Introduction
Cleft lip with or without palate (CL/P) is one of
the most common birth defects, affecting 0.5–1% of newborns
worldwide. Nonsyndromic CL/P (NSCLP) accounts for 70% of CL/P
cases, and is genetically distinct from syndromic CL/P. NSCLP is a
complex disorder with a multifactorial etiology that involves
genetic and environmental factors (1). Alcohol consumption (2), maternal active smoking (3), passive smoking (4), anti-epileptic drugs (5) and radiation exposure (6) are considered to be environmental risk
factors for NSCLP; however, the biological mechanisms remain
unknown. A number of studies have shown that the risk of NSCLP is
associated with variants of genes that regulate detoxification
pathways (7).
The human arylamine N-acetyltransferases, NAT1 and
NAT2, are important xenobiotic-metabolizing enzymes involved in the
detoxification and metabolic activation of numerous drugs and
chemicals. NATs have been identified to transfer an acetyl group
from acetyl-CoA to the amino group of aryl hydrazines (including
isoniazid and hydralazine) or arylamines (including
p-aminosalicylate) (8). NAT1 and
NAT2 are enzymatically distinct, with certain aromatic amine or
hydrazine drugs being preferentially acetylated by NAT1 (e.g.,
p-aminosalicylic acid and p-aminobenzoic acid) and others by NAT2
(e.g., isoniazid and sulfamethazine). Human NAT1 is ubiquitously
expressed at various stages of development (9). NAT1 protein and/or mRNA have been
identified in every fetal and adult tissue examined to date, from
as early as the blastocyst stage of development (10); NAT2 activity has also been reported
in the placenta (11). Folic acid
is an essential vitamin in humans and is important in the
prevention of neural tube defects. Several studies have shown that
NATI is inhibited by folic acid (12). Folic acid is catabolized to
p-aminobenzoylglutamate (pABG), which is an endogenous substrate
for arylamine NAT (13).
Transgenics for human NAT1 and knockouts for NAT2 have been
investigated for susceptibility to Dilantin, hydrocortisone and
6-aminonicotinamide-induced orofacial clefting. Erickson et
al(14) found that Nat2
significantly influences teratogen-induced orofacial clefting.
The aim of the present study was to characterize
genetic variations in NAT1 and NAT2 genes, and to utilize data from
case-control studies to investigate the association of single
nucleotide polymorphisms (SNPs), haplotypes and gene-gene
interactions with the risk of NSCLP in the Chinese population.
Materials and methods
Study population
A total of 204 NSCLP patients and 226 normal
controls were included in this case-control study. Subjects in the
control group had no congenital malformations of the body and no
family history of genetic disease, were born in the same region as
the NSCLP patients and had a male:female ratio that was as similar
to that of the NSCLP group as possible. All the study participants
were recruited between January 2010 and January 2012 from the Cleft
Surgery Department of the Plastic Surgery Hospital, Chinese Academy
of Medical Sciences (Beijing, China). Informed consent was obtained
from each participant prior to enrollment in the study, and the
study was approved by the ethics committee of Plastic Surgery
Hospital, Chinese Academy of Medical Sciences, Beijing, China.
General characteristics including age, gender, ethnicity, health
status and birth defects were documented.
SNP identification and selection
Using the HapMap genome browser (http://www.hapmap.org/cgi-perl/gbrowse/hapmap3r2_B36),
based on the Han Chinese population in Beijing (CHB), eight tag
SNPs (r2 coefficient cut-off, 0.80; with a minor allele
frequency of 0.05) were selected to capture the NAT1 region of
chromosome 8. Five tag SNPs were selected for the NAT2 region of
chromosome 8.
Genotyping
Peripheral blood samples (2 ml) were collected from
each participant and frozen. Genomic DNA was extracted from a
200-μl aliquot of each sample using a Tiangen™ Genomic DNA kit
(Tiangen, Beijing, China) according to the manufacturer’s
instructions, and was stored at −70°C. All the SNPs were genotyped
using the Sequenom MassARRAY matrix-assisted laser
desorption/ionization time-of-flight mass spectrometry platform
(Sequenom, Inc., San Diego, CA, USA). Primers were designed using a
semi-automated method (Assay Design 3.1; Sequenom, Inc.); primer
sequences are available on request. The call rate for each assay
was set at >90%.
Statistical analysis
Data from the control and NSCLP groups were compared
and analyzed using SPSS 17.0 software (SPSS Inc., Chicago, IL,
USA), and HWE software (http://www.broad.mit.edu/mpg/haploview/) was used for
assessment of the Hardy-Weinberg equilibrium. Based on the
multivariable logistic regression method, the case-control
association of genotypes in three inheritance models (dominant,
recessive and additive) was examined, and these models were coded
as follows for genotypes AA, AB and BB, respectively (assuming B is
the minor allele): dominant 0 1 1 (AA vs. AB + BB), recessive 0 0 1
(AA + AB vs. BB) and additive 0 1 2 (trend test on B allele count).
The odds ratios (ORs) and 95% confidence intervals (CIs) were
provided. The association between each SNP and haplotype with NSCLP
was estimated using Haploview software (http://www.broad.mit.edu/mpg/haploview/). For
haplotype construction, genotype data from the case and control
groups were used to estimate intermarker linkage disequilibrium
(LD) by measuring pairwise D′ and r2, and defining LD
blocks. We used the CI method in the Haploview software to define
an LD block with an extended spine when r2 was 0.8.
P-values were corrected for multiple tests with 10,000
permutations. To identify higher order gene-gene interactions in
our samples, we used logistic regression analysis to calculate ORs
and 95% CIs to search for gene-gene interactions in the case and
control groups. The interactions between SNPs were classified in
four groups using a dominant model. In each case, a wild-type
genotype was evaluated as a reference. Since the research is
exploratory and Bonferroni corrections are too conservative,
Bonferroni corrections were not used.
Results
Study subject characteristics
A total of 13 SNPs were genotyped from NSCLP
patients and normal controls of a Han Chinese origin. The mean age
was 3.23±0.91 years old and the male:female gender ratio was 1:0.76
in the NSCLP group, while the mean age was 3.96±1.03 years old and
the male:female gender ratio was 1:0.70 in the control group. The
genomic position, nucleic acid composition and minor allele
frequencies of the SNPs are shown in Table I. The HWE was calculated for all
the SNPs; none of the SNPs among these groups deviated from the
HWE.
 | Table ICharacteristics of NAT1 and NAT2 gene
polymorphisms. |
Table I
Characteristics of NAT1 and NAT2 gene
polymorphisms.
| Gene symbol | rs number | SNP function | Location | Allelesa | MAF | P-HWE |
|---|
| NAT1 | rs2410545 | UTR-5 | Chr8 18068835 | a/G | 0.412 | 0.1809 |
| rs4921580 | Intron | Chr8 18071000 | C/g | 0.174 | 0.8829 |
| rs17693097 | Intron | Chr8 18072809 | c/G | 0.113 | 0.9299 |
| rs13278990 | Intron | Chr8 18073354 | a/T | 0.328 | 0.4586 |
| rs6586714 | Intron | Chr8 18073942 | a/G | 0.087 | 1 |
| rs17126350 | Intron | Chr8 18074322 | A/g | 0.047 | 0.7727 |
| rs4921880 | Intron | Chr8 18075906 | a/T | 0.45 | 0.0651 |
| rs8190845 | Intron | Chr8 18078628 | a/G | 0.037 | 0.218 |
| NAT2 | rs11780272 | Intron | Chr8 18249652 | c/T | 0.033 | 1 |
| rs11996129 | Intron | Chr8 18254575 | c/T | 0.224 | 0.4344 |
| rs1961456 | Intron | Chr8 18255709 | a/G | 0.297 | 0.8645 |
| rs1041983 | cds-synon | Chr8 18257795 | C/t | 0.385 | 0.6728 |
| rs1799931 | Missense | Chr8 18258370 | a/G | 0.159 | 1 |
Single SNP analysis
To evaluate the association between genetic variants
and the risk of NSCLP, we compared NAT1 and NAT2 genotype frequency
distributions in the NSCLP and control groups (Table II). For NAT1, there was a
significant difference in the allele frequency of rs4921580 between
the NSCLP and control groups (P=0.0128); however, no significant
difference was identified following a correction with 10,000
permutations (P=0.0872). The dominant model and Cochran-Armitage
trend tests of rs4921580 demonstrated a significant difference
(P=0.010 and 0.034, respectively). For NAT2, a Cochran-Armitage
trend test of rs1041983 remained significant (P=0.040) between the
NSCLP and control groups.
 | Table IIAssociation of polymorphisms of NAT1
and NAT2 genes with nonsyndromic cleft lip and palate (NSCLP). |
Table II
Association of polymorphisms of NAT1
and NAT2 genes with nonsyndromic cleft lip and palate (NSCLP).
| | | | | Genotypeb | Dominant model | Recessive model | |
|---|
| | | | |
|
|
| |
|---|
| Gene symbol | rs number | Allelea | P-value |
Ppermutation | Patients | Control | OR | 95% CI | P-value | OR | 95% CI | P-value |
PCochran-Armitage trend
test |
|---|
| NAT1 | rs2410545 | a/G | 0.3829 | 0.9495 | 66/113/24 | 74/108/41 | 1.031 | 0.688–1.545 | 0.883 | 0.595 | 0.345–1.026 | 0.062 | 0.130 |
| rs4921580 | C/g | 0.0128 | 0.0872 | 126/71/7 | 166/55/5 | 1.713 |
1.138–2.577 | 0.010 | 1.571 | 0.491–5.028 | 0.447 | 0.034 |
| rs17693097 | c/G | 0.2822 | 0.8706 | 156/45/3 | 183/40/3 | 1.309 | 0.824–2.082 | 0.254 | 1.109 | 0.221–5.559 | 0.899 | 0.516 |
| rs13278990 | a/T | 0.4863 | 0.9828 | 96/87/21 | 102/95/29 | 0.925 | 0.633–1.353 | 0.689 | 0.780 | 0.429–1.416 | 0.413 | 0.708 |
| rs6586714 | a/G | 0.408 | 0.9685 | 166/37/1 | 192/32/2 | 1.293 | 0.778–2.147 | 0.321 | 0.552 | 0.050–6.130 | 0.628 | 0.481 |
| rs17126350 | A/g | 0.3269 | 0.9252 | 182/22/0 | 208/18/0 | 1.397 | 0.726–2.686 | 0.316 | | | | 0.315 |
| rs4921880 | a/T | 0.9562 | 1 | 60/104/40 | 40/129/37 | 0.867 | 0.569–1.323 | 0.509 | 1.246 | 0.761–2.041 | 0.383 | 0.432 |
| rs8190845 | a/G | 0.5117 | 0.9876 | 188/15/1 | 212/13/1 | 1.289 | 0.613–2.711 | 0.504 | 1.108 | 0.069–17.835 | 0.942 | 0.795 |
| NAT2 | rs11780272 | c/T | 0.1527 | 0.4672 | 187/17/0 | 215/11/0 | 1.777 | 0.812–3.889 | 0.150 | | | | 0.146 |
| rs11996129 | c/T | 0.2485 | 0.627 | 124/74/5 | 129/81/13 | 0.874 | 0.593–1.288 | 0.497 | 0.408 | 0.143–1.165 | 0.094 | 0.219 |
| rs1961456 | a/G | 0.626 | 0.9785 | 101/88/15 | 112/89/24 | 1.011 | 0.692–1.477 | 0.956 | 0.665 | 0.338–1.305 | 0.236 | 0.443 |
| rs1041983 | C/t | 0.7754 | 0.9943 | 71/111/22 | 89/98/39 | 1.217 | 0.822–1.802 | 0.327 | 0.580 | 0.331–1.016 | 0.057 | 0.040 |
| rs1799931 | a/G | 0.3504 | 0.7848 | 140/58/6 | 164/57/5 | 1.209 | 0.798–1.833 | 0.370 | 1.339 | 0.403–4.457 | 0.634 | 0.647 |
Haplotype analysis
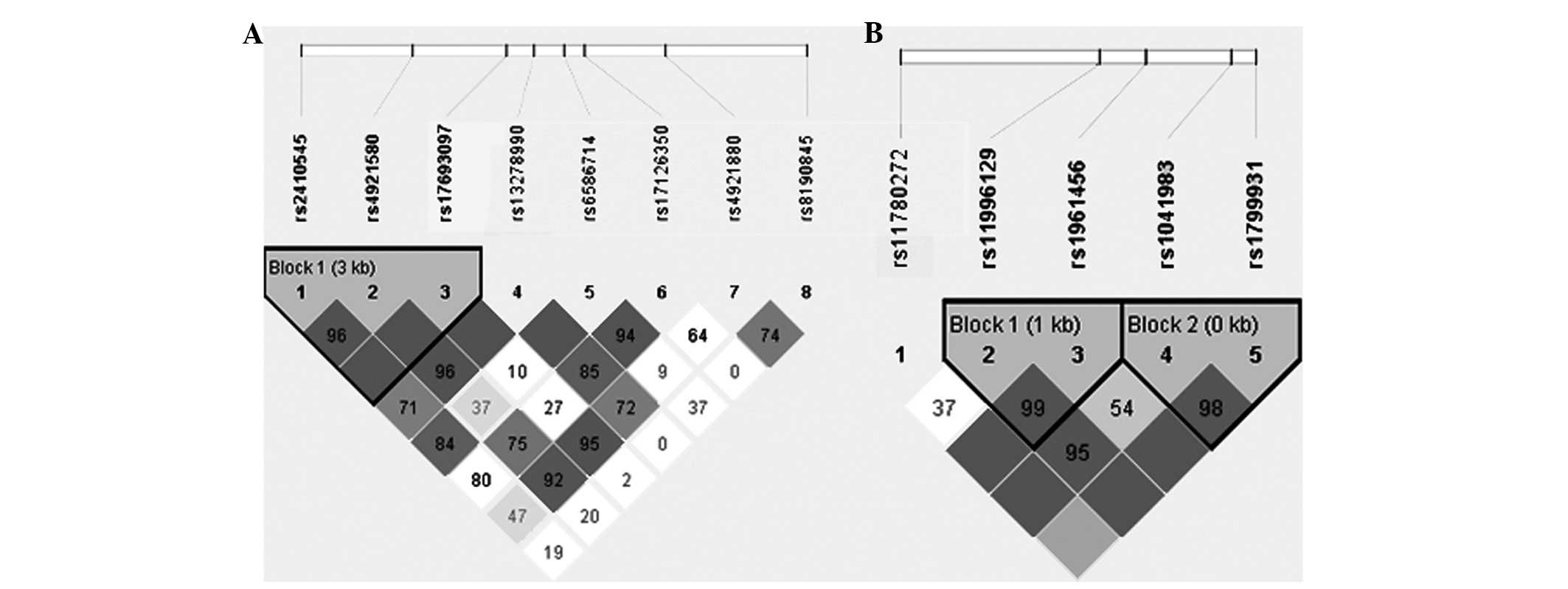
For NAT1, the Haploview program revealed that
rs2410545, rs4921580 and rs17693097 were in the same LD block
(Fig. 1A). Haplotype analysis of
polymorphisms in the NAT1 gene showed that SNP combinations were
associated with the risk of NSCLP (Table III). The GGG haplotypes were
shown to have an increased risk of NSCLP (OR, 2.832; 95% CI,
1.437–5.584; P=0.0093), which remained significant after correction
with 10,000 permutations (P=0.0191). For NAT2, rs11996129 and
rs1961456 were in the same LD block (block 1), while rs1041983 and
rs1799931 were also in the same LD block (block 2; Fig. 1B; Table III). There was no difference at
block 1 or block 2 between the NSCLP and controls groups.
 | Table IIIHaplotype analysis of NAT1 and NAT2
gene polymorphisms. |
Table III
Haplotype analysis of NAT1 and NAT2
gene polymorphisms.
| | Frequency | | 95% CI | | |
|---|
| |
| |
| | |
|---|
| Gene symbol | Haplotype | Case | Control | OR | 95% Low | 95% High | Nominal
P-value | Permuted
P-value |
|---|
| NAT1 | GCG | 0.399 | 0.436 | 1 | Ref | Ref | 0.2735 | 0.5209 |
| ACG | 0.393 | 0.421 | 1.041 | 0.7731 | 1.401 | 0.41 | 0.7265 |
| GGC | 0.125 | 0.102 | 1.349 | 0.8599 | 2.115 | 0.2822 | 0.53 |
| GGG | 0.080 | 0.039 | 2.832 | 1.437 | 5.584 | 0.0093 | 0.0191 |
| NAT2 |
| Block 1 | TG | 0.708 | 0.694 | 1 | Ref | Ref | 0.6573 | 1.0000 |
| CA | 0.205 | 0.24 | 0.8394 | 0.6039 | 1.167 | 0.2217 | 0.5009 |
| TA | 0.084 | 0.066 | 1.262 | 0.7496 | 2.125 | 0.3026 | 0.6667 |
| Block 2 | CG | 0.620 | 0.608 | 1 | Ref | Ref | 0.7276 | 1.0000 |
| TG | 0.209 | 0.244 | 0.8367 | 0.6011 | 1.165 | 0.2223 | 0.5046 |
| TA | 0.171 | 0.146 | 1.157 | 0.7929 | 1.688 | 0.3068 | 0.6806 |
Gene-gene interaction analysis
A logistic regression model was built to analyze
rs4921580 in NAT1 and rs1041983 in NAT2. The combinations of
rs4921580 (Cg+gg) × rs1041983 (Ct+tt) were significantly associated
with NSCLP (OR, 1.983; 95% CI, 1.150–3.418; P=0.014; Table IV).
 | Table IVResults of gene-gene interactions
using a logistic regression method. |
Table IV
Results of gene-gene interactions
using a logistic regression method.
| Frequency | | | |
|---|
|
| | | |
|---|
| SNP in gene (NAT1 ×
NAT2) | Cases | Controls | OR | 95% CI | P-value |
|---|
| rs4921580 ×
rs1041983 |
| CC × CC | 0.25 | 0.300885 | 1 | Reference | |
| Cg+gg × CC | 0.098039 | 0.09292 | 1.270 | 0.623–2.588 | 0.511 |
| CC × Ct+tt | 0.367647 | 0.433628 | 1.020 | 0.637–1.635 | 0.933 |
| Cg+gg × Ct+tt | 0.284314 | 0.172566 | 1.983 | 1.150–3.418 | 0.014 |
Discussion
The results of the present study revealed a
significant association of NSCLP with rs4921580 in the NAT1 gene
and rs1041983 in the NAT2 gene. For the NAT1 gene, the GGG
haplotype, including rs2410545, rs4921580 and rs17693097, was
significantly associated with NSCLP. Analysis using a logistic
regression model also showed that gene-gene interactions between
NAT1 and NAT2 increased the risk of NSCLP.
Previous studies on NAT1 mainly focused on rs1057126
(T1088A) and rs15561 (C1095A). In a Californian population-based
case-control study, the OR for isolated CL/P was increased (OR=1.7;
95% CI=0.97–2.9) among infants homozygous for 1095A compared with
genotype 1095CC, while not for heterozygotes. There was an ~4-fold
increased risk for isolated CL/P for the 1088AA genotype in the
presence of maternal smoking. Similar to the 1088 polymorphism
analysis, the AA genotype of the NAT1 1095 polymorphism modified
risks for isolated CL/P and was associated with smoking with a
4-fold increased risk. The risk with the NAT1 genotype 1088AA +
1095AA in the presence of maternal smoking was higher compared with
1088TT + 1095CC (15). A study on
NAT1 1095 genotypes by Lammer et al(16) found a 2-fold higher risk for
isolated CL/P among infants who were homozygous for the variant
allele and whose mothers did not take multivitamins during early
pregnancy. However, there were not any differential risks for
clefts associated with maternal multivitamin consumption for the
NAT1 1088 genotypes. Lie et al(17) did not identify an association
between NAT1 and CL.
For NAT2, children heterozygous for the rs1799930
(G590A) variant were shown to have a 2-fold risk of NSCLP.
Furthermore, the C-A-G haplotype of NAT2 (referring to the variants
of the SNPs rs1799929/C481T, rs1799930 and rs1799931/G857A,
respectively) has been associated with isolated CL in a Norwegian
population. However, there was no indication of interaction between
the NAT2 C-A-G haplotype in children and maternal smoking (17). In case-parent triads, the
overtransmission of NAT2 G590A was observed in an oral cleft group,
while not in the control group (18). A number of additional studies did
not find an association between NAT2 and NSCLP (15,19)
To the best of our knowledge, this is the first
study on the association of NAT1 and NAT2 genes with NSCLP using
tagging SNP methods, and also the first study on a Chinese
population. For NAT1, the results of the present study revealed
that rs4921580 was associated with NSCLP, which has not been
previously reported. Additionally, the
rs2410545-rs4921580-rs17693097 haplotype (GGG) was found to have a
2.8-fold risk for NSCLP. According to the HapMap CHB database,
rs15561 is located in the same LD block as rs4921880. However, an
association between rs4921880 and NSCLP was not identified. This
may be due to the use of different geographical populations, sample
sizes or study methods. For NAT2, rs1041983 was found to be
associated with NSCLP, which has not been previously reported.
rs1041983 function is synonymous (cds-synon); no change occurs in
the amino acid sequence of the protein. DNA mutations do not result
in a change in the amino acid sequence of a protein; however, a
recent study showed that the ‘silent’ polymorphism may lead to the
synthesis of a protein product with the same amino acid sequence
but with different structural and functional properties (20). Therefore, rs1041983 should not be
neglected and further studies are required. rs1799930 is located in
the same LD block as rs11996129 in the HapMap CHB database.
However, an association between rs11996129 and NSCLP was not
identified. rs1799931 was not shown to be associated with NSCLP,
which was consistent with the results of Lie et al(17).
NAT1 and NAT2 are enzymatically distinct, with
certain aromatic amines being preferentially acetylated by NAT1
(e.g., p-aminosalicylic acid and p-aminobenzoic acid) and others by
NAT2 (e.g., isoniazid and sulfamethazine). Cornish et
al(21) confirmed that NAT
activity levels are strictly gene-dose-dependent and that NAT
deficiency is not compensated for by other enzymes of xenobiotic
metabolism. The deficiency of both NAT1 and NAT2 may impair the
detoxification and metabolic activation of drugs (22). In the present study, the
combination of rs4921580 (Cg+gg) × rs1041983 (Ct+tt) was shown to
increase the risk of NSCLP, which has not been previously reported.
A possible explanation for this result may be that the mutation of
the two genes results in metabolic abnormalities that are
associated with NSCLP. Erickson et al(14) found that Nat2 significantly
influenced teratogen-induced orofacial clefting using transgenic
and knockout animals. However, Sugamori et al(22) showed that Nat1/Nat2 double-knockout
mice develop normally and do not suffer from any obvious phenotypic
defects. This indicates that the mechanism of NAT1 and NAT2
requires further investigation.
The present study had several limitations. Firstly,
the sample size used was relatively small and other ethnic
populations were not included. Secondly, NAT1 and NAT2 are
associated with the detoxification and metabolic activation of
numerous chemicals. In the present study, environmental factors
during pregnancy, including alcohol consumption, maternal active
smoking, passive smoking, anti-epileptic drugs and radiation
exposure, were not investigated. Therefore, further studies are
required.
In conclusion, despite limitations caused by the use
of a small sample size, evidence in support of the association of
rs4921580 in the NAT1 gene and rs1041983 in the NAT2 gene with
NSCLP in a Chinese population was identified; haplotype analysis of
the gene supported these findings, and gene-gene interactions
between NAT1 and NAT2 were also demonstrated to play a role in the
susceptibility to NSCLP. Further studies are required to
investigate the association between NAT1 and NAT2, and NSCLP.
Acknowledgements
The authors thank all the participants who donated
samples for this study. This study was supported by the National
Natural Science Foundation of China (30901569).
Abbreviations:
|
NSCLP
|
nonsyndromic cleft lip with or without
cleft palate
|
|
NAT
|
N-acetyltransferase
|
|
HWE
|
Hardy-Weinberg equilibrium
|
|
LD
|
linkage disequilibrium
|
|
MAF
|
minor allele frequency
|
|
OR
|
odds radio
|
|
CI
|
confidence interval
|
References
|
1
|
Stuppia L, Capogreco M, Marzo G, et al:
Genetics of syndromic and nonsyndromic cleft lip and palate. J
Craniofac Surg. 22:1722–1726. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Munger RG, Romitti PA, Daack-Hirsch S,
Burns TL, Murray JC and Hanson J: Maternal alcohol use and risk of
orofacial cleft birth defects. Teratology. 54:27–33. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Little J, Cardy A, Arslan MT, Gilmour M
and Mossey PA: Smoking and orofacial clefts: a United Kingdom-based
case-control study. Cleft Palate Craniofac J. 41:381–386. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jia ZL, Shi B, Chen CH, Shi JY, Wu J and
Xu X: Maternal malnutrition, environmental exposure during
pregnancy and the risk of non-syndromic orofacial clefts. Oral Dis.
17:584–589. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hernández-Diaz S, Smith CR, Shen A, et al:
Comparative safety of antiepileptic drugs during pregnancy.
Neurology. 78:1692–1699. 2012.
|
|
6
|
Cech I, Burau KD and Walston J: Spatial
distribution of orofacial cleft defect births in Harris County,
Texas, 1990 to 1994, and historical evidence for the presence of
low-level radioactivity in tap water. South Med J. 100:560–569.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ramirez D, Lammer EJ, Iovannisci DM,
Laurent C, Finnell RH and Shaw GM: Maternal smoking during early
pregnancy, GSTP1 and EPHX1 variants, and risk of isolated orofacial
clefts. Cleft Palate Craniofac J. 44:366–373. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sim E, Fakis G, Laurieri N and Boukouvala
S: Arylamine N-acetyltransferases - from drug metabolism and
pharmacogenetics to identification of novel targets for
pharmacological intervention. Adv Pharmacol. 63:169–205. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mortensen HM, Froment A, Lema G, et al:
Characterization of genetic variation and natural selection at the
arylamine N-acetyltransferase genes in global human populations.
Pharmacogenomics. 12:1545–1558. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Boukouvala S and Fakis G: Arylamine
N-acetyltransferases: what we learn from genes and genomes. Drug
Metab Rev. 37:511–564. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Smelt VA, Mardon HJ and Sim E: Placental
expression of arylamine N-acetyltransferases: evidence for linkage
disequilibrium between NAT1*10 and NAT2*4 alleles of the two human
arylamine N-acetyltransferase loci NAT1 and NAT2. Pharmacol
Toxicol. 83:149–157. 1998.PubMed/NCBI
|
|
12
|
Ward A, Hickman D, Gordon JW and Sim E:
Arylamine N-acetyltransferase in human red blood cells. Biochem
Pharmacol. 44:1099–1104. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Minchin RF: Acetylation of
p-aminobenzoylglutamate, a folic acid catabolite, by recombinant
human arylamine N-acetyltransferase and U937 cells. Biochem J.
307(Pt 1): 1–3. 1995.PubMed/NCBI
|
|
14
|
Erickson RP, Cao W, Acuña DK, et al:
Confirmation of the role of N-acetyltransferase 2 in
teratogen-induced cleft palate using transgenics and knockouts. Mol
Reprod Dev. 75:1071–1076. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lammer EJ, Shaw GM, Iovannisci DM, Van
Waes J and Finnell RH: Maternal smoking and the risk of orofacial
clefts: susceptibility with NAT1 and NAT2 polymorphisms.
Epidemiology. 15:150–156. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lammer EJ, Shaw GM, Iovannisci DM and
Finnell RH: Periconceptional multivitamin intake during early
pregnancy, genetic variation of acetyl-N-transferase 1 (NAT1), and
risk for orofacial clefts. Birth Defects Res A Clin Mol Teratol.
70:846–852. 2004. View Article : Google Scholar
|
|
17
|
Lie RT, Wilcox AJ, Taylor J, et al:
Maternal smoking and oral clefts: the role of detoxification
pathway genes. Epidemiology. 19:606–615. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shi M, Christensen K, Weinberg CR, et al:
Orofacial cleft risk is increased with maternal smoking and
specific detoxification-gene variants. Am J Hum Genet. 80:76–90.
2007. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
van Rooij IA, Groenen PM, van Drongelen M,
Te Morsche RH, Peters WH and Steegers-Theunissen RP: Orofacial
clefts and spina bifida: N-acetyltransferase phenotype, maternal
smoking, and medication use. Teratology. 66:260–266.
2002.PubMed/NCBI
|
|
20
|
Komar AA: Silent SNPs: impact on gene
function and phenotype. Pharmacogenomics. 8:1075–1080. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cornish VA, Pinter K, Boukouvala S, et al:
Generation and analysis of mice with a targeted disruption of the
arylamine N-acetyltransferase type 2 gene. Pharmacogenomics J.
3:169–177. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sugamori KS, Wong S, Gaedigk A, et al:
Generation and functional characterization of arylamine
N-acetyltransferase Nat1/Nat2 double-knockout mice. Mol Pharmacol.
64:170–179. 2003. View Article : Google Scholar : PubMed/NCBI
|