Introduction
Ribosomes, the organelles that catalyze protein
synthesis, consist of a small 40S subunit and a large 60S subunit
each, and are the components of cells that produce proteins from
all amino acids (1,2). Ribosomal protein S23 (RPS23)
gene encodes a ribosomal protein, as a component of the 40S
subunit. This protein belongs to the S12P family of ribosomal
proteins, is located in the cytoplasm and is crucial in the
development of human diseases. Multiple processed pseudogenes of
this gene are dispersed throughout the genome, which is typical for
genes encoding ribosomal proteins (3).
Ribosomal protein gene mutations or disturbance in
their expression levels have been observed in a number of inherited
genetic diseases such as Tuner syndrome, Diamond-Blackfan anemia
syndrome, Camurati-Engelmann disease, Noonan syndrome, and
Bardet-Biedl syndrome (4). The
physiological function of ribosomal proteins in human diseases has
been progressively clarified due to the progress in the scientific
techniques as well as the continuous and in-depth research
(4). RPS23 gene is involved
in cell growth and regulation. The small ribosomal protein RPS23
has been found to be located in the vicinity of the mRNA entry
channel and to play an important role in identifying specific
interactions of the human eukaryotic translation initiation factor
(eIF) 3 (5). The eukaryotic
translation initiation factor eIF5B is a ribosome-dependent GTPase
that mediates the displacement of initiation factors from the 40S
ribosomal subunit, and RPS23 has been shown to be located in
its domain 2 (6). A mouse gene
recently identified, RPS23 retroposed gene 1
(RPS23rg1), was found to regulate β-amyloid (Aβ) levels and
tau phosphorylation, two major pathological hallmarks of
Alzheimer’s disease (AD), and RPS23rg1 was also found to
originate through retroposition of the mouse RPS23 mRNA
(7).
Cancer is one of the leading causes of mortality
worldwide and is medically known as a malignant neoplasm. This is a
term for a large group of different diseases, all involving
unregulated cell growth (8).
Cancer cells divide rapidly, grow in an uncontrolled manner, form
malignant tumors and invade neighbouring parts of the body. Cancer
may also spread to more distant parts of the body through the
lymphatic system or bloodstream (9). Cancer is one of the most fatal
diseases in the human population and constitutes a common cause of
death worldwide (10,11). However, many proteins possess
antitumor activities. The aim of the present study was to evaluate
the anticancer activity of RPS23 protein from the giant panda.
Ailuropoda melanoleuca (A.
melanoleuca), considered a National treasure of China, is one
of the oldest species belonging to the group of endangered mammals.
Macrograph of giant panda has been achieved. The investigation of
gene function constitutes an important field in current giant panda
research (12–17). The ribosomal protein RPS23 of the
giant panda has received increasing attention due to its various
biological functions, particularly in relation to anticancer
activity.
In the present study, we used reverse
transcription-polymerase chain reaction (RT-PCR) to amplify the
cDNA of RPS23 gene from the total RNA of the skeleton muscle
of giant panda, based on relative information regarding the
RPS23 gene of the designed primers of some mammals,
including Homo sapiens (H. sapiens), Bos
Taurus (B. Taurus), Felis catus (F.
catus), Mus musculus (M. musculus) and Rattus
norvegicus (R. norvegicus). The sequence characteristics
of the protein encoded by the cDNA was also analyzed and compared
with those of human and other animals reported. A recombinant
expression vector containing RPS23 cDNA was constructed and
overexpressed in Escherichia coli (E. coli) using
pET28a plasmids. Under the optimized expression conditions, many
recombinant proteins of RPS23 were obtained from the giant panda,
which were then purified using Ni chelate affinity chromatography.
The aim of the present study was to evaluate the anticancer
activity of RPS23 protein from the giant panda in vivo.
Materials and methods
Materials
Skeletal muscle was collected from a dead giant
panda provided from the Wolong Conservation Center of the Giant
Panda (Sichuan, China). The collected skeletal muscle was frozen in
liquid nitrogen and then used for RNA isolation. Total Tissue/Cell
RNA Extraction kits were purchased from Waton Inc. (Shanghai,
China). Reverse transcription kits were purchased from Promega,
Beijing, China. Gel Extraction Mini kits were purchased from Omega
(Kanpur, India). pMD19-T Vector Systems and the restriction enzymes
EcoRI and HindIII were purchased from Takara Bio
(Dalian, China). DNA polymerases were purchased from Sangon Co.,
Ltd. (Shanghai, China). Host bacteria E. coli DH5α were
stored in the Key Laboratory of Southwest China Wildlife Resources
Conservation (Nanchong, China). CW0009 Ni-Agarose His-tag Protein
purification kits were purchased from Beijing Ealysino Biological
Technology Co., Ltd. (Beijing, China). Bradford Protein Assay kits
were purchased from Majorbio BioTech Co., Ltd. (Shanghai, China).
Penicillin/streptomycin (penicillin 10,000 U/ml, streptomycin
10,000 μg/ml) and Dulbecco’s modified Eagle’s medium (DMEM) reagent
were purchased from Gibco-BRL (Grand Island, NY, USA). Fetal bovine
serum (FBS) was obtained from Sijiqing Biological Engineering
Materials Co., Ltd. (Huangzhou, China).
DNA and RNA isolation
A total of 500 mg of muscle tissue was ground in
liquid nitrogen to a fine powder, and the powder was suspended
completely in 15 ml lysis buffer containing 10 mM Tris-HCl, pH 8.0,
100 mM EDTA and 0.5% SDS. Following treatment with proteinase K
(final concentration, 100 mg/ml) at 55°C for 3 h, the mixture was
cooled to room temperature and mixed with an equal volume of
saturated phenol (pH 8.0) prior to centrifugation at 5,000 × g at
4°C for 20 min. The supernatant was pooled and mixed with an equal
volume of 1:1 (v:v) phenol-chloroform, followed by centrifugation
as described above. The supernatant was then collected, from which
the DNA was precipitated by ethanol. The DNA obtained was then
dissolved in TE buffer and maintained at −20°C. Total RNAs were
isolated from ~400 mg muscle tissue using the Total Tissue/Cell RNA
Extraction kits (Waton Inc.) according to the manufacturer’s
instructions. The total RNAs extracted were dissolved in diethyl
pyrocarbonate (DEPC) water and maintained at −70°C.
Primer design and RT-PCR
The PCR primers were designed using Primer Premier
5.0, according to the mRNA sequence of RPS23 gene from H.
sapiens (NM_001025), B. Taurus (NM_001034690), M.
musculus (NM_024175), S. scrofa (NM_213764) and R.
norvegicus (NM_078617), as follows: RPS23 forward,
5′-AGGGAATTCATGGGTAAGTGTC-3′ and reverse,
5′-ATTCTCGAGTTATGATCTTGGC-3′.
Total RNAs were synthesized into first-stranded
cDNAs using a reverse transcription kit with oligo(dT) as the
primers, according to the manufacturer’s instructions (Promega,
Madison, WI, USA). Twenty microliters of the first-strand cDNA
synthesis reaction system were included in 1 mg total RNAs, 5 mM
MgCl2, 1 mM dNTPs, 0.5 mg oligo(dT)15, 10 U/ml RNase
inhibitor and 15 units of AMV reverse transcriptase, and incubated
at 42°C for 60 min. The first-strand cDNA synthesized was used as a
template. The total reaction volume for DNA amplification was 25
μl. Reaction mixtures contained 1.5 mM MgCl, 200 μM of each dATP,
dGTP, dCTP and dTTP (Omega, China), 0.3 μM of each primer, 5.0
units of Taq plus DNA polymerase (Sangon Co., Ltd.). DNA
amplification was performed using an MJ Research PTC-200 Thermal
Cycler (Watertown, MA, USA) with a program of 4 min at 94°C,
followed by 30 cycles of 1 min at 94°C, 0.5 min at 48°C and 1.5 min
at 72°C, with the final extension for 10 min at 72°C. Following
amplification, PCR products were separated by electrophoresis on
1.5% agarose gel with 1X TAE (Tris-acetate-EDTA) buffer, stained
with ethidium bromide and visualized under ultraviolet (UV) light.
The expected fragments of PCR products were harvested and purified
from gel using a DNA harvesting kit (Omega) and stored at
−20°C.
Cloning and identification of the cDNA
sequence
The harvested PCR products were ligated into a
pMD19-T vector at 4°C for 12 h. The recombinant molecules were
transformed into E. coli complete cells (DH5α), and then
spread on the LB-plate containing 50 μg/ml ampicillin, 200 mg/ml
isopropyl-β-D-thiogalactopyranoside (IPTG) and 20 mg/ml X-gal.
Plasmid DNA was isolated and digested by PstI and
ScaII to verify the insert size. Plasmid DNA was sequenced
by Huada Zhongsheng Scientific Corporation (Beijing, China).
Cloning of the genomic sequence of
RPS23
The PCR primers used were the RPS23 forward
and reverse mentioned above. The genomic sequence of the
RPS23 gene was amplified using touchdown PCR under the
following conditions: 94°C for 30 sec, 62°C for 45 sec, 72°C for 4
min in the first cycle and the annealing temperature was decreased
by 0.5°C/cycle; after 20 cycles, the conditions were changed to
94°C for 30 sec, 52°C for 45 sec, 72°C for 4 min for an additional
20 cycles. The fragment amplified was also purified, ligated into
the clone vector and transformed into E. coli competent
cells. The recombinant fragment was then sequenced by Invitrogen
(Carlsbad, CA, USA).
Construction of the expression vector and
overexpression of recombinant RPS23
PCR fragment corresponding to the RPS23 polypeptide
was amplified from the RPS23 cDNA clone with the forward,
5′-TGGAATTCATGGGTAAGTG TCG-3′ (EcoRI) and reverse,
5′-TTAAGCTTTTATGA TCTTGGCC-3′ (HindIII) primers,
respectively. PCR was performed at 94°C for 3 min; 35 cycles of 30
sec at 94°C, 45 sec at 53°C and 1 min at 72°C; 10 min at 72°C. The
amplified PCR product was cut and ligated into the corresponding
site of the pET28a vector (Stratagene, La Jolla, CA, USA). The
resulting construct was transformed into the E. coli BL21
(DE3) strain (Novagen, Madison, WI, USA) and used for the induction
by adding IPTG at an OD600 of 0.6 and culturing further for 4 h at
37°C, using the empty vector-transformed BL21 (DE3) as a control.
The recombinant protein samples were induced after 0, 1, 1.5, 2,
2.5, 3 and 4 h, separated by SDS-PAGE and stained with Commassie
Blue R-250.
Purification of recombinant protein
RPS23
The acquired genetically modified recombinant
protein has a tag comprising of six histidines (His-tag), thus Ni
chelate affinity chromatography was conducted as described below.
The ultrasonic product was centrifuged to collect sediments, which
were then suspended with soluble binding buffer (20 mM Tris-HCl,
0.5 M NaCl, 10 mM imidazole, pH 7.9) until the inclusion body was
clean. This was followed by centrifugation and suspension of the
sediments with inclusion body binding buffer (20 mM Tris-HCl, 0.5 M
NaCl, 5 mM imidazole, urea 8 M, pH 7.9) on ice, until the inclusion
body was thoroughly dissolved. The supernatant was centrifuged and
transferred to the chromatography column with nickel. After the
above supernatant was completely transferred, it was left to stand
for 2 min so that the six His-tag and nickel in the padding were
fully combined. Inclusion body binding buffer of 15 times the
column volume was used to flush the column, in order to wash the
uncombined protein. The excurrent liquid was collected.
Subsequently, inclusion body elution buffer of 5 times the column
volume was used to wash the combined protein. The outflow liquid
was collected according to the amount of the column volume.
SDS-PAGE was then used to detect the effect of purification. The
concentration of recombinant protein was determined using Bradford
Protein Assay kits. To purify the elution protein, dialysis was
available. After 48 h of desalination, the purified protein with
10% glycerol was maintained at −20°C.
Animals
S180 tumor cells were maintained in the peritoneal
cavities of male Kunming strain mice obtained from the Institute of
Biochemistry and Molecular Immunology of North Sichuan Medical
College (NSMC) (Nanchong, China). Male Kunming strain mice,
weighing 25.0±1.0 g, purchased from NSMC, were housed 6/plastic
cage with wood chip bedding in an animal room with a 12-h
light/dark cycle at room temperature (25±2°C) and allowed free
access to standard laboratory diet (purchased from the Institute of
Biochemistry and Molecular Immunology, NSMC). The animal
experiments were conducted according to the ‘Guidelines for Animal
Experimentation’ of the NSMC.
Assessment of in vivo antitumor
activity
S180 tumor cells (3×106) were implanted
subcutaneously into the right hind groin of the male Kunming strain
mice. The mice were randomly allocated into 5 groups (n=6/group).
One day after inoculation, RPS23 was dissolved in distilled water
and administered intraperitoneally (i.p.) to mice at doses of
0.025, 0.05 and 0.1 μg/ml (S1, S2 and S3 groups, respectively).
Positive and negative controls were set for comparison. Positive
control mice (M group) were administered 0.2 ml mannatide (2 mg/ml)
and negative control mice (N group) were administered physiological
saline instead of the test solution. The mice were sacrificed after
2 weeks and the body weights were measured. Tumors, spleens and
livers were excised and the inhibitory ratio of tumor growth was
calculated using the formula: Inhibition ratio (%) = [(A−B)/A]
×100, where A and B were the average tumor weights of the negative
control and treated groups, respectively.
Histopathological and morphological
observations
After treating the mice with RPS23 as described
above, a portion of the tissues were cut into small sections, fixed
in Heidenhain’s Susa Fluid (HgCl2, 4.5 g; NaCl, 0.5 g;
distilled water, 80.0 ml; formalin, 20.0 ml; acetic acid, 4.0 ml;
trichloroacetic acid, 2.0 ml), stained with hematoxylin and eosin
(H&E), examined and photographed using an Olympus
microscope.
Data analysis
Sequence data were analyzed by GenScan software
(http://genes.mit.edu/GENSCAN.html).
Homology investigations of the giant panda RPS23 compared
with the gene sequences of other species was performed using BLAST
2.1 (http://www.ncbi.nlm.nih.gov/blast/). Open reading
frame (ORF) of the DNA sequence was searched using ORF Finder
software (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Molecular
weight (Mw) and isoelectric point (pI) values were computed using
the Compute pI/Mw tool (http://www.expasy.org/tools/pi_tool.html). The protein
structure of the RPS23 sequence cloned was analyzed using
PredictProtein software (http://cubic.bioc.columbia.edu/predictprotein/).
Multiple Sequence Alignment was performed by DNASTAR Lasergene and
DNAman 6.0 software.
Results and Discussion
Analysis of RPS23 cDNA from the giant
panda
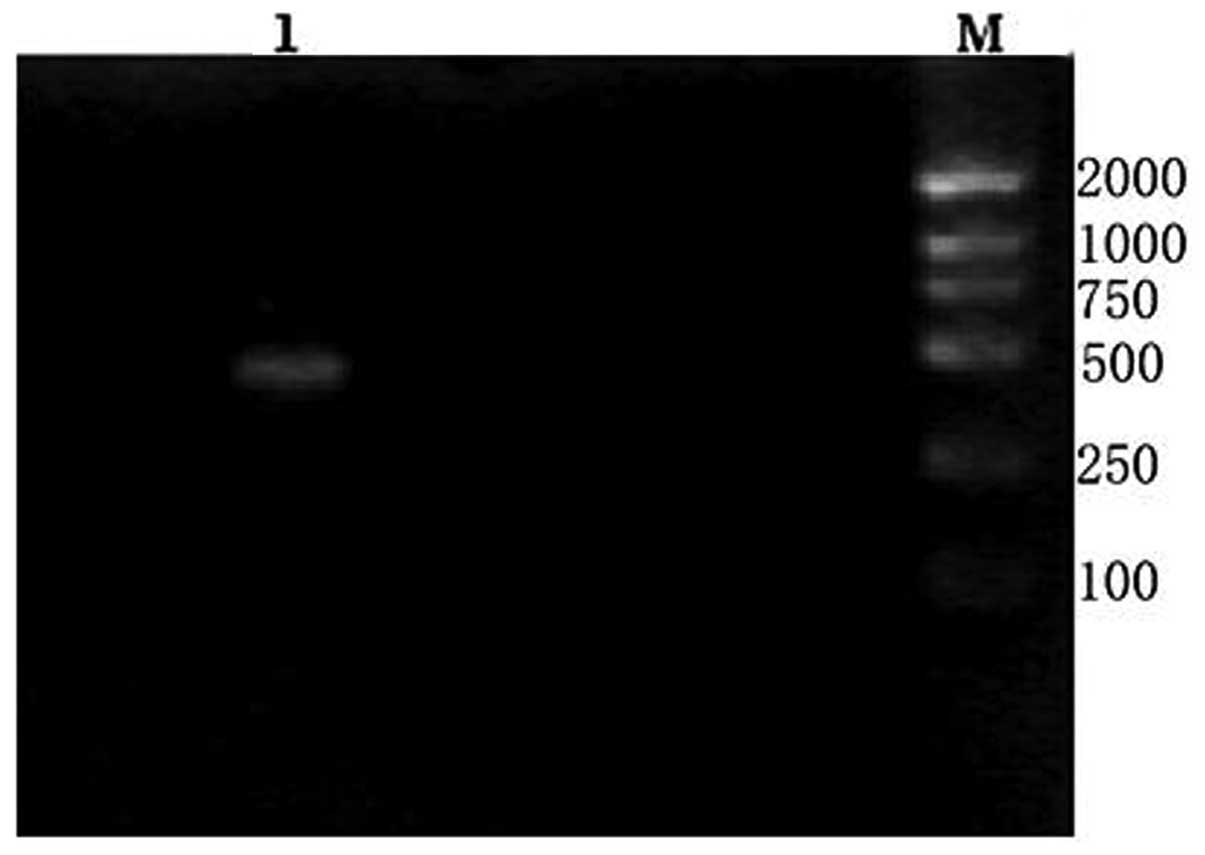
Approximately 500 bp of the cDNA fragment was
amplified from the giant panda. The length of the cDNA cloned was
472 bp (Fig. 1). On the basis of
the high identity, we concluded that the cDNA isolated is the cDNA
encoding the giant panda RPS23 protein. The RPS23 sequence
was submitted to GenBank (accession no. HQ318026). The 472 bp of
the giant panda RPS23 sequence contains a 20-bp
5′-untranslated sequence and a 20-bp 3′-unstranslated region. An
ORF of 432 bp encoding 142 amino acids was found in the cDNA
sequence (Fig. 2). Alignment
analysis of RPS23 from the giant panda and those of H.
sapiens, M. musculus, R. norvegicus and M.
mulatta, indicated that both the nucleotide and the deduced
amino acid sequences are highly conserved. There was no deletion or
insertion of nucleotide and amino acid residues. As determined by
BLAST analysis, the nucleotide sequence RPS23 cloned from the giant
panda shares a high degree of homology with those of H.
sapiens, B. taurus, R. norvegicus, M.
musculus and S. scrofa at 92.59, 92.82, 88.66, 88.43 and
93.29%, respectively. The homologies for amino acid sequences were
all 100%, with the exception of S. scrofa, which was 99.30%,
compared with the remaining 6 species (Table I). This marked pattern of
evolutionary conservation is considered reasonable, since ribosomal
protein genes are a group of highly conserved housekeeping genes.
The average levels of the base sequence was: A, 29.2; C, 20.6; G,
27.5; and T, 22.7%.
 | Table IComparison of nucleotide, amino acid
sequences and physicochemical properties of RPS23 between A.
melanoleuca and other five mammal species. |
Table I
Comparison of nucleotide, amino acid
sequences and physicochemical properties of RPS23 between A.
melanoleuca and other five mammal species.
| Species | | |
|---|
| | |
|---|
| Characteristics | H.
sapiens | B. taurus | R.
norvegicus | M.
musculus | S. scrofa |
|---|
| CDS similarity
(%) | 92.59 | 92.82 | 88.66 | 88.43 | 93.29 |
| aa similarity
(%) | 100 | 100 | 100 | 100 | 99.30 |
| Molecular weight
(kDa) | 15.80 | 15.80 | 15.80 | 15.80 | 15.81 |
| pI | 11.23 | 11.23 | 11.23 | 11.23 | 11.23 |
Analysis of the genomic sequence of RPS23
from the giant panda
A fragment of ~2,000 bp was amplified from the
genonic DNA of the giant panda using RPS23 forward and
reverse primers (Table II). The
length of the DNA fragment cloned was 2,105 bp. Comparison between
the cDNA sequence and this DNA fragment indicated that the cDNA
sequence is a full cDNA corresponding to 4 exons in the
RPS23 genomic sequence of the giant panda. The genomic
sequence of the RPS23 gene has been submitted to GenBank
(accession no. HQ_318027).
 | Table IIComparison of RPS23 genomic sequences
among 6 mammal species. |
Table II
Comparison of RPS23 genomic sequences
among 6 mammal species.
| Species | Length (bp) | No. of exons | Join sites in the
CDS | Accession nos. |
|---|
| A.
melanoleuca | 2,105 | 4 |
21..24,499..658,1729..1849,1959..2105 | HQ_318027 |
| H.
sapiens | 5,097 | 4 |
94..97,565..724,1899..2019,2162..2308 | NC_000005.9 |
| B.
taurus | 1,860 | 4 |
32..35,487..646,1320..1440,1559..1705 | NC_007305.4 |
| M.
musculus | 1,611 | 4 |
75..78,466..625,1205..1325,1416..1562 | NC_000079.5 |
| R.
norvegicus | 1,571 | 4 |
40..43,447..606,1154..1274,1386..1532 | NC_005101.2 |
| S.
scrofa | 1,645 | 4 |
8..11,454..613,1257..1377,1494..1640 | NC_010444.2 |
Prediction and analysis of protein
functional sites in the RPS23 protein of the giant panda
The molecular weight of the putative RPS23 protein
was 15.80 kDa with a theoretical pI of 11.23, containing 37
positively charged (Arg, Lys and His), 11 negatively charged (Asp
and Glu) and 95 uncharged amino acid residues. Among them, Lys was
the optimum, and Trp was the least. Topology prediction showed
there are 7 different patterns of functional sites: two
N-myristoylation sites, one N-glycosylation site, two protein
kinase C phosphorylation sites, one cAMP- and cGMP-dependent
protein kinase phosphorylation site, three amidation sites, one
tyrosine kinase phosphorylation site and one ribosomal protein S12
signature site in the RPS23 protein of the A. melanoleuca.
Comparison was conducted for functional sites of the RPS23 amino
acid sequences among the different mammals, including H.
sapiens, B. taurus, M. musculus, R.
norvegicus and S. scrofa (Fig. 3). There was no deletion and
insertion of nucleotide and amino acid residues observed. Further
analysis indicated that there were polymorphic sites in these
deduced amino acid sequences of RPS23 proteins. These polymorphic
sites were located irregularly in the amino acid sequences all of
which resulted from the transversion or transition of the
corresponding codons without any deletion and insertion of bases.
Additionally, most base transitions of the gene coding sequence in
these mammals were synonymous mutations. These synonymous mutations
did not result in any changes in the corresponding DNA information
and did not change the amino acid sequence of the expressed
product. Consequently, the spatial structure of the corresponding
protein was not affected.
Although some functional sites were not conserved
since some polymorphic sites were located inside the functional
sites, while others were located outside the functional sites,
secondary and tertiary structure analysis showed that amino acid
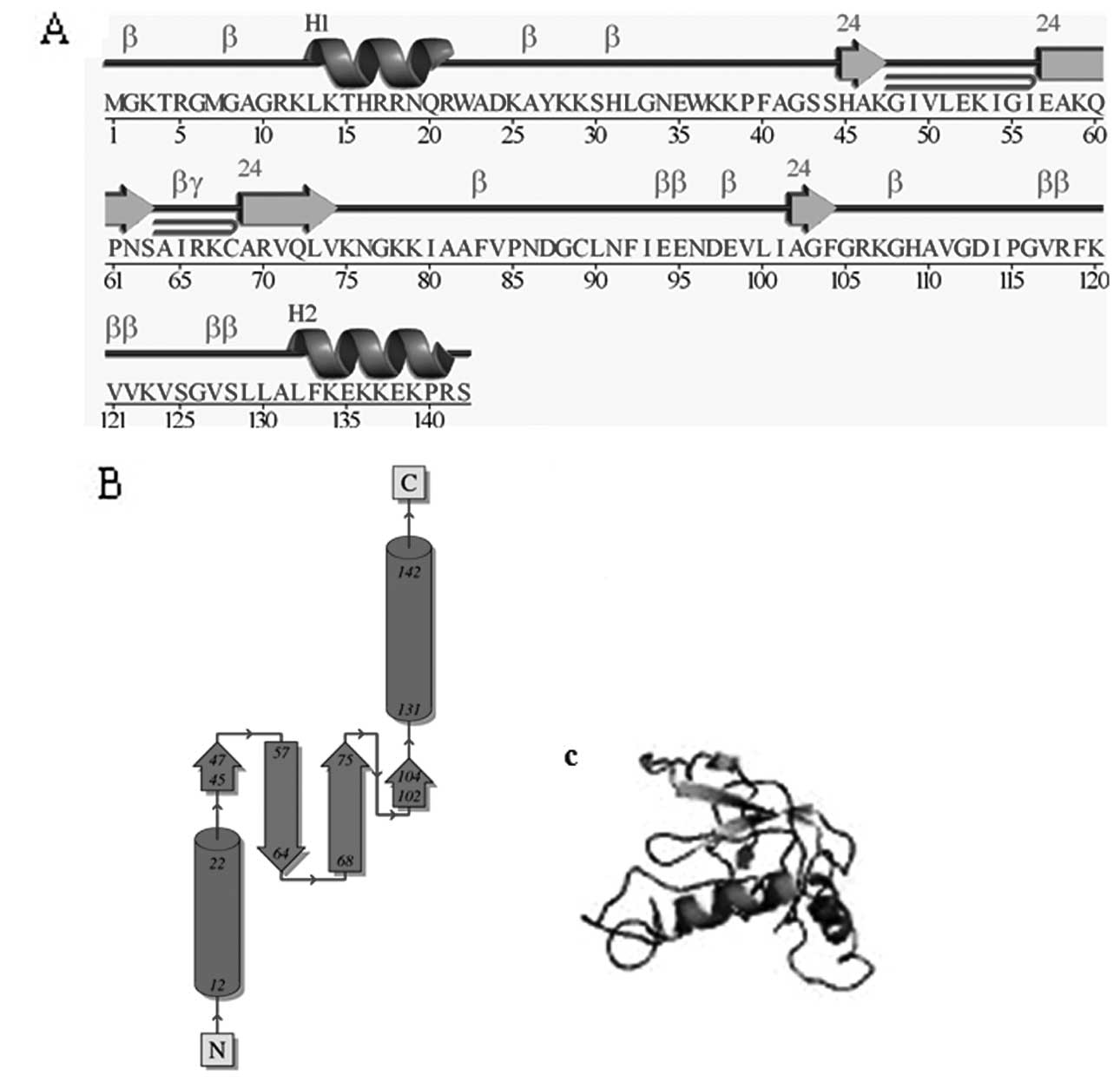
sequences formed the same main structures (Fig. 4). For RPS23, residues spanned from
8 to 134 to form the same main structure: 3 helices (8–15, 25–31,
129–134) and 5 β strands (49–59, 69–75, 80–85, 99–103, 119–125).
These results show that the variation of sites has no effect on the
structure and function of RPS23 protein. This finding may reflect
previous evolutionary association between these proteins and the
main, ancient structural and functional features of the ribosomal
protein (RPS) family.
Overexpression and purification of the
RPS23 gene in E. coli
The RPS23 gene was overexpressed in E.
coli, using pET28a plasmids carrying strong promoter and
terminator sequences derived from phage T7. For this purpose, the
RPS23 gene was amplified individually using PCR and cloned
in a pET28a plasmid, resulting in a gene fusion coding for a
protein bearing a His-tag extension at the N-terminal. Expression
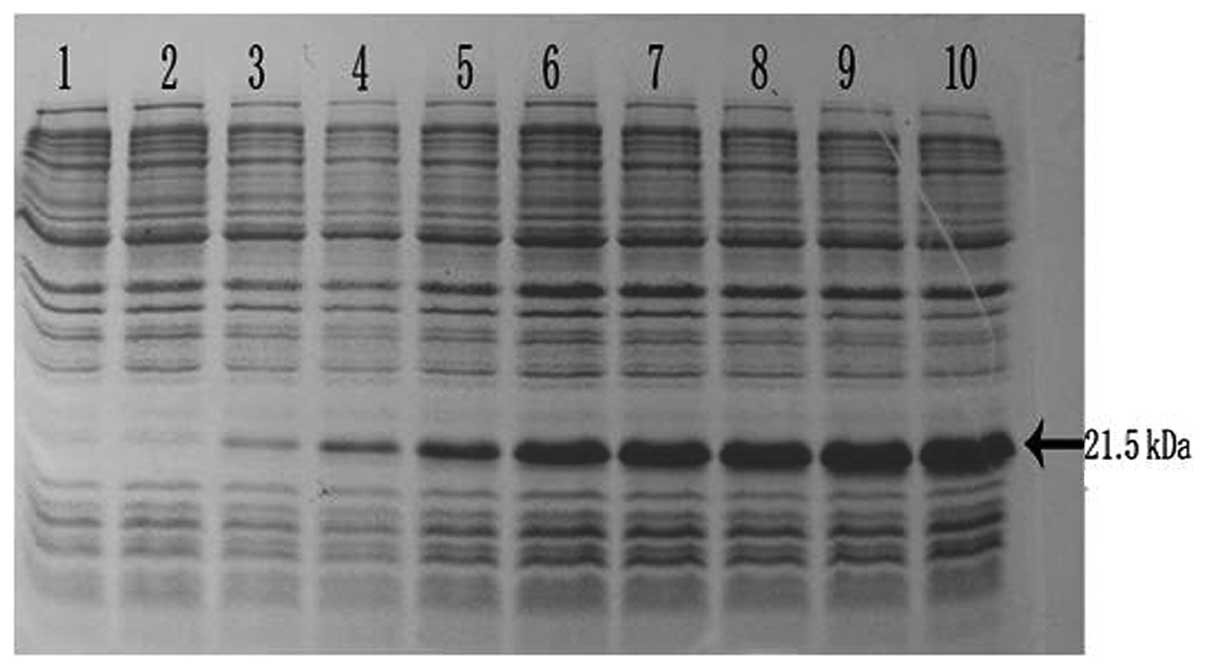
was assessed by SDS-PAGE analysis of protein extracts from
recombinant in E. coli BL21 strains (Fig. 5). These results indicate that the
protein RPS23 fusion with the N-terminal His-tag form
triggered the accumulation of an expected 21.5-kDa polypeptide that
formed inclusion bodies. The recombinant protein was expressed
after half an hour of induction and after 3 h reached the highest
level. The expression product obtained could be used for
purification and further investigation of its function. Under the
optimized expression conditions, a number of recombinant proteins
were obtained, which were then purified using Ni-NTA chelate
affinity chromatography. Protein separation and purification are
key steps in genetic engineering technology. Consequently, purified
protein was obtained through affinity chromatography. During
affinity chromatography, the pH of the protein solution was altered
twice which enabled us to achieve a highly purified protein.
Particularly, the protein solution initially went through the
column under acid conditions, and then finally outflowed from the
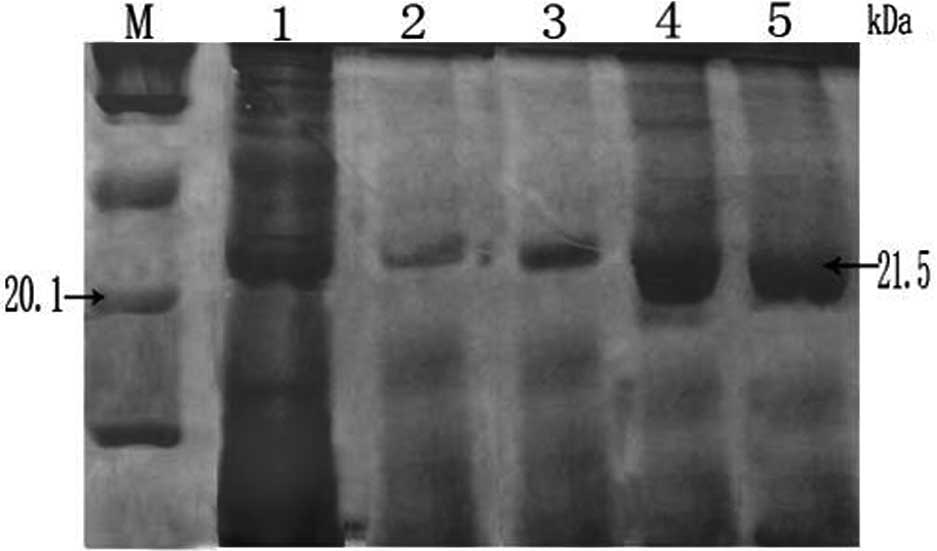
column by changing the pH of the effluent liquid. SDS-PAGE analysis
clearly indicated that there were ~21.5-kDa polypeptides in the
lanes 3–5 with a theoretical pI of 10.57 (Fig. 6). Size consistency of the purified
and unpurified RPS23 protein suggest that this is the only protein
encoded by the RPS23 gene from the giant panda.
Antitumor activity of RPS23 in vivo
The antitumor activity of RPS23 in vivo was
subsequently examined. Mice transplanted with S180 were used to
investigate the effects of RPS23 in vivo (Table I). The weight and histological
preparations of vital organs in each male mouse of the control
group were compared to those in the mice of the RPS23-treated
group. RPS23 was found to inhibit the growth of the tumors
(P<0.01) in a dose-dependent manner (Table III). The inhibitory rate of tumor
growth in mice treated with 0.1 μg/ml RPS23 was 49.45%, which was
the highest in the three doses used. Furthermore, during the
experiments, the appetite, activity and coat luster of each animal
in the RPS23-treated groups were improved compared with the mice
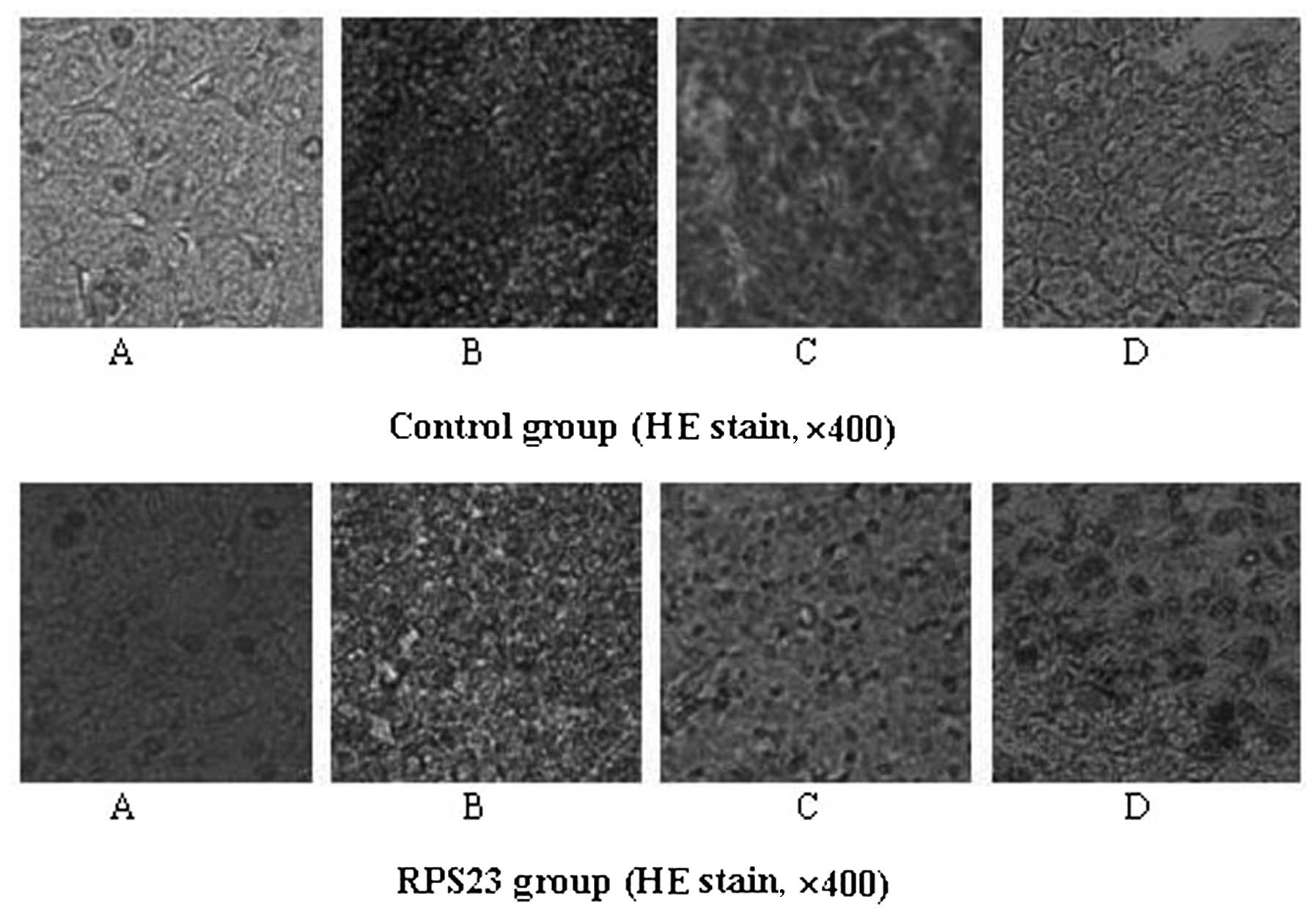
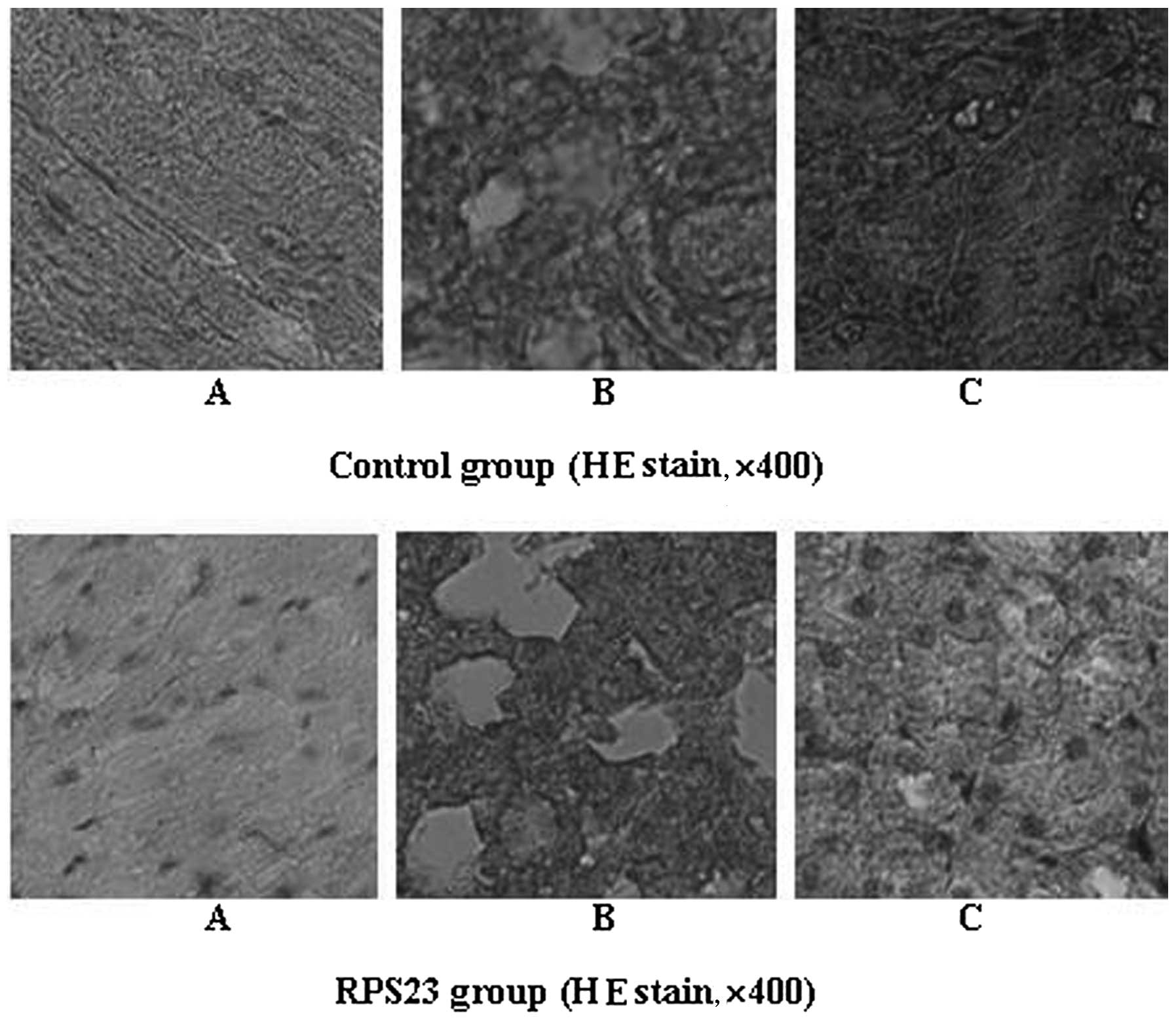
treated with mannatide. Histology of immune organs including liver,
spleen and thymus showed that tissues were characterized by regular
and tight arrangement, while the tumor tissue of the mice in the
RPS23-treated group exhibited a loose arrangement compared with the
control group. No obvious damage to other organs, such as the
heart, lung and kidney was evident (Figs. 7 and 8). The results also indicated a slight
change in the average liver weight in the mice of the test groups,
showing that RPS23 did not cause serious liver damage (Table III). On the 14th day, the average
tumor weight of negative control mice was 1.52 g, while it was 0.77
g in the mice of the RPS23-treated group (0.1 μg/ml). The average
tumor weight was also significantly reduced when doses of 0.05 and
0.025 μg/ml were used, resulting in 0.89 and 1.00 g, respectively.
Notably, the average weight of the spleens and thymus in the mice
of the test groups was significantly increased when a dose of 0.05
μg/ml was used compared with the negative control, and the
mannatide-treated mice. These results indicated that RPS23
increased the weight of immune organs when moderate doses were used
(Table III). Consequently,
activation of immune responses in the host might constitute one of
the mechanisms underlying the antitumor activity of RPS23. Studies
on the mechanism and signal transduction pathways of RPS23 are
currently being conducted.
 | Table IIIAntitumor activities of RPS23 on S180
tumors (mean ± SD, n=6/group). |
Table III
Antitumor activities of RPS23 on S180
tumors (mean ± SD, n=6/group).
| Group | Spleen index
(mg/g) | Liver index
(mg/g) | Thymus index
(mg/g) | Average tumor
weight (g) | Inhibitory rate of
tumor growth (%) |
|---|
| N | 5.74±2.18 | 60.30±9.35 | 1.32±0.71 | 1.52±0.36 | - |
| S1 | 7.61±2.14 | 55.46±4.50 | 4.72±0.31 | 1.00±0.74 | 34.36a |
| S2 | 7.87±3.27 | 65.31±8.82 | 7.71±3.23 | 0.89±0.30 | 41.32b |
| S3 | 5.02±0.89 | 57.54±4.76 | 0.90±0.41 | 0.77±0.28b | 49.45b |
| M | 7.39±3.05 | 56.23±5.39 | 2.84±1.76 | 0.29±0.14b | 81.03b |
Acknowledgements
This study was supported by the Application
Foundation Project of Sichuan Province (no. 2011JY0135), the
Foundation Project of Educational Committee of Sichuan Province
(no. 10ZC120), the National Natural Science Foundation of China
(no. 31200012) and the Doctor Startup Foundation Project of China
West Normal University (nos. 11B019 and 11B020).
References
|
1
|
Wool IG: Extraribosomal functions of
ribosomal proteins. Trends Biochem Sci. 21:164–165. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wool IG, Chan YL and Glück A: Structure
and evolution of mammalian ribosomal proteins. Biochem Cell Biol.
73:933–947. 1995. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fan W, Christensen M, Eichler E, Zhang X
and Lennon G: Cloning, sequencing, gene organization, and
localization of the human ribosomal protein RPL23A gene. Genomics.
46:234–239. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yang F and Liu WP: The progress of
ribosomal protein genes and human diseases. J Clin Exp Pathol.
20:354–356. 2005.
|
|
5
|
Elantak L, Wagner S, Herrmannová A,
Karásková M, Rutkai E, Lukavsky PJ and Valásek L: The indispensable
N-terminal half of eIF3j/HCR1 cooperates with its structurally
conserved binding partner eIF3b/PRT1-RRM and with eIF1A in
stringent AUG selection. J Mol Biol. 396:1097–1116. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Unbehaun A, Marintchev A, Lomakin IB,
Didenko T, Wagner G, Hellen CU and Pestova TV: Position of
eukaryotic initiation factor eIF5B on the 80S ribosome mapped by
directed hydroxyl radical probing. EMBO J. 26:3109–3123. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Huang X, Chen Y, Li WB, Cohen SN, Liao FF,
Li L, Xu H and Zhang YW: The Rps23rg gene family originated through
retroposition of the ribosomal protein s23 mRNA and encodes
proteins that decrease Alzheimer’s beta-amyloid level and tau
phosphorylation. Hum Mol Genet. 19:3835–3843. 2010.PubMed/NCBI
|
|
8
|
Anand P, Kunnumakkara AB, Sundaram C,
Harikumar KB, Tharakan ST, Lai OS, Sung B and Aggarwal BB: Cancer
is a preventable disease that requires major lifestyle changes.
Pharm Res. 25:2097–2116. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
10
|
Tolar J and Neglia JP: Transplacental and
other routes of cancer transmission between individuals. J Pediatr
Hematol Oncol. 25:430–434. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Croce CM: Oncogenes and cancer. N Engl J
Med. 358:502–511. 2008. View Article : Google Scholar
|
|
12
|
Wu GF, Hou YL, Hou WR, Song Y and Zhang T:
Giant pandaribosomal protein s14: cDNA, genomic sequence cloning,
sequence analysis, and overexpression. Genet Mol Res. 9:2004–2015.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Du YJ, Luo XY, Hao YZ, Zhang T and Hou WR:
Cloning and overexpression of acidic ribosomal phosphoprotein P1
gene (RPLP1) from the giant panda. Inter J Biol Sci. 3:428–433.
2007.PubMed/NCBI
|
|
14
|
Hou WR, Chen Y, Peng ZS, Wu X and Tang ZX:
cDNA cloning and sequences analysis of ubiquinol-cytochrome
c reductase complex ubiquinone-binding protein (QP-C) from
giant panda. Acta Theriol Sin. 27:190–194. 2007.
|
|
15
|
Hou YL, Du YJ, Hou WR, Zhou CQ, Hao YZ and
Zhang T: Cloning and sequence analysis of translocase of inner
mitochondrial membrane 10 homolog (yeast) gene (TIMM10) from the
giant panda. J Cell Anim Biol. 3:9–14. 2009.
|
|
16
|
Liao MJ, Zhu MY, Zhang ZH, Zhang AJ, Li GH
and Sheng FJ: Cloning and sequence analysis of FSH and LH in the
giant panda (Ailuropoda melanoleuca). Anim Reprod Sci.
77:107–116. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu ZA, Liu WX, Murphy C and Gall J:
Satellite 1 DNA sequence from genomic DNA of the giant panda
Ailuropoda melanoleuca. Nucleic Acids Res. 18:10541990.
View Article : Google Scholar : PubMed/NCBI
|