Introduction
The liver has the ability to regenerate following
injury, allowing it to rapidly and efficiently restore lost mass
without jeopardizing the viability of the entire organism (1). The phenomenon of liver regeneration
(LR) may be induced by various methods, including liver damage by
hepatotoxic chemicals (2), the
rodent model of 70% partial hepatectomy (PHx) (3) and small-for-size liver
transplantation (4). LR is a
complicated process involving the secretion of numerous cytokines
and growth factors and the activation of metabolic networks
(5). Although this process is
histologically well described, the genes that orchestrate LR have
only been partially characterized (6). Investigating the mechanisms involved
in LR will offer new therapeutic strategies to rationally regulate
and control hepatic function in a diseased state, which has caused
this physiological process to gain increasing attention.
LR following PHx develops in three stages; the
priming/initiation stage, the proliferation stage and the
termination/inhibition stage (7,8). A
large number of growth-response genes are involved in these phases
of LR. Furthermore, it is necessary to elucidate the complex
interplay of numerous cellular events, particularly between the
hepatocytes and liver sinusoidal endothelial cells (LSECs)
(9). Angiogenesis is a fundamental
requirement for organ development, function restoration and tumor
growth. As a potent, diffusible and specific pro-angiogenic factor
among several growth factors, vascular endothelial growth factor
(VEGF) is important in liver regeneration, due in part to its
effect on neovascularization (10). VEGF expression in the regenerating
liver is mainly detected in the periportal hepatocytes, and
promotes the proliferation of hepatocytes through the
reconstruction of liver sinusoids by the proliferation of LSECs
(11).
MicroRNAs (miRNAs or miRs) are a family of small
non-coding RNAs that regulate target gene expression at the
post-transcriptional level by blocking protein translation or
inducing the degradation of gene messenger RNA (mRNA) after
hybridization to its 3′-untranslated region (UTR) (12). miRNAs are capable of regulating
every aspect of cellular activity, including differentiation and
development, metabolism, proliferation, apoptotic cell death, viral
infection and tumorigenesis (13).
miRNAs are critical regulators of hepatocyte proliferation during
LR (14). The response of the
vascular endothelium to angiogenic stimuli is modulated by miRNAs.
Thus, targeting the expression of miRNAs may be a novel therapeutic
approach for diseases involving excess or insufficient vasculature
(15). Hypoxia is a potent
stimulant for angiogenesis, and hypoxia-inducible factor-1α
(HIF-1α) is an essential transcriptional factor for adaptation
against hypoxia (16). Many miRNAs
also demonstrate dynamic changes in response to cellular
stimulation, including exposure to hypoxia. Whether HIF-1α is
involved in the transcriptional regulation of miRNAs related to
angiogenesis during LR, particularly miRNAs targeting VEGF, remains
unknown.
In this study, we hypothesized that the expression
of a specific miRNA in the regenerating liver may correlate closely
with angiogenesis. To identify this miRNA, we screened seven
candidate miRNAs, obtained from related databases, and correlated
their expression with the VEGF-A gene. Furthermore, to investigate
whether the expression of the selected miRNAs were able to be
regulated by HIF-1α mRNA, we established hypoxic conditions via the
administration of cobalt chloride (CoCl2) in isolated
hepatocytes and evaluated the expression and association of HIF-1α
mRNA, the selected microRNA and VEGF-A mRNA under hypoxia.
Materials and methods
Bioinformatics analysis
miRNA target site prediction for the VEGF-A gene was
performed using the starBase database (http://starbase.sysu.edu.cn) (17). StarBase links to five commonly used
miRNA target prediction programs including TargetScan (www.targetscan.org), picTar (www.mdc-berlin.de/en/research/research_teams/systems_biology_of_gene_regulatory_elements/projects/pictar),
RNA22 (http://cbcsrv.watson.ibm.com/rna22.html), PITA
(http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html)
and miRanda (http://www.microrna.org/microrna/home.do), which
provide a comprehensive search for intersections among the putative
miRNAs predicted by the programs and target gene VEGF-A.
Animals and operative procedures
Specific pathogen-free eight-week-old male C57BL/C
mice were purchased from the Experimental Animal Medicine Centre of
Sichuan University (Chengdu, China). The mice were housed at room
temperature and 55% humidity and given access to food and water
ad libitum throughout the experiment. After quarantine for
one week, 12 mice, with a mean weight of 18.2±2.7 g, were subjected
to 70% PHx using the procedure described previously (18) under chloral hydrate (50 mg/kg,
i.p.) anesthesia. Following surgery, the rats were further divided
into three groups (12, 24 and 48 h following PHx, n=4/group). At
the indicated times, four mice were randomly sacrificed and the
remnant regenerating liver tissues were collected and stored at
−80°C. All experiments were approved by the Committee of the
Experimental Use of Animals, Sichuan University, and performed in
accordance with institutional animal care instructions approved by
the Ethics Committee for Animals, Sichuan University.
Primary hepatocyte culture
Primary hepatocytes were obtained from mouse liver
using in situ two-step collagenase liver perfusion (19) with minor modifications. The mean
viability of hepatocytes was 90%, as determined by trypan blue
exclusion. The isolated hepatocytes were suspended in DMEM
(Hyclone, Logan, UT, USA) supplemented with 10% fetal calf serum
(Invitrogen, Carlsbad, CA, USA), 1 mM insulin and 1%
penicillin/streptomycin (Sigma-Aldrich, St. Louis, MO, USA) and
seeded in 6-well culture plates precoated with rat tail tendon
collagen type I (Shengyou Biotech, Co., Ltd., Hangzhou, China) at
1×106 cells/ml. Subsequently, the primary hepatocytes
were maintained for 4 h at 37°C in a 95% O2 and 5%
CO2 atmosphere to allow for stabilization. Four hours
later, the unattached cells were removed.
Hypoxia induction in primary
hepatocytes
Hypoxia induction was achieved using DMEM medium
containing HIF-1α-stabilizing agent CoCl2
(Sigma-Aldrich) at a final concentration of 100 μM (20). In order to evaluate the gene
expression levels, the cells were divided into three groups. In
group A, the hepatocytes were cultured without CoCl2 and
used as a control. In groups B and C, hepatocytes were cultured
with CoCl2 for 12 and 48 h, respectively.
miR-150 inhibitor transfection
Hepatocytes were seeded in 96-well plates at a
density of 5×103 cells/well and transfected with
micrOFF™ miR-150 inhibitor or a negative control (RiboBio Biotech.,
Co., Ltd., Guangzhou, China) at a final concentration of 100 nM
using Lipofectamine™ 2000 (Invitrogen), according to the
manufacturer's instructions. Total RNA and protein were extracted
24 and 48 h after transfection for quantitative PCR (qPCR) and
western blot analysis.
qPCR for miRNA detection
Total RNA was extracted from prepared samples using
TRIzol reagent (Invitrogen), according to the manufacturer's
instructions. The quality and quantity of each RNA sample were
measured spectrophotometrically using a NanoDrop 1000 (Thermo
Scientific, Waltham, MA, USA) and the integrity of the RNA was
assessed by 1% ethidium bromide agarose gel electrophoresis
(Invitrogen). Equal amounts of 1 μg RNA were reverse transcribed
into cDNA using the miRcute miRNA first-strand cDNA synthesis kit
(Tiangen Biotech Co., Ltd., Beijing, China), according to the
manufacturer's instructions. Subsequently, qPCR was performed using
the miRcute miRNA qPCR detection kit (Tiangen). The 20 μl qPCR
mixture consisted of 10 μl 2X miRcute miRNA premix, 2 μl forward
primer (2 μM), 2 μl reverse primer (2 μM), 1 μl cDNA and 5 μl
RNase-free ddH2O. The primers used are listed in
Table I. PCR amplification was
performed using a 1000-Series Thermal Cyclers real-time PCR
detection system (Bio-Rad, Hercules, CA, USA) under the following
conditions: 94°C for 2 min followed by 40 cycles of 94°C for 20
sec, 62°C for 30 sec and 72°C for 30 sec. The production of
specific products was confirmed using melting curve analysis
(65–95°C) at the end of the amplification cycle. All experiments
were performed in triplicate for each miRNA to obtain Ct values,
and a non-template control was included on the same plate. A small
stably expressed housekeeping RNA molecule, 5s rRNA (Rn5s), was
included as an internal control to normalize gene expression levels
and its primer product was purchased from Tiangen Biotech Co., Ltd.
Data were analyzed using Bio-Rad CFX Manager software (Bio-Rad) and
the relative expression (fold difference) of candidate genes
(miR126–5p, 134, 150, 185, 29a, 29b and 29c) was calculated using
the 2−ΔΔCt method.
 | Table IPrimer sequences for qPCR. |
Table I
Primer sequences for qPCR.
| Name | Sequence |
|---|
| miR-126–5p |
5′-GAGGCATTATTACTTTTGGTACG-3′ |
| miR-134 |
5′-GTGACTGGTTGACCAGAGGG-3′ |
| miR-150 |
5′-TCTCCCAACCCTTGTACCAGT-3′ |
| miR-185 |
5′-TGGAGAGAAAGGCAGTTCCTG-3′ |
| miR-29a |
5′-TAGCACCATCTGAAATCGGTTAA-3′ |
| miR-29b |
5′-TAGCACCATTTGAAATCAGTGTTA-3′ |
| miR-29c |
5′-GTAGCACCATTTGAAATCGGTTA-3′ |
| HIF-1α | F:
5′-ATCGCGGGGACCGATT-3′
R: 5′-CGACGTTCAGAACTTATCTTTTTCTT-3′ |
| VEGF-A | F:
5′-CTGTACCTCCACCATGCCAAGT-3′
R: 5′-AGATGTCCACCAGGGTCTCAAT-3′ |
| β-actin | F: 5′-AGAG
GGAAATCGTGCGTGAC-3′
R: 5′-CAATAGTGATGACCTGGCCGT-3′ |
qPCR for mRNA detection
Treated hepatocytes were harvested at the indicated
times and total RNA was extracted using TRIzol reagent. RNA was
reverse transcribed using the PrimeScript® RT reagent
kit with gDNA Eraser (Takara, Dalian, China). cDNA was amplified by
qPCR with a 1000-Series Thermal Cyclers real-time PCR detection
system (Bio-Rad) and SYBR® Premix Ex Taq™ II (Tli RNaseH
Plus) (Takara), according to the manufacturer's instructions. PCR
was performed in a 10 μl reaction mixture containing 5 μl SYBR
premix, 1 μl forward primer and reverse primer, 1 μl cDNA template
and 3 μl RNase-free ddH2O. The housekeeping gene β-actin
mRNA normalized the levels of cDNA. Primers for PCR are listed in
Table I. The PCR conditions were
as follows: HIF-1α, 95°C for 3 min followed by 45 cycles of
amplification at 95°C for 5 sec and 57°C for 15 sec; VEGF-A, 95°C
for 3 min followed by 40 cycles of amplification at 95°C for 10
sec, 56°C for 10 sec and 72°C for 10 sec; and β-actin, 95°C for 3
min followed by 40 cycles of amplification at 95°C for 5 sec and
59°C for 10 sec. All the samples were amplified in triplicate and
each experiment was repeated three times.
Western blot analysis
Liver tissues, or cultured hepatocytes, were
homogenized and lysed in cell lysis buffer (Beyotime institute of
Biotechnology, Nantong, China) containing phenylmethanesulfonyl
fluoride (PMSF; Beyotime) for 30 min on ice. The lysates were
centrifuged for 10 min at 10,612 × g at 4°C. Protein concentrations
were determined using the BCA protein assay kit (Beyotime) with BSA
as a standard. Equal amounts of protein were loaded onto a 10%
SDS-polyacrylamide gel and separated intermittently for 1 h at 100
V on an electrophoresis system (Bio-Rad). Proteins were then
transferred to a nitrocellulose membrane (Pall, Port Washington,
NY, USA) for 2.5 h at 0.35 mA. Membranes were blocked for 1 h with
a blocking buffer (Beyotime) and subsequently incubated overnight
at 4°C with a 1:100 dilution of primary rabbit anti-HIF-1α or
anti-VEGF polyclonal antibody (Boster Bioengineering Co., Ltd.,
Wuhan, China). A mouse anti-GAPDH polyclonal antibody (Boster) was
used as a loading control to normalize protein levels. After
washing four times with TBS-T for 15 min, the immunoblots were
incubated for 1 h with a 1:5,000 dilution of secondary horseradish
peroxidase-conjugated goat anti-rabbit IgG (Boster), followed by
detection with enhanced chemiluminescence (Beyotime) according to
the manufacturer's instructions. Protein levels were visualized on
the gel imaging and analysis system and quantified using the
Quantity One 4.5 software (Bio-Rad). The gray ratio value between
the target protein and GAPDH was used to calculate the relative
expression level of target protein for the subsequent statistical
analysis.
Statistical analysis
All data are expressed as the mean ± SD. Statistical
analysis was performed using one-way ANOVA and a Newman-Keuls test
between groups using Statistical Package for the Social Science
(SPSS) for Windows (SPSS, Inc., Chicago, IL, USA). Graphs were
created with Origin 7.5 (OriginLab data analysis and graphing
software, Northampton, MA, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Computational predictions of miRNAs
targeting VEGF-A
Since this study investigated the effect of miRNA
profile change in the proliferating hepatocytes upon VEGF-A gene
expression and aimed to identify an miRNA that was associated with
the VEGF-A gene with relative specificity, we first analyzed target
site intersections between mouse miRNAs and the VEGF-A gene using a
prediction assay of miRNA target genes. Initially, in silico
analysis predicted 22 putative miRNAs based on three reference
sequences of the mouse VEGF-A gene (GenBank accession numbers:
NM_009505, NM_001025257 and NM_001025250) using five bioinformatics
algorithms (Table II). The animal
miRNA target genes were roughly estimated since there was
incomplete base pairing on binding sites between the miRNA and mRNA
target sequences (21). In order
to make full use of all the algorithm programs with different
characteristics and to improve the accuracy rate of target
prediction, we selected miRNAs that were simultaneously predicted
by two or more of the five bioinformatics algorithms as putative
regulators of VEGF-A. In total, 7 of the 22 putative miRNAs were
selected for further screening in order to identify a specific
miRNA with close correlation to the VEGF-A gene.
 | Table II22 miRNAs predicted to target the
VEGF-A gene. |
Table II
22 miRNAs predicted to target the
VEGF-A gene.
| | | | CLIP-Seq read
number |
|---|
| | | |
|
|---|
| Name | Gene name | Gene reference
accession | miRNA position | TargetScan sites | picTar sites | RNA22 sites | PITA sites | miRanda sites |
|---|
| mmu-miR-1224 | VEGF-A | NM_001025250 |
chr17:46155316-46155336 | 0 | 0 | 0 | 0 | 57 |
| mmu-miR-126–5p | VEGF-A | NM_009505 |
chr17:46154100-46154106 | 0 | 59 | 0 | 59 | 59 |
| mmu-miR-130b | VEGF-A | NM_001025250 |
chr17:46154521-46154542 | 0 | 0 | 0 | 0 | 35 |
| mmu-miR-134 | VEGF-A | NM_009505 |
chr17:46154429-46154435 | 0 | 31 | 0 | 31 | 31 |
| mmu-miR-138 | VEGF-A | NM_009505 |
chr17:46154424-46154430 | 0 | 31 | 0 | 0 | 0 |
| mmu-miR-150 | VEGF-A | NM_001025250 |
chr17:46155331-46155337 | 0 | 0 | 0 | 57 | 57 |
| mmu-miR-15a | VEGF-A | NM_009505 |
chr17:46154116-46154122 | 0 | 59 | 0 | 0 | 0 |
| mmu-miR-15b | VEGF-A | NM_009505 |
chr17:46154116-46154122 | 0 | 59 | 0 | 0 | 0 |
| mmu-miR-16 | VEGF-A | NM_009505 |
chr17:46155349-46155355 | 0 | 57 | 0 | 0 | 0 |
| mmu-miR-185 | VEGF-A | NM_009505 |
chr17:46154446-46154452 | 0 | 31 | 0 | 31 | 31 |
| mmu-miR-185 | VEGF-A | NM_009505 |
chr17:46154532-46154538 | 0 | 35 | 0 | 0 | 0 |
| mmu-miR-195 | VEGF-A | NM_009505 |
chr17:46154116-46154122 | 0 | 59 | 0 | 0 | 0 |
|
mmu-miR-199a–3p | VEGF-A | NM_009505 |
chr17:46154114-46154120 | 0 | 59 | 0 | 0 | 0 |
|
mmu-miR-199a–5p | VEGF-A | NM_001025250 |
chr17:46155362-46155368 | 57 | 0 | 0 | 0 | 0 |
| mmu-miR-214 | VEGF-A | NM_009505 |
chr17:46155346-46155352 | 0 | 57 | 0 | 0 | 0 |
| mmu-miR-29a | VEGF-A | NM_001025250 |
chr17:46154118-46154125 | 59 | 59 | 0 | 59 | 59 |
| mmu-miR-29b | VEGF-A | NM_001025250 |
chr17:46154118-46154125 | 59 | 59 | 59 | 59 | 59 |
| mmu-miR-29c | VEGF-A | NM_001025250 |
chr17:46154118-46154125 | 59 | 59 | 0 | 59 | 59 |
|
mmu-miR-450b–3p | VEGF-A | NM_001025250 |
chr17:46154457-46154480 | 0 | 0 | 0 | 0 | 31 |
|
mmu-miR-466c–3p | VEGF-A | NM_009505 |
chr17:46154476-46154482 | 0 | 31 | 0 | 0 | 0 |
| mmu-miR-466i | VEGF-A | NM_001025250 |
chr17:46154477-46154483 | 0 | 0 | 0 | 31 | 0 |
| mmu-miR-592 | VEGF-A | NM_001025250 |
chr17:46155697-46155721 | 0 | 0 | 0 | 0 | 6 |
Screening of miRNAs targeting VEGF-A in
the early phase of LR
Since PHx is considered to be the most potent
reproducible stimulus for hepatocyte proliferation (22), we analyzed the liver tissues
following PHx to explore changes in the expression of miRNAs in
hepatocytes of the regenerating liver. We investigated which miRNA
was regulated upon hypoxia in order to modulate the expression of
VEGF-A in the seven miRNAs predicted to target VEGF-A. We measured
the miRNA expression levels according to the variation tendency of
VEGF-A expression in the early phase of LR. Alterations in the
expression profiles of the miRNAs were examined using an miRNA
microarray assay. However, the significantly altered miRNAs
observed in the microarray results were selected for further
validation by qPCR, as the microarray detects changes in gene
expression without the need for high specificity and the microarray
product of different sources may have inconsistent results. Thus,
we directly detected substantial levels of seven miRNAs by qPCR
instead of miRNA microarray assay and observed changes in their
expression at three time-points (12, 24 and 48 h) once the total
RNA was extracted from prepared liver samples. We identified that
six of the miRNAs, with the exception of miR-29b, were
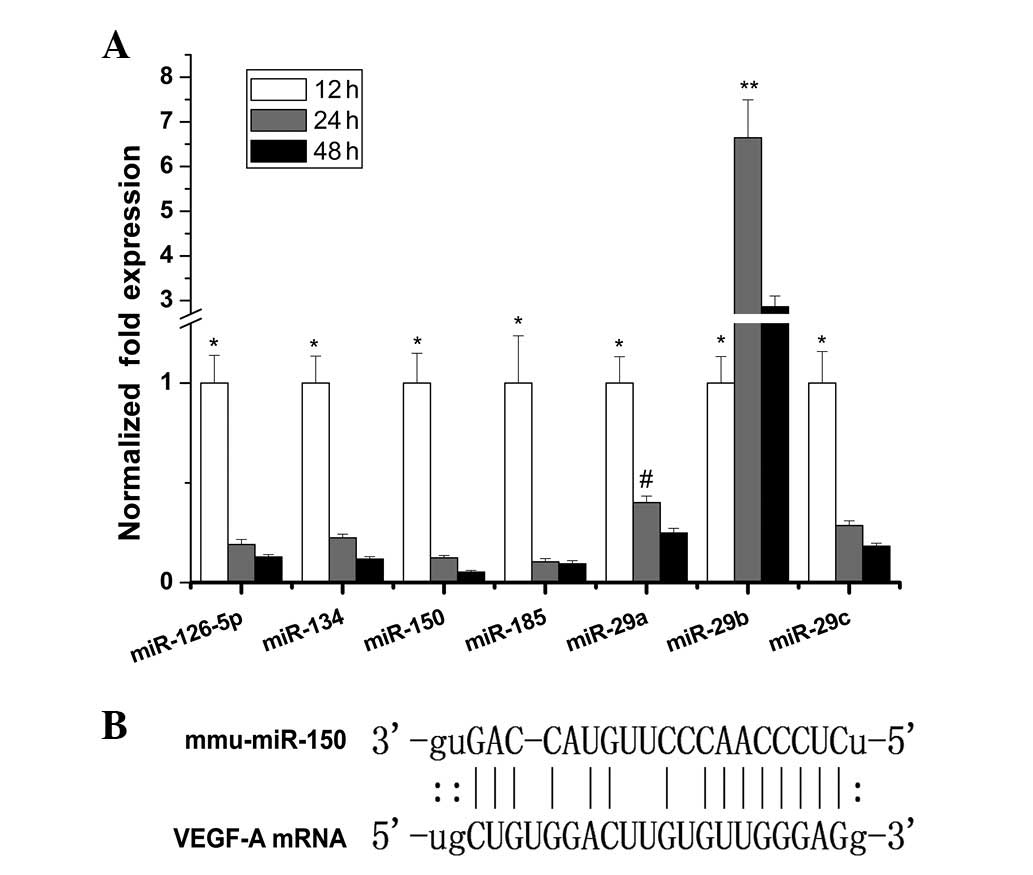
downregulated >2-fold between 12 and 48 h (Fig. 1A). Among them, miR-150 was
downregulated ~19-fold from 12–48 h and demonstrated the most
marked downwards trend. The direct binding site of miR-150 to the
VEGF target sequence is shown in Fig.
1B.
Effect of miR-150 inhibitors on VEGF-A
gene expression
miRNAs with significant alterations in their
expression levels, as determined by the microarray results, were
generally considered to correlate closely with different
physiological or pathological phenomena. We infer that the
increased production of VEGF in the early stage of LR is due to a
global downregulation in the production of certain miRNAs. Of all
the miRNAs predicted to target VEGF-A, miR-150 was selected for
further investigation as it was the most downregulated. In view of
an interaction between miR-150 and VEGF-A mRNA with the highest
degree of correlation, we evaluated the role of miR-150-mediated
regulation of VEGF-A gene expression. The miR-150 inhibitors and a
negative control were transfected into the hepatocytes for 24 and
48 h. As a chemosynthetic and modified inhibitor, the micrOFF miRNA
inhibitor specifically bound the mature target miRNA and suppressed
its expression. The efficacy of downregulating the expression of
the miR-150 gene and upregulating the expression of the VEGF-A gene
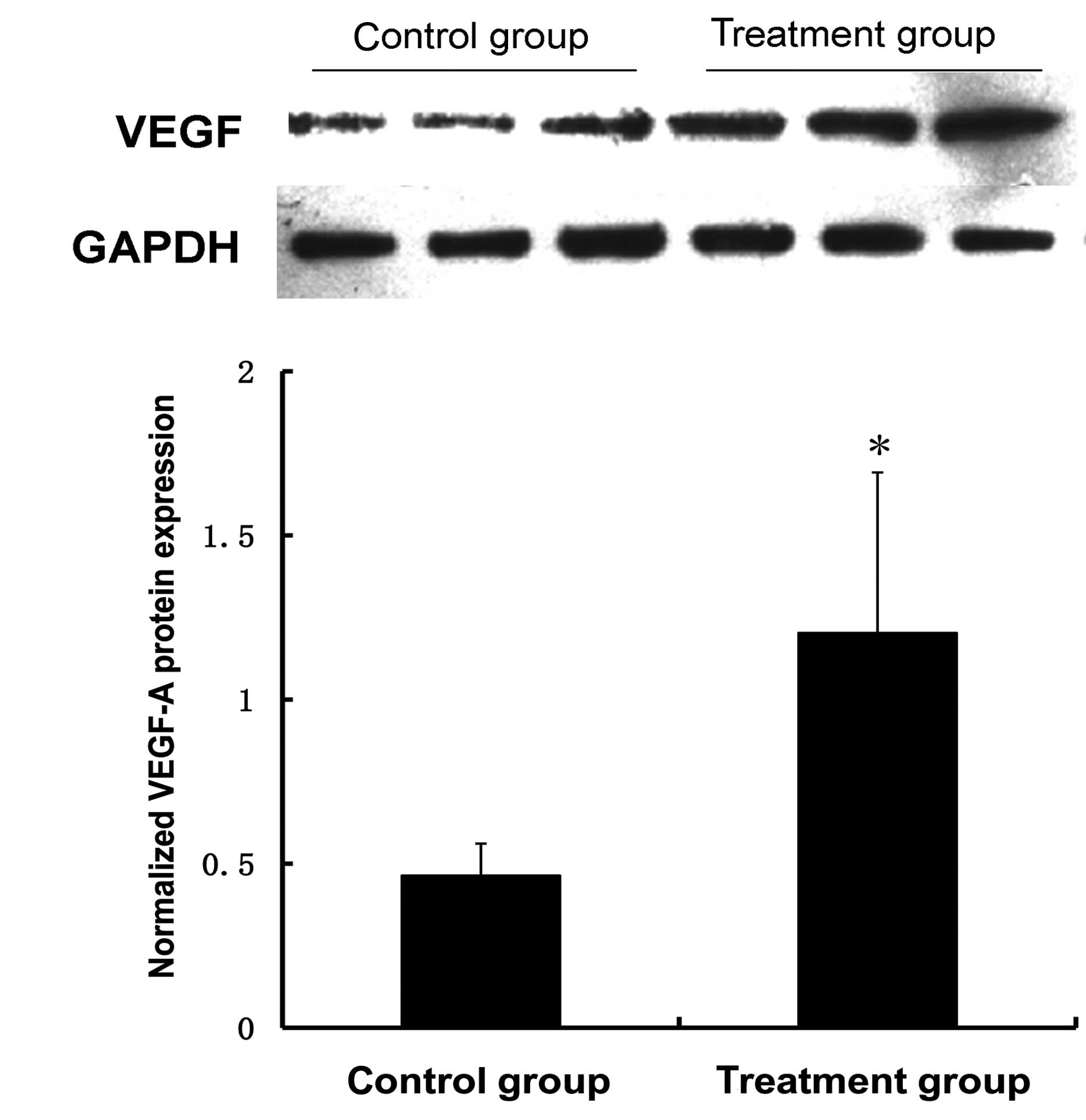
was examined by qPCR and western blot analysis. Fig. 2 demonstrates that the miR-150
inhibitors effectively suppressed the expression of miR-150
2.4-fold 48 h after transfection and promoted the corresponding
transcription and translation of the VEGF-A gene. The mRNA and
protein levels of VEGF-A increased 3.1-fold and 2.6-fold,
respectively, 48 h after transfection with the miR-150 inhibitors
compared with the control group. The differences were not
statistically significant at 24 h when compared with the control
group (Fig. 3).
HIF-1α protein expression in the early
phase of LR
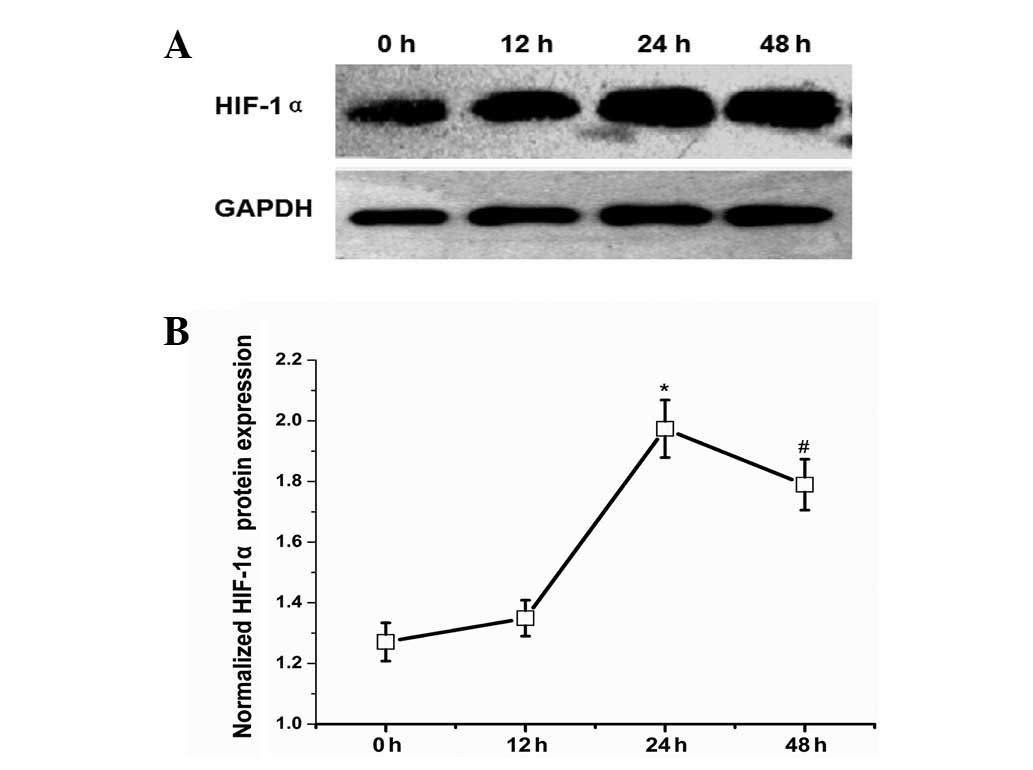
In order to evaluate the hypoxic conditions in the
early phase of LR, we selected the HIF-1α protein as a detection
index to indicate the degree of hypoxia. Intraoperatively excised
liver tissues served as the normal control (0 h). Western blot
analysis demonstrated that HIF-1α protein expression levels were
increased 12 h after PHx and continued increasing until the highest
expression level was achieved 24 h after PHx with an ~4-fold
increase relative to the control group (Fig. 4).
HIF-1α, miR-150 and VEGF-A mRNA
expression in hypoxia-induced hepatocytes
To determine whether miR-150 was regulated by
hypoxia, primary hepatocytes were isolated and hypoxia was induced
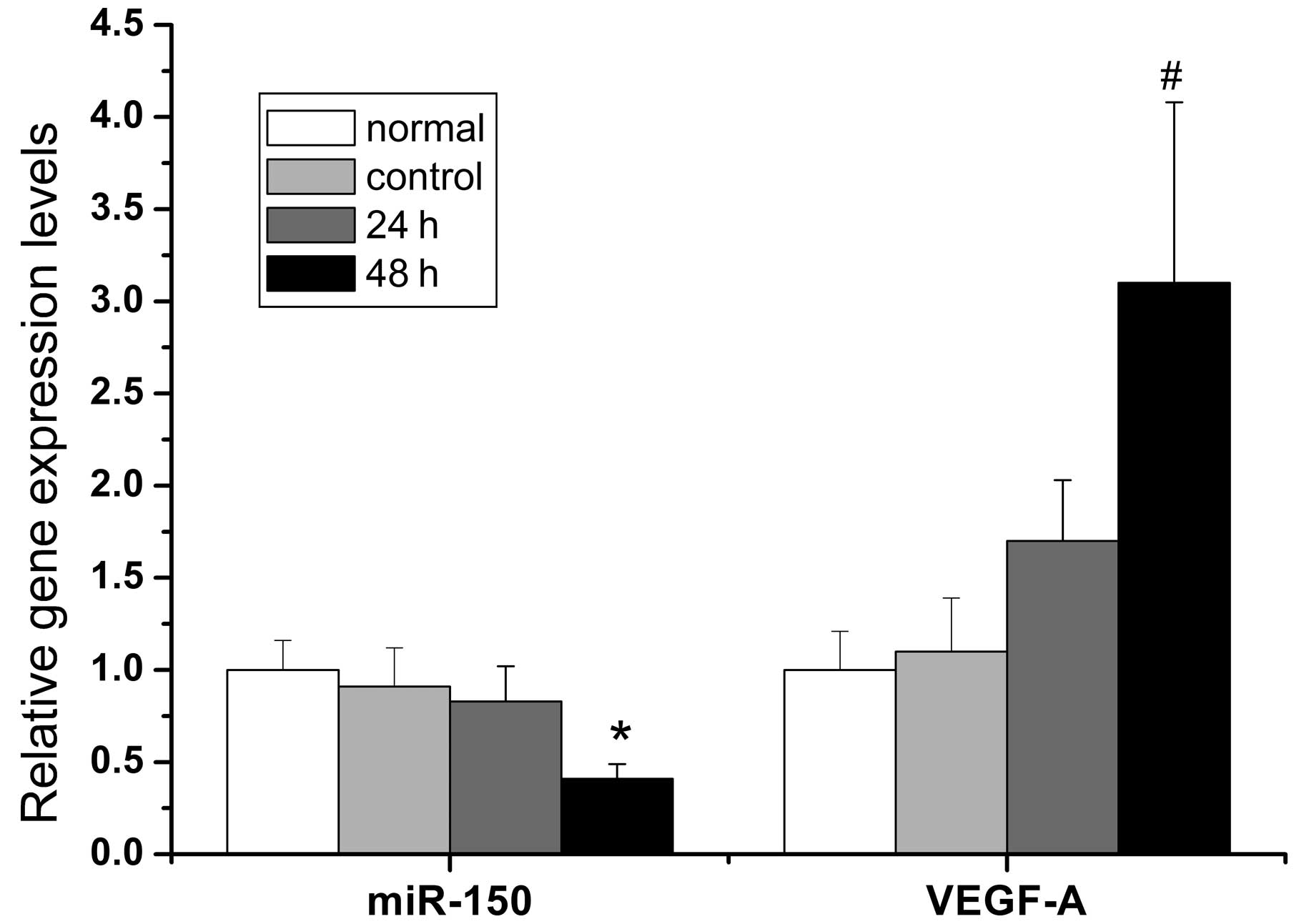
with CoCl2. As shown in Fig. 5, the expression of HIF-1α mRNA was
low in the normoxic state, but increased at 12 h (2.7-fold vs.
control) and increased further at 48 h (4-fold vs. control)
following the administration of CoCl2. Moreover, the
expression of miR-150 declined and VEGF-A mRNA increased 48 h after
hypoxia with differences of 4.2-fold and 3.1-fold, respectively,
relative to the normal control. However, the expression of miR-150
and VEGF-A mRNA was not significantly different from the control 12
h after hypoxia.
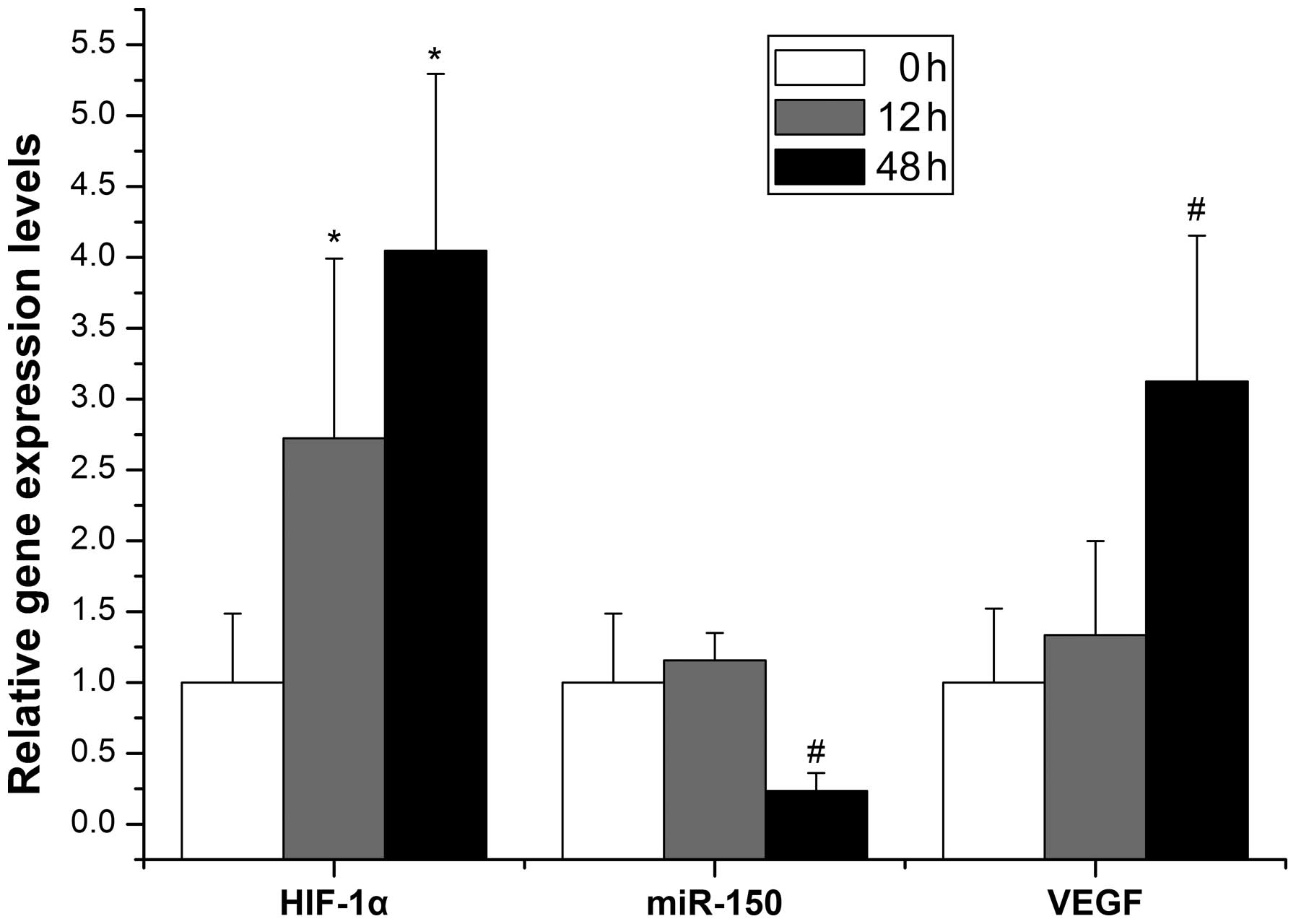 | Figure 5Evolution of miR-150, HIF-1α mRNA and
VEGF-A mRNA levels in primary hepatocytes treated with
CoCl2 at the indicated time points (0, 12 and 48 h,
respectively), as determined by qPCR. A gradual increase in HIF-1α
mRNA expression was observed with increasing time (2.7-fold at 12 h
and 4-fold at 48 h vs. control, *P<0.05). By
contrast, elevated HIF-1α mRNA levels resulted in a decrease in
miR-150 expression and an increase in VEGF-A mRNA expression at 48
h compared with the control (#P<0.05). All results
were normalized to 5s rRNA or β-actin mRNA expression. All data are
presented as the mean ± SD of three independent experiments (n=3
per condition). HIF-1α, hypoxia-inducible factor-1α; VEGF-A,
vascular endothelial growth factor-A; qPCR, quantitative PCR; mRNA,
messenger RNA; miR, microRNA. |
Discussion
In the early phase of LR, proliferating hepatocytes
show hypoxia-induced VEGF expression, which initiates a process
that aims to achieve the proper flow of blood through the liver
(23). However, changes in the
miRNA profile that promote the expression of the VEGF gene upon
hypoxia remain unknown. In order to determine these changes, the
present study focused on miRNAs targeting VEGF-A. Initially, we
examined the dynamic change of miRNAs predicted to target VEGF-A in
the early phase of LR and selected miR-150 as the miRNA with the
highest potential to modulate VEGF-A gene expression. Additionally,
we observed the effect of miR-150 knockdown on VEGF-A gene
expression in primary hepatocytes. Finally, we demonstrated the
kinetics of HIF-1α expression in mouse regenerating liver and the
hypoxia responsiveness of miR-150 by assessing HIF-1α and VEGF-A
gene expression upon hypoxic exposure.
miRNAs have multiple gene targets and each target
may be regulated by multiple miRNAs (24). Thus, we performed computational
predictions with five algorithms in order to guarantee the accuracy
rate. Based on the computer analysis of putative binding sites on
the target gene, the interactions of the investigated miRNAs with
VEGF are purely speculative. In principle, miRNAs are hypothesized
to negatively regulate target gene expression by inhibiting their
stability or translation. Thus, the expression of the target gene
is inversely proportional to its upstream miRNAs if their
regulating relationship is highly specific. A previous report
demonstrated that VEGF was detected as early as 12 h following PHx,
peaked at 48–72 h and gradually declined to the baseline level
after 8–10 days (25). Our results
suggest that miR-150 exhibited the most marked decrease from 12–48
h following PHx among the seven miRNAs predicted to target VEGF-A.
Furthermore, we revealed that the expression of VEGF-A mRNA was
negatively correlated with miR-150 expression in hepatocytes 48 h
after exposure to hypoxia. These results suggest that there is an
interaction between miR-150 and VEGF-A mRNA for their inverse
co-expression with a high degree of correlation. Shen et
al(26) observed a decrease in
the levels of miR-150 in the ischemia-induced retinal
neovascularization model compared with normal retinas, and the
luciferase levels of a VEGF reporter were downregulated by
co-transfection with pre-miR-150. In the present study, our results
demonstrated that the knockdown of miR-150 using the micrOFF
miR-150 inhibitor promoted the expression of VEGF-A mRNA and
protein in primary hepatocytes. Evidence from the validation of
gene structure and function demonstrates that VEGF-A may be a
target of miR-150 and miR-150 may be crucial in the regulation of
VEGF-A gene expression and angiogenesis during mouse LR.
Following cell division in the initial stage of LR,
hepatocytes are found in clusters and no longer associate with
sinusoids. Ninomiya et al(27) hypothesized that the abrupt
regenerative response of hepatocytes to resection suppresses
sinusoids, resulting in local hypoxia and induction of the HIF-1α
genes in the regenerating hepatocytes. In other words, hypoxia
occurs due to the incongruous replication of hepatocytes and LSECs.
Our results demonstrated that expression of the HIF-1α protein in
mouse tissues in LR increased and peaked at 24 h, similar to the
result observed by Maeno et al(28). We also revealed that HIF-1α
expression increased in primary hepatocytes following the
administration of CoCl2, which indicates that a
hypoxia-induced hepatocyte model was established. This is due to
the fact that cobalt has been widely used to mimic a hypoxic
response and induce HIF-1α production in cultured cells (29). Although no marked change in miR-150
and VEGF-A mRNA expression was observed 12 h after hypoxia
treatment in hypoxia-induced hepatocytes when HIF-1α mRNA levels
had already started to increase, we assume that HIF-1α mRNA had not
yet translocated from the cytoplasm into the nucleus where HIF-1α
functions in combination with its target gene. These results
suggested that miR-150 may be identified as a downregulated miRNA
by hypoxia, not only during LR, but also in the anoxic model of
primary hepatocytes, since its production is dependent on low
levels of HIF-1α expression. Thus, we conclude that HIF-1α
participates in the transcriptional regulation of miR-150.
Previous studies have focused on the role of VEGF in
LR. However, little is known with regards to the underlying
molecular mechanisms by which the VEGF gene is produced and
modulated. The discovery of miRNAs offers a unique insight into
their ability to regulate target genes.
In conclusion, we provide evidence that the
expression of miR-150 is specifically related to VEGF-A and is
subject to negative regulation by HIF-1α during the early phase of
LR. Therefore, miR-150 serves as an indirect bridge between HIF-1α
and VEGF-A, suggesting that there is an alternative pathway for the
HIF-1α regulation of VEGF-A. The phenomenon identified in this
study will extend our knowledge of factors controlling liver
regeneration.
Acknowledgements
This work was supported by grants from the Sichuan
provincial science and technology support program
(No.2010FZ0016).
References
|
1
|
Michalopoulos GK: Liver regeneration:
alternative epithelial pathways. Int J Biochem Cell Biol.
43:173–179. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Donahower B, McCullough SS, Kurten R, et
al: Vascular endothelial growth factor and hepatocyte regeneration
in acetaminophen toxicity. Am J Physiol Gastrointest Liver Physiol.
291:G102–G109. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Michalopoulos GK: Liver regeneration after
partial hepatectomy: critical analysis of mechanistic dilemmas. Am
J Pathol. 176:2–13. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hua ZY, Song J, Cheng F, et al: The effect
of hepatocyte growth factor on the initiation phase of liver
regeneration after cold ischemia in a rat model of small-for-size
liver transplantation. Hepatogastroenterology. 59:1548–1552.
2012.PubMed/NCBI
|
|
5
|
Fausto N, Campbell JS and Riehle KJ: Liver
regeneration. Hepatology. 43:S45–S53. 2006. View Article : Google Scholar
|
|
6
|
Böhm F, Köhler UA, Speicher T and Werner
S: Regulation of liver regeneration by growth factors and
cytokines. EMBO Mol Med. 2:294–305. 2010.PubMed/NCBI
|
|
7
|
Chen JA, Shi M, Li JQ and Qian CN:
Angiogenesis: multiple masks in hepatocellular carcinoma and liver
regeneration. Hepatol Int. 4:537–547. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tarlá MR, Ramalho FS, Ramalho LN, et al: A
molecular view of liver regeneration. Acta Cir Bras. 21(Suppl 1):
58–62. 2006.
|
|
9
|
Crawford SE: Vascular interference: a
blockade to tumor epithelial growth. Hepatology. 39:1491–1494.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bockhorn M, Goralski M, Prokofiev D, et
al: VEGF is important for early liver regeneration after partial
hepatectomy. J Surg Res. 138:291–299. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Taniguchi E, Sakisaka S, Matsuo K,
Tanikawa K and Sata M: Expression and role of vascular endothelial
growth factor in liver regeneration after partial hepatectomy in
rats. J Histochem Cytochem. 49:121–130. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen XM: MicroRNA signatures in liver
diseases. World J Gastroenterol. 15:1665–1672. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Song G, Sharma AD, Roll GR, et al:
MicroRNAs control hepatocyte proliferation during liver
regeneration. Hepatology. 51:1735–1743. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fish JE and Srivastava D: MicroRNAs:
opening a new vein in angiogenesis research. Sci Signal.
2:pe12009.PubMed/NCBI
|
|
16
|
Chaudhuri S, McCullough SS, Hennings L, et
al: Acetaminophen hepatotoxicity and HIF-1α induction in
acetaminophen toxicity in mice occurs without hypoxia. Toxicol Appl
Pharmacol. 252:211–220. 2011.
|
|
17
|
Yang JH, Li JH, Shao P, et al: starBase: a
database for exploring microRNA-mRNA interaction maps from
Argonaute CLIP-Seq and Degradome-Seq data. Nucleic Acids Res.
39:D202–D209. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mitchell C and Willenbring H: A
reproducible and well-tolerated method for 2/3 partial hepatectomy
in mice. Nat Protoc. 3:1167–1170. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li WC, Ralphs KL and Tosh D: Isolation and
culture of adult mouse hepatocytes. Methods Mol Biol. 633:185–196.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fradette C and du Souich P:
Hypoxia-inducible factor-1 and activator protein-1 modulate the
upregulation of CYP3A6 induced by hypoxia. Br J Pharmacol.
140:1146–1154. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yu CH, Xu CF and Li YM: Association of
MicroRNA-223 expression with hepatic ischemia/reperfusion injury in
mice. Dig Dis Sci. 54:2362–2366. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao XM, Zhao J and Li Y and Li Y: Effects
of bicyclol on liver regeneration after partial hepatectomy in
rats. Dig Dis Sci. 54:774–781. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kajdaniuk D, Marek B, Foltyn W and
Kos-Kudła B: Vascular endothelial growth factor (VEGF) - part 1: in
physiology and pathophysiology. Endokrynol Pol. 62:444–455.
2011.PubMed/NCBI
|
|
24
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mohammed FF and Khokha R: Thinking outside
the cell: proteases regulate hepatocyte division. Trends Cell Biol.
15:555–563. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Shen J, Yang X, Xie B, et al: MicroRNAs
regulates ocular neovascularization. Mol Ther. 16:1208–1216. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ninomiya M, Shirabe K, Terashi T, et al:
Deceleration of regenerative response improves the outcome of rat
with massive hepatectomy. Am J Transplant. 10:1580–1587. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Maeno H, Ono T, Dhar DK, et al: Expression
of hypoxia inducible factor-1alpha during liver regeneration
induced by partial hepatectomy in rats. Liver Int. 25:1002–1009.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Battaglia V, Compagnone A, Bandino A, et
al: Cobalt induces oxidative stress in isolated liver mitochondria
responsible for permeability transition and intrinsic apoptosis in
hepatocyte primary cultures. Int J Biochem Cell Biol. 41:586–594.
2009. View Article : Google Scholar
|