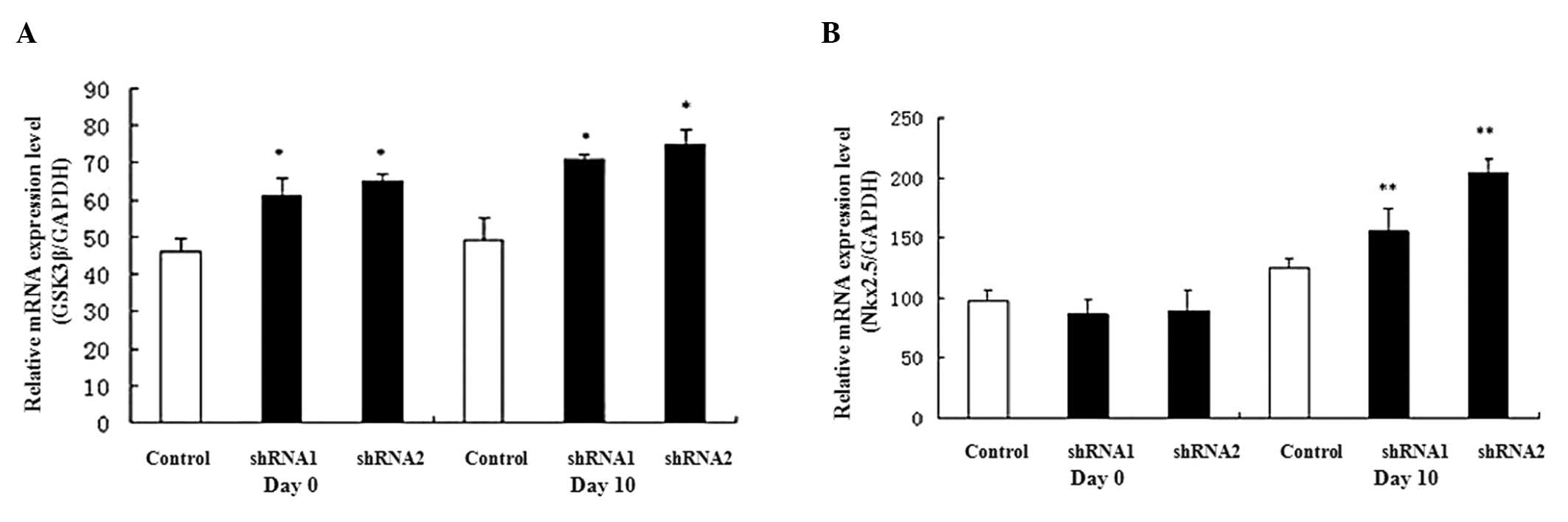
Expression of marker gene during P19 cell
differentiation
The GATA4 and Nkx2.5 genes are
expressed in cardiomyocytes as cardiac-specific genes; therefore,
their expression was examined during P19 cell differentiation.
GATA4 and Nkx2.5 gene expression levels were
upregulated during heart development (from day 0 to day 10) and the
expression levels were increased on day 10 in the AHR-silenced P19
cells compared with the control cells (Fig. 4).
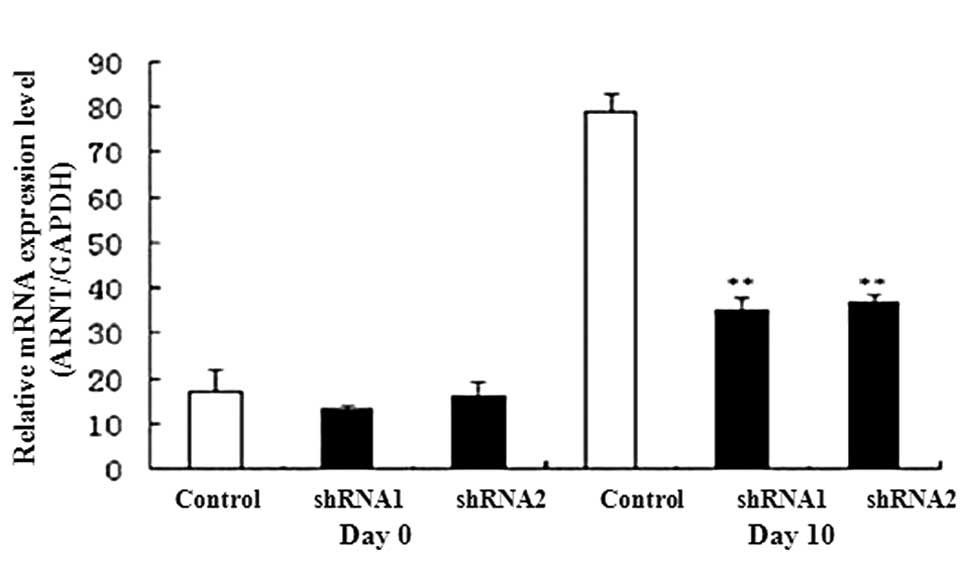
Expression of AHR and Wnt signal
transduction genes during P19 cell differentiation
We detected the expression levels of AHR signal
transduction-related genes, including ARNT and CYP1A,
using qPCR. On day 10 expression levels of ARNT and
CYP1A1 were lower in the AHR-silenced P19 cells than in the
controls (Fig. 5). We also
determined the expression levels of genes GSK3β and
β-catenin involved in the Wnt signal transduction pathway.
Results from qPCR revealed that β-catenin was suppressed,
whereas GSK3β was increased, in the AHR-silenced P19 cells
(Fig. 6).
Discussion
RNA interference (RNAi) is one of the most powerful
technologies for the specific blocking of gene expression (13). In this study, we successfully
transfected two shRNAs that targeted the AHR gene into P19
cells. The results showed that AHR mRNA expression examined in the
shRNA1 and shRNA2 groups was inhibited by approximately 64 and 52%,
respectively, compared with the control group.
Using shRNA, we found that the gene silencing of AHR
caused the expression levels of two myocardial cell
differentiation-related genes (GATA4 and Nkx2.5) to
be elevated. In addition, real-time qPCR revealed that the
expression of ARNT, CYP1A1 and β-catenin was
suppressed, but the expression of GSK3β was increased in the
AHR-silenced P19 cells. These results suggest that the silencing
effect of AHR promotes the differentiation of P19 mouse embryonic
carcinoma cells into cardiomyocytes. The Wnt signal transduction
pathway may be responsible for the effect of silencing AHR in P19
cells.
Findings of studies have shown that PCBs are one of
the most ubiquitous contaminants that are important in the
development of cardiovascular disease (14,15).
It is generally believed that the effects of PCBs are mediated
through the AHR pathway (16).
Lund et al found that knockout of the AHR gene in
mice disrupted cardiovascular homeostasis, which caused significant
cardiac hypertrophy and elevated levels of expression of
cardiovascular markers (17). In
this study, we also found that the expression of cardiac
development-specific genes was induced in the AHR-silenced P19
cells. These results suggested that the AHR signaling pathway is
crucial in cardiovascular development programs.
Wnt protein and its signaling pathways are among the
most intensely studied pathways in biology (18). Previous studies have suggested that
the Wnt proteins are capable of inducing cell proliferation,
differentiation and maturation (19). The canonical Wnt signaling pathway
may be manipulated to regulate the expansion and differentiation of
cardiac progenitor cells, and Wnt signaling was found to be
essential to the development of the heart in mammals (20,21).
In this study, the expression of the critical AHR signaling pathway
genes (AHR, ARNT and CYP1A1) was reduced in the
AHR-silenced P19 cells. In addition, the β-catenin
expression level was decreased but GSK3β was increased in
the AHR-silenced P19 cells during heart development. Taking into
account the fact that AHR activation may inappropriately activate
the Wnt signaling pathway and that there is crosstalk between AHR
and Wnt signaling (22), we
suggest that the silencing of AHR inhibited the differentiation of
embryonic carcinoma P19 cells, possibly through the Wnt signaling
transduction pathway.
In conclusion, we constructed a shRNA expression
vector for AHR. Findings of the present study showed that the
expression levels of GATA4 and Nkx2.5 genes were
increased in the AHR-silenced P19 cells. We also found that
ARNT, CYP1A1 and β-catenin were suppressed, whereas
GSK3β was elevated, in the AHR-silenced P19 cells. However,
the exact mechanisms of how the AHR and Wnt signaling pathways
affect the differentiation of P19 cells into cardiac myocytes
should be investigated.
Acknowledgements
This study was supported by a grant from the
National Natural Science Foundation of China (no. 30973213).
References
|
1
|
Olson EN: Gene regulatory networks in the
evolution and development of the heart. Science. 313:1922–1927.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hsiao SM, Wu MH, Jou HJ, Lee CN, Shyu MK,
Shih JC and Hsieh FJ: Outcome for fetuses with prenatally detected
congenital heart disease and cardiac arrhythmias in Taiwan. J
Formos Med Assoc. 106:423–431. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hennig B, Meerarani P, Slim R, Toborek M,
Daugherty A, Silverstone AE and Robertson LW: Proinflammatory
properties of coplanar PCBs: in vitro and in vivo evidence. Toxicol
Appl Pharmacol. 181:174–183. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pelclová D, Urban P, Preiss J, Lukás E,
Fenclová Z, Navrátil T, Dubská Z and Senholdová Z: Adverse health
effects in humans exposed to 2, 3, 7,
8-tetrachlorodibenzo-p-dioxin (TCDD). Rev Environ Health.
21:119–138. 2006.
|
|
5
|
DeWitt JC, Millsap DS, Yeager RL, Heise
SS, Sparks DW and Henshel DS: External heart deformities in
passerine birds exposed to environmental mixtures of
polychlorinated biphenyls during development. Environ Toxicol Chem.
25:541–551. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Majkova Z, Smart E, Toborek M and Hennig
B: Up-regulation of endothelial monocyte chemoattractant protein-1
by coplanar PCB77 is caveolin-1-dependent. Toxicol Appl Pharmacol.
237:1–7. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hong Y, Chunhong Z and Xiaoxiong Z:
Investigation of pollution characteristics of polychlorinated
biphenyls in the typical drinking water sources in Jiangsu
Province, China. Environ Monit Assess. 158:573–579. 2009.
|
|
8
|
Shen H, Shen H, Han J, Tie X, Xu W, Ren Y
and Ye C: Polychlorinated dibenzo-p-dioxins/furans and
polychlorinated biphenyls in human adipose tissue from Zhejiang
Province, China. Chemosphere. 74:384–388. 2009.
|
|
9
|
Suzuki G, Nakano M and Nakano S:
Distribution of PCDDs/PCDFs and co-PCBs in human maternal blood,
cord blood, placenta, milk, and adipose tissue: dioxins showing
high toxic equivalency factor accumulate in the placenta. Biosci
Biotechnol Biochem. 69:1836–1847. 2005. View Article : Google Scholar
|
|
10
|
Shen H, Shen H, Main KM, Virtanen HE,
Damggard IN, Haavisto AM, Kaleva M, Boisen KA, Schmidt IM,
Chellakooty M, Skakkebaek NE, Toppari J and Schramm KW: From mother
to child: investigation of prenatal and postnatal exposure to
persistent bioaccumulating toxicants using breast milk and placenta
biomonitoring. Chemosphere. 67:S256–S262. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu S and Piatigorsky J: Regulation of
mouse small heat shock protein αb-crystallin gene by aryl
hydrocarbon receptor. PLoS One. 6:e179042011.
|
|
12
|
Shimada T, Sugie A, Shindo M, Nakajima T,
Azuma E, Hashimoto M and Inoue K: Tissue-specific induction of
cytochromes P450 1A1 and 1B1 by polycyclic aromatic hydrocarbons
and polychlorinated biphenyls in engineered C57BL/6J mice of
arylhydrocarbon receptor gene. Toxicol Appl Pharmacol. 187:1–10.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Manoharan M: RNA interference and
chemically modified small interfering RNAs. Curr Opin Chem Biol.
8:570–579. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Han SG, Eum SY, Toborek M, Smart E and
Hennig B: Polychlorinated biphenyl-induced VCAM-1 expression is
attenuated in aortic endothelial cells isolated from caveolin-1
deficient mice. Toxicol Appl Pharmacol. 246:74–82. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Hennig B, Reiterer G, Majkova Z,
Oesterling E, Meerarani P and Toborek M: Modification of
environmental toxicity by nutrients: implications in
atherosclerosis. Cardiovasc Toxicol. 5:153–160. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Grimes AC, Erwin KN, Stadt HA, Hunter GL,
Gefroh HA, Tsai HJ and Kirby ML: PCB126 exposure disrupts zebrafish
ventricular and branchial but not early neural crest development.
Toxicol Sci. 106:193–205. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lund AK, Peterson SL, Timmins GS and
Walker MK: Endothelin-1-mediated increase in reactive oxygen
species and NADPH oxidase activity in hearts of aryl hydrocarbon
receptor (AhR) null mice. Toxicol Sci. 88:265–273. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shevtsov SP, Haq S and Force T: Activation
of beta-catenin signaling pathways by classical G-protein-coupled
receptors: mechanisms and consequences in cycling and non-cycling
cells. Cell Cycle. 5:2295–2300. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakamura T, Sano M, Songyang Z and
Schneider MD: A Wnt- and beta-catenin-dependent pathway for
mammalian cardiac myogenesis. Proc Natl Acad Sci USA.
100:5834–5839. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kwon C, Arnold J, Hsiao EC, Taketo MM,
Conklin BR and Srivastava D: Canonical Wnt signaling is a positive
regulator of mammalian cardiac progenitors. Proc Natl Acad Sci USA.
104:10894–10899. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ai D, Fu X, Wang J, Lu MF, Chen L, Baldini
A, Klein WH and Martin JF: Canonical Wnt signaling functions in
second heart field to promote right ventricular growth. Proc Natl
Acad Sci USA. 104:9319–9324. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mathew LK, Sengupta SS, Ladu J, Andreasen
EA and Tanguay RL: Crosstalk between AHR and Wnt signaling through
R-Spondin1 impairs tissue regeneration in zebrafish. FASEB J.
22:3087–3096. 2008. View Article : Google Scholar : PubMed/NCBI
|