Introduction
Tuberculosis is a chronic infectious disease caused
by Mycobacterium tuberculosis (MTB) infection (1,2).
Although drugs are capable of effectively treating tuberculosis,
misuse or mismanagement of drug courses may lead to drug-resistant
tuberculosis (DR-TB) or multidrug-resistant tuberculosis (MDR-TB).
These drug-resistant disease forms are difficult to treat and
underlie a new global prevalence of tuberculosis (3,4). A
better understanding of MTB pathogenesis is required to ameliorate
this threat.
Two-component signal transduction systems (TCSSs)
are pathogen junction systems that monitor external environmental
stimuli and coordinate cellular responses, particularly the
modulation of gene expression. TCSSs are important bacterial
mechanisms for pathogenesis (5)
and typically consist of two proteins; a transmembrane histidine
kinase and a corresponding cytoplasmic response regulator. The MTB
genome encodes 30 TCSS proteins (6), several of which are strongly
associated with MTB drug resistance (4,5).
Thus, inhibiting TCSSs of resistant bacteria may be an effective
strategy to combat DR-TB and MDR-TB. SenX3/RegX3 was the first MTB
TCSS to be reported, where SenX3 is a transmembrane histidine
kinase and RegX3 is the corresponding DNA-binding response
regulator (6). SenX3 is associated
with MTB virulence and, thus, its investigation may further
elucidate the molecular mechanisms of MTB drug resistance (7–9).
In the present study, we purified the MTB strain
H37Rv recombinant SenX3 following expression in Escherichia
coli (E. coli) in order to provide reagents for the
future investigation of SenX3 and its role in MTB pathogenesis.
Materials and methods
Cloning of MTB strain H37Rv SenX3 into
pMD18-T
According to the published sequence (GenBank
accession no. Y13628.1), the primers were designed to incorporate
an NcoI restriction site upstream and an XhoI
restriction site downstream of SenX3 in the genomic DNA of
the MTB strain H37Rv, which was provided by Dr Wen-Tao Shen
(Chinese Academy of Tropical Agricultural Sciences, Hainan, China).
The primers were synthesized by Yingjun Life Technologies
(Shanghai, China) and the following sequences were used, with the
restriction sites underlined: P1, 5′-CA CCATGGCAACTGTGTTCTCGGCGCTGTTGC-3′
and P2, 5′-TCCTCGAGTCGGCTCAGCTCTTCCTCTCGTTG-3′.
SenX3 was amplified from the genomic DNA of the MTB strain
H37Rv by polymerase chain reaction (PCR) using Pyrobest DNA
polymerase (Takara Biotechnology, Dalian, China) as well as forward
and reverse primers. PCR cycling conditions were as follows: 94°C
for 2 min; 30 cycles at 94°C for 30 sec, 55°C for 30 sec, 72°C for
90 sec and 72°C for 10 min. The amplicon was separated by gel
electrophoresis. Correctly sized products were excised and purified
with a gel extraction kit (CoWin Biotech Co., Ltd., Beijing,
China). The purified and confirmed PCR product was ligated into the
pMD18-T vector using a DNA ligation kit (D6023; Takara
Biotechnology). The resulting pMD18-T-SenX3 construct was
transformed into E. coli DH5α-competent cells. The cells
were then cultured at 37°C overnight on LB-ampicillin (100 mg/l)
plates. Individual colonies were selected and screened by colony
PCR using a vector sequencing primer.
Site-directed mutagenesis of an internal
NcoI site in SenX3
Site-directed mutagenesis was used as previously
described (10) to mutate an
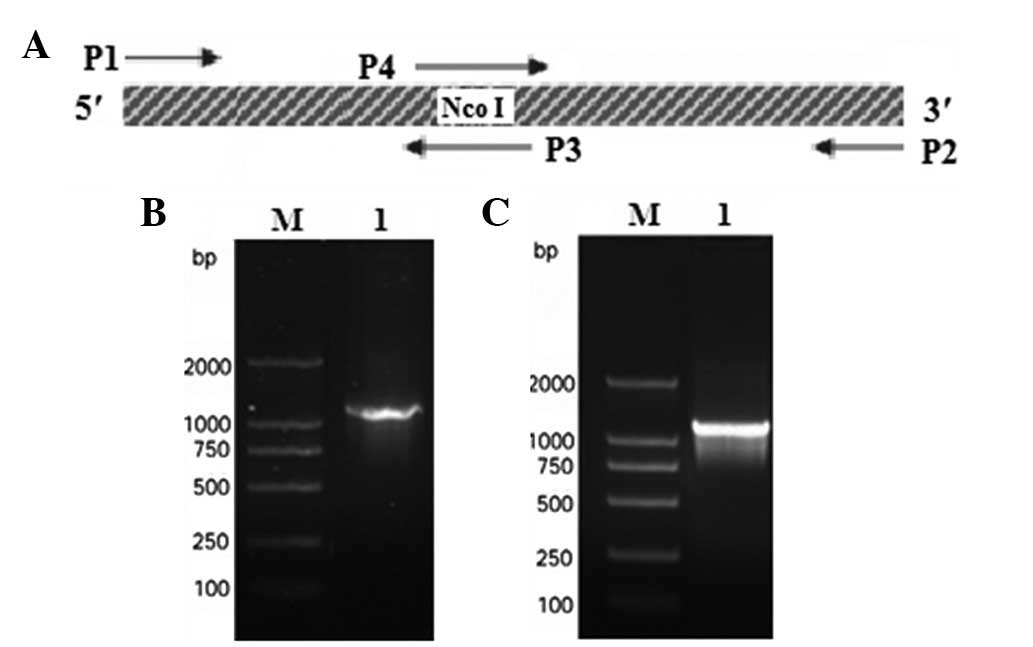
internal NcoI site within SenX3 (Fig. 1A). The primers were synthesized by
Yingjun Life Technologies and the sequences used were as follows:
P3, 5′-GAGTAGAGCCATCGCACCGACGGG-3′ and P4,
5′-CCCGTCGGTGCGATGGCTCTACTC-3′. The PCR cycling conditions for
P1–P3 primer amplification from pMD18-T-SenX3 were as follows: 94°C
for 2 min; 30 cycles at 94°C for 30 sec, 55°C for 30 sec, 72°C for
30 sec and 72°C for 10 min. The cycling conditions for P2–P4 primer
amplification from pMD18-T-SenX3 were as follows: 30 cycles of 94°C
for 30 sec, 55°C for 30 sec, 72°C for 35 sec and 72°C for 10 min.
The P1–P2 primers were then used to amplify a 1:1 mixture of the
P1–P3/P2–P4 PCR products in the following conditions: 94°C for 2
min; 30 cycles at 94°C for 30 sec, 55°C for 30 sec, 72°C for 90 sec
and 72°C for 10 min. The final PCR products were separated by gel
electrophoresis and purified with a gel extraction kit (CoWin
Biotech Co., Ltd.). The purified product was ligated back into
pMD18-T to yield pMD18-T-mSenX3, which was then transformed into
E. coli DH5α-competent cells (11). Individual colonies were screened by
colony PCR.
Cloning SenX3 into the prokaryotic
expression vector pET-28b
pMD18-T-mSenX3 and the prokaryotic expression vector
pET-28b(+) were digested with NcoI and XhoI
restriction endonucleases (Takara Biotechnology) to produce
compatible ends. The products were separated by gel
electrophoresis, purified with a gel extraction kit (CoWin Biotech
Co., Ltd.) and ligated together with T4 DNA ligase
(Takara Biotechnology) at 16°C overnight. The ligated vector was
transformed into E. coli DH5α-competent cells and grown
overnight on LB-kanamycin (50 mg/l) plates at 37°C. Individual
colonies were selected and the recombinant pET-28b-mSenX3 plasmid
was purified and confirmed by restriction digestion. Following
identification, pET-28b-mSenX3 was transformed into E. coli
BL21 (DE3) and grown overnight on LB-kanamycin (50 mg/l) plates at
37°C. Individual colonies were selected and confirmed by PCR with
P1 and P2 primers (11).
Expression of recombinant SenX3
Recombinant expression strains were inoculated in 20
ml of LB-kanamycin (50 mg/l) broth and grown at 37°C overnight.
Overnight cultures were diluted 1:100 in 50 ml of fresh
LB-kanamycin (50 mg/l) broth and cultured with agitation at 37°C
until the OD600 was 0.6–0.8. The cultures were then
divided: Protein expression was induced in one half with 0.1 mM
isopropyl β-D-thiogalactoside (IPTG) the other half was an
uninduced control. The cultures were induced at either 28 or 37°C
to optimize protein expression. For the analysis of protein
induction, 1-ml samples were collected after 2 h and centrifuged at
5,000 × g at room temperature for 5 min. Pellets were resuspended
in buffer [50 mM Tris-HCl, 5 mM EDTA, 100 mM NaCl (pH 8.0)] and the
cells were lysed by ultrasonic homogenization. The lysates were
centrifuged at 12,000 × g at 4°C for 10 min. The supernatant and
precipitate were collected, denatured at 100°C for 5 min and
analyzed by SDS-PAGE and Coomassie Blue.
Purification of recombinant SenX3
For protein purification, pellets were resuspended
in buffer [20 mM Tris-HCl, 150 mM NaCl (pH 7.4)] and the cells were
lysed by ultrasonic homogenization. Inclusion bodies were collected
by high-speed centrifugation (12,000 rpm) and dissolved in 8 M
carbamide buffer, followed by centrifugation at 12,000 rpm for 1 h.
The supernatant was removed and purified using Ni-NTA affinity
chromatography (Invitrogen Life Technologies, Carlsbad, CA, USA).
The proteins were eluted with 50, 150 or 250 mM imidazole and
fractions were collected and analyzed by SDS-PAGE to determine the
optimal elution conditions. Eluted proteins were diluted in
renaturation buffer [50 mM Tris-HCl, 200 mM carbamide, 1% glycerol,
0.5 mM EDTA (pH 7.9)], incubated at 4°C overnight and centrifuged
at 12,000 rpm for 10 min. Precipitates were removed and the
solution was concentrated by ultrafiltration.
Western blot analysis
Purified recombinant SenX3 was separated by SDS-PAGE
and transferred onto nitrocellulose membranes for western blot
analysis with an anti-6X His Tag primary antibody (CoWin Biotech
Co., Ltd.). Signals were detected by conjugation to a horseradish
peroxidase (HRP)-labeled goat anti-mouse IgG secondary antibody
(CoWin Biotech Co., Ltd.) and visualized with an
HRP-3,3′-diaminobenzidine tetrahydrochloride (DAB) substrate
chromogenic kit (CoWin Biotech Co., Ltd.).
Results
Site-directed mutagenesis to eliminate an
internal NcoI site within SenX3
In accordance with GenBank (accession no. Y13628.1),
PCR amplification of the genomic DNA of the MTB strain H37Rv
generated a 1,233-bp SenX3 DNA fragment (Fig. 1B). This product was cloned into the
pMD-18T vector to generate pMD18-T-SenX3, which was confirmed by
colony PCR identification and sequencing. Analysis of the
SenX3 fragment with Vector NIT 9 software revealed an
NcoI restriction site at 524–529 bp; to clone the
SenX3 fragment within the pET-28b(+) NcoI and
XhoI sites, the internal NcoI site within
SenX3 had to be destroyed by site-directed mutagenesis.
Overlapping PCRs were designed to induce a synonymous mutation into
the internal NcoI site (Fig.
1C). Diagnostic cloning and sequencing results demonstrated
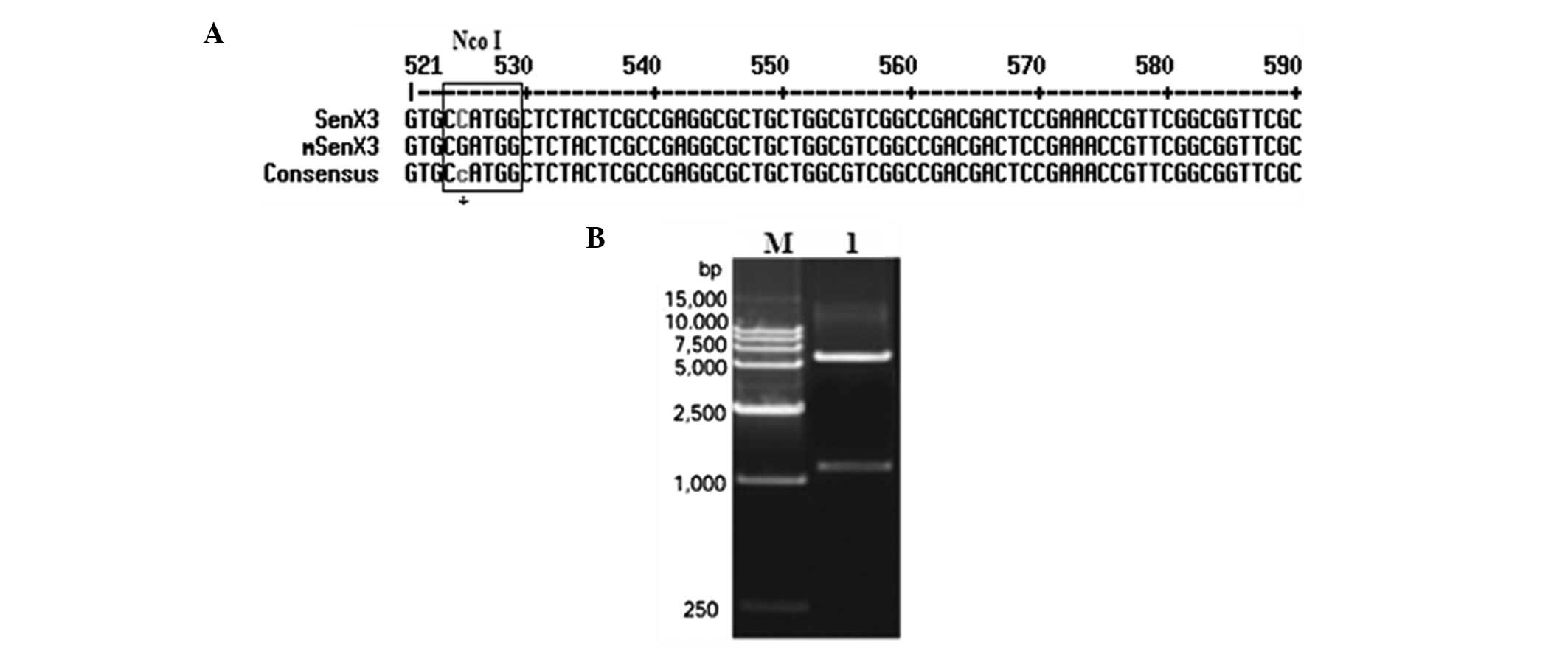
that the restriction site was mutated (Fig. 2A); the mutated vector was named
pMD18-T-mSenX3.
Identification of the prokaryotic
expression vector pET-28b-mSenX3
After mutating the internal NcoI site,
mSenX3 was digested from the pMD18-T-mSenX3 vector by double
digestion with NcoI and XhoI restriction
endonucleases. The isolated fragment was then ligated into the
prokaryotic expression vector pET-28b(+) following similar
digestion with NcoI and XhoI to create compatible
ends, generating pET-28b-mSenX3. Diagnostic digestion with
NcoI and XhoI confirmed this construct (Fig. 2B). Sequencing results also
confirmed that the open reading frame of mSenX3 was correct.
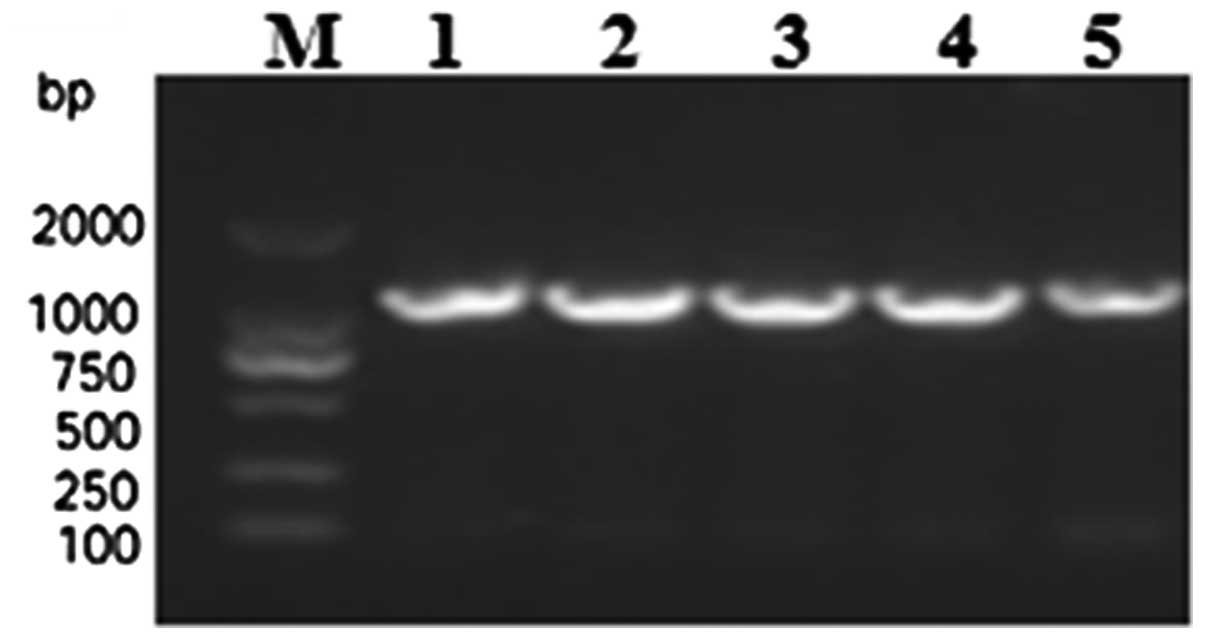
Finally, the confirmed pET-28b-mSenX3 vector was transformed into
E. coli BL21 (DE3) and confirmed by colony PCR
identification (Fig. 3).
Expression of recombinant SenX3
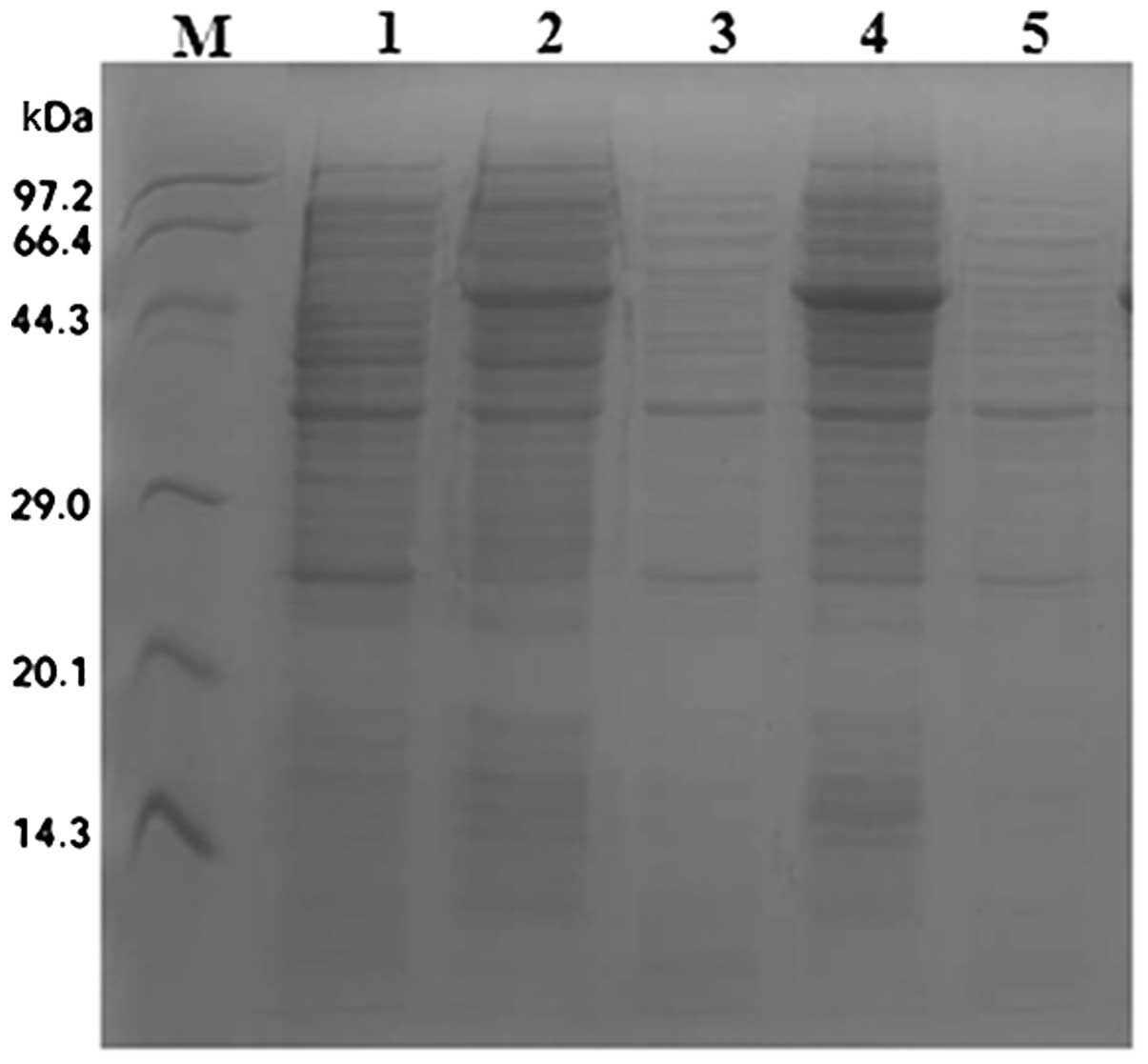
Expression of SenX3 was induced from E. coli
by treatment with IPTG, and soluble (supernatant) and insoluble
(precipitate) proteins were separated by ultrasonication. Analysis
by SDS-PAGE and Coomassie staining indicated the appearance of a
band ~46 kDa (expected size) following IPTG induction in the
precipitate lanes only (Fig.
4).
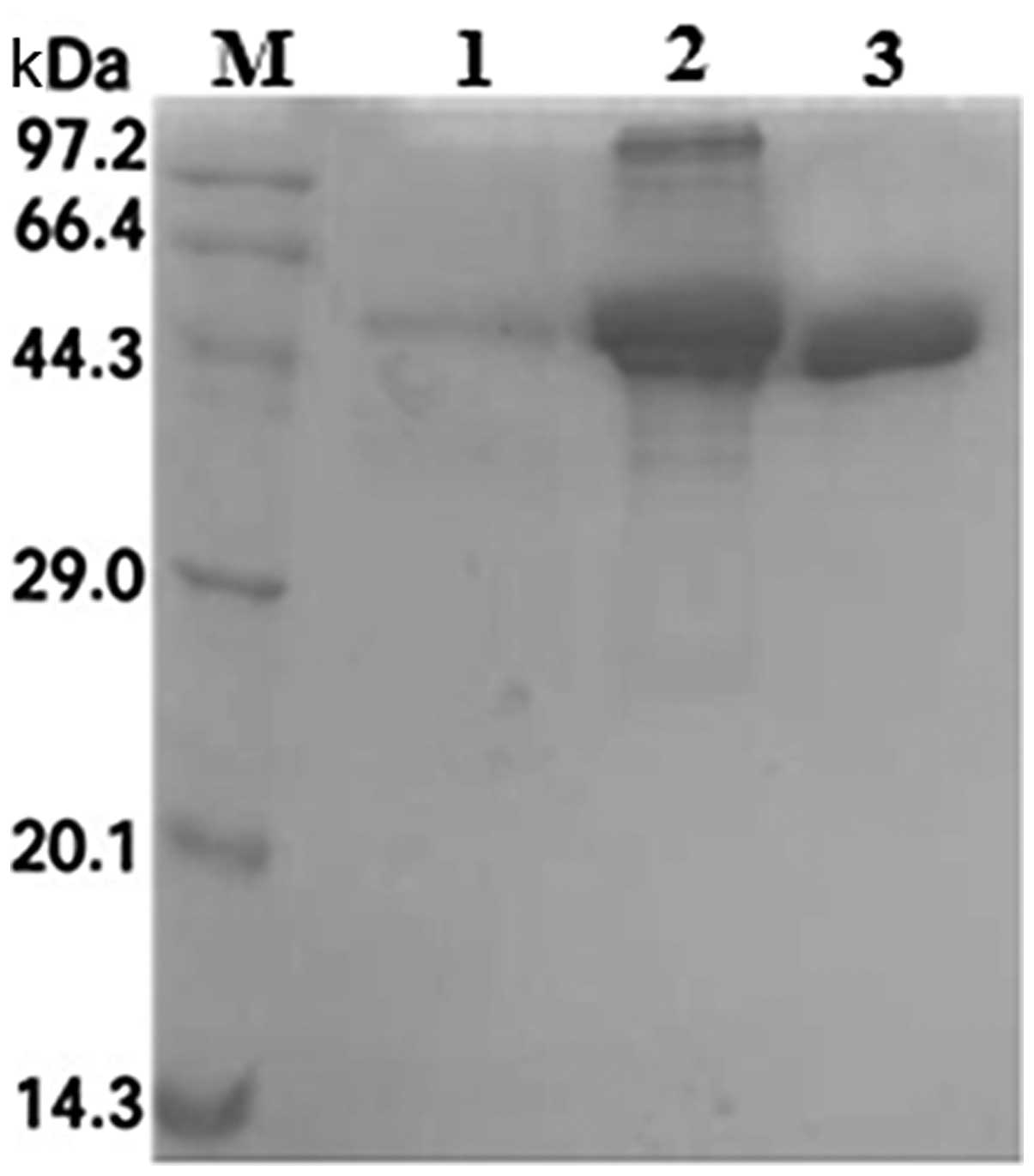
Purification of recombinant SenX3
Following induction, the ultrasonicated precipitate
was collected, solubilized and purified by Ni-sepharose affinity
chromatography, from which protein fractions were collected and
analyzed by SDS-PAGE. SenX3 was eluted at an optimal concentration
of 250 mM imidazole (Fig. 5).
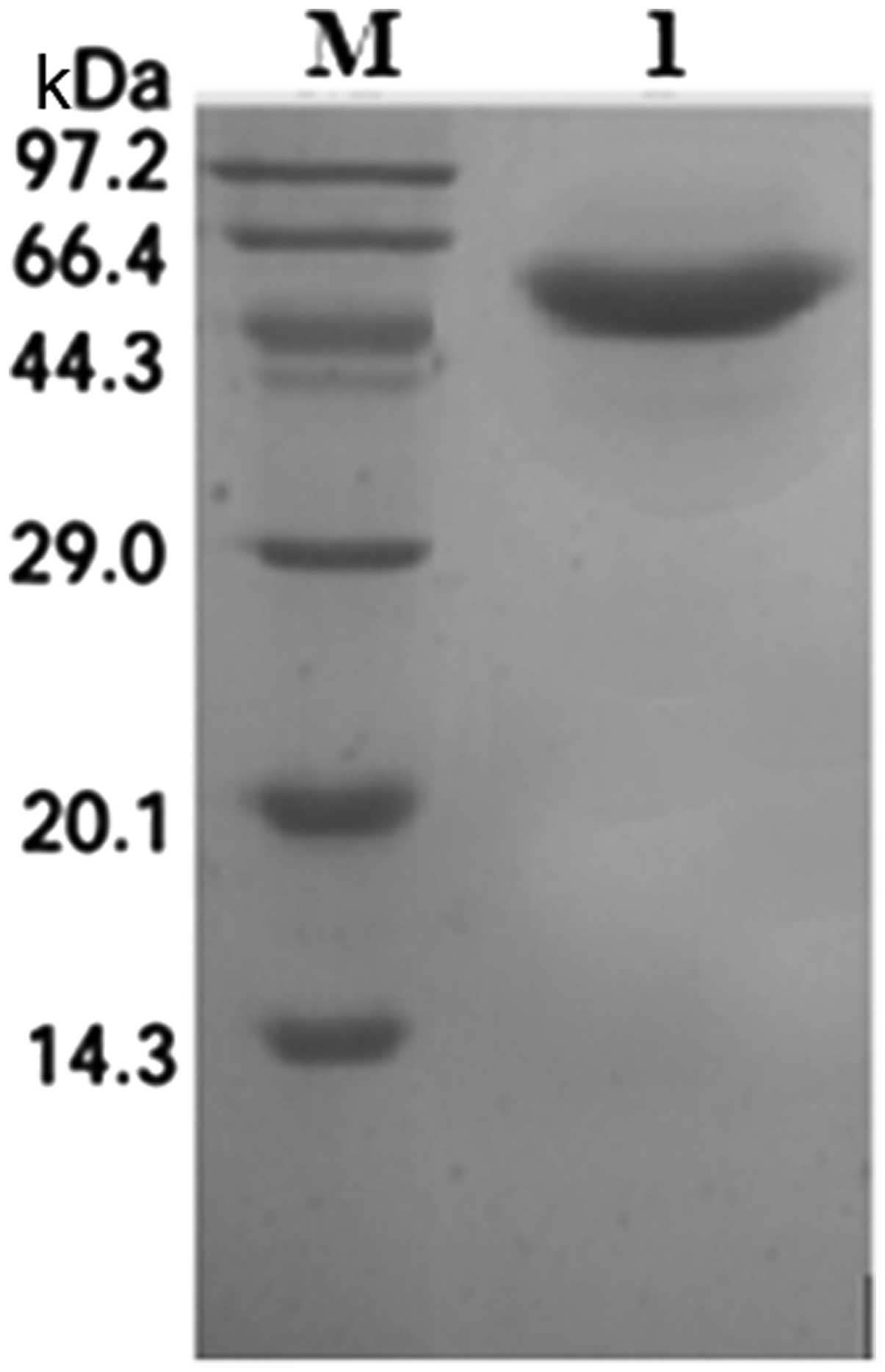
SDS-PAGE analysis and Coomassie staining indicated one band of the
expected size of 46 kDa (Fig. 6).
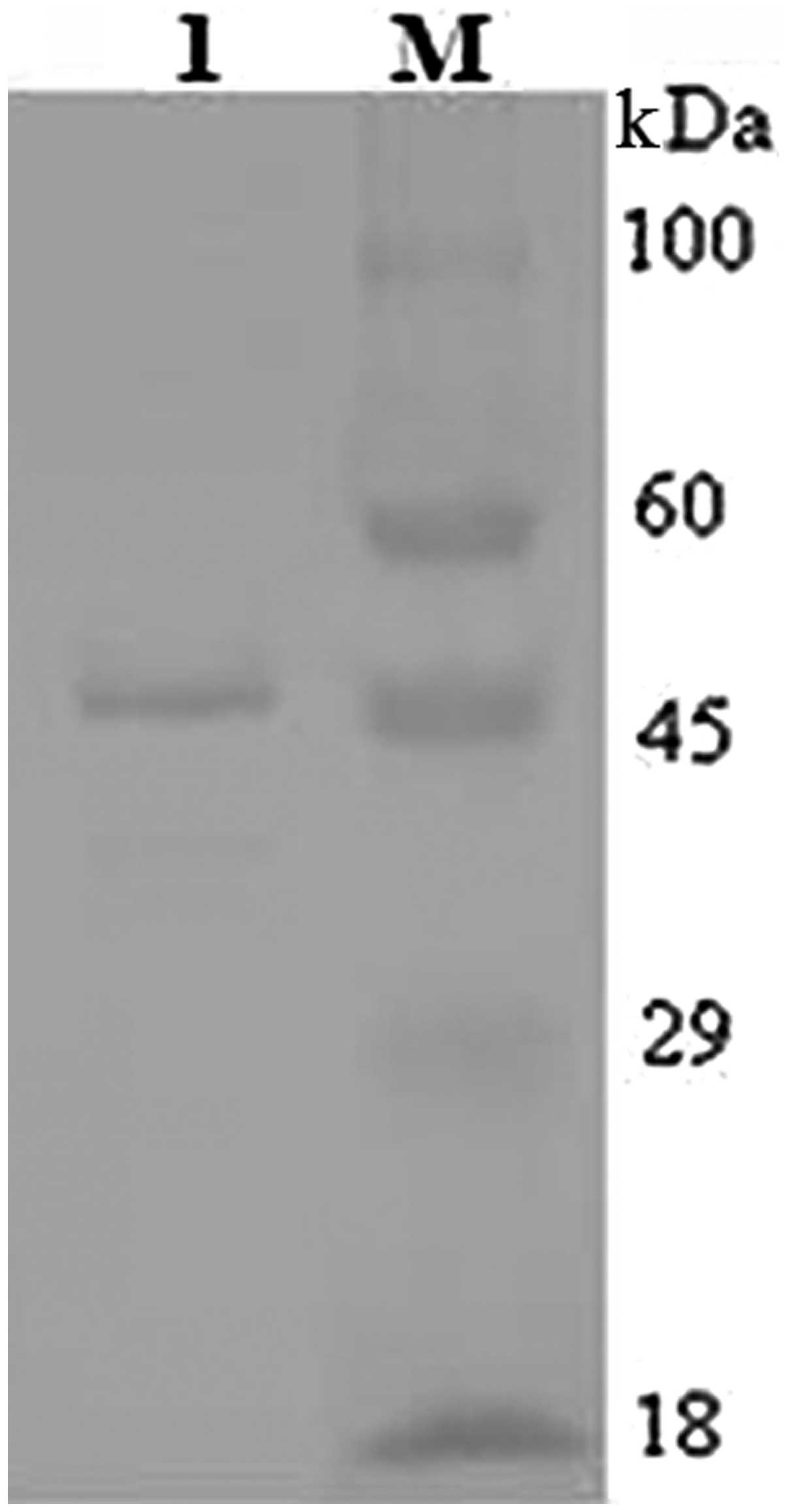
Western blot analysis with an anti-6X His Tag antibody also
detected a band of ~46 kDa (Fig.
7).
Discussion
SenX3/RegX3 is a TCSS transmembrane sensor that
coordinates pathogen environmental responses. Parish et
al(7) constructed MTB strains
mutated for SenX3/RegX3 that were shown to infect human THP-1
macrophage-like cells and to gradually grow in
γ-interferon-activated, bone marrow-derived murine macrophages.
These strains were also shown to infect the lungs of mice and
significantly decrease splenic function.
MTB strains mutated for either SenX3 or RegX3 have
been shown to exhibit decreased virulence similar to MTB strains
mutated for SenX3 and RegX3 (8),
suggesting that regulatory mechanisms of the SenX3/RegX3 signaling
pathway are important for MTB virulence. Furthermore, understanding
the functions of SenX3/RegX3 and additional MTB TCSS proteins is of
great significance for enabling the identification of the
underlying molecular mechanisms of MTB resistance, the clinical
treatment of tuberculosis, the identification of novel drugs and
controlling the prevalence of drug-resistant strains (12,13).
According to Himpens et al(13), cytoplasmic SenX3 from
Mycobacterium smegmatis was expressed using the pQE30
prokaryotic expression system. To the best of our knowledge, this
was the first study to report the exogenous expression of MTB
SenX3.
In the present study, PCR techniques were used to
clone the genomic sequence of MTB SenX3, and SenX3 was
purified from E. coli using affinity chromatography. The
results of the present study provide important data for the future
investigation of SenX3 and the molecular mechanisms of MTB
resistance.
Acknowledgements
This study was supported by the Zhejiang Provincial
Natural Science Foundation (grant no. Y2100445).
References
|
1
|
Maxmen A: TB drugs chalk up rare win.
Nature. 487:413–414. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
World Health Organization. Multidrug and
extensively drug-resistant TB (M/XDR-TB): 2010 global report on
surveillance and response. World Health Organization; Geneva,
Switzerland: 2010
|
|
3
|
Crunkhorn S: Trial watch: Novel
antimicrobial fights TB resistance. Nat Rev Drug Discov.
11:5902012. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kalokhe AS, Shafiq M, Lee JC, Ray SM, Wang
YF, Metchock B, Anderson AM and Nguyen ML: Multidrug-resistant
tuberculosis drug susceptibility and molecular diagnostic testing.
Am J Med Sci. 345:143–148. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bretl DJ, Demetriadou C and Zahrt TC:
Adaptation to environmental stimuli within the host: two-component
signal transduction systems of Mycobacterium tuberculosis.
Microbiol Mol Biol Rev. 75:566–582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cole ST, Brosch R, Parkhill J, et al:
Deciphering the biology of Mycobacterium tuberculosis from
the complete genome sequence. Nature. 393:537–544. 1998.
|
|
7
|
Parish T, Smith DA, Roberts G, Betts J and
Stoker NG: The senX3-regX3 two-component regulatory system of
Mycobacterium tuberculosis is required for virulence.
Microbiology. 149:1423–1435. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rickman L, Saldanha JW, Hunt DM, et al: A
two-component signal transduction system with a PAS
domain-containing sensor is required for virulence of
Mycobacterium tuberculosis in mice. Biochem Biophys Res
Commun. 314:259–267. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rifat D, Bishai WR and Karakousis PC:
Phosphate depletion: a novel trigger for Mycobacterium
tuberculosis persistence. J Infect Dis. 200:1126–1135. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
An Y, Ji J, Wu W, Lv A, Huang R and Wei Y:
A rapid and efficient method for multiple-site mutagenesis with a
modified overlap extension PCR. Appl Microbiol Biotechnol.
68:774–778. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Green MR and Sambrook J: Molecular
Cloning: A Laboratory Manual. 4th edition. Cold Spring Harbor
Laboratory Press; Cold Spring Harbor, New York: 2012
|
|
12
|
Carroll P, Faray-Kele MC and Parish T:
Identifying vulnerable pathways in Mycobacterium
tuberculosis by using a knockdown approach. Appl Environ
Microbiol. 77:5040–5043. 2011.PubMed/NCBI
|
|
13
|
Himpens S, Locht C and Supply P: Molecular
characterization of the mycobacterial SenX3-RegX3 two-component
system: evidence for autoregulation. Microbiology. 146:3091–3098.
2000.PubMed/NCBI
|