Introduction
Tuberous sclerosis (TS), an autosomal dominant
disease, is caused by mutation of one of the two tumor suppressor
genes, tuberous sclerosis complex 1 (TSC1) or 2
(TSC2), encoding hamartin (TSC1) and tuberin (TSC2),
respectively. TSC1, located on chromosome 9q34, contains 23
exons and an 8.6 kb mRNA transcript, while TSC2 is located
on chromosome 16p13.3 and contains 41 exons. Two thirds of the TS
cases are sporadic. Based on the relatively high incidence rate of
1:6,000, the disease is considered common in the general
population. However, no studies comparing the frequency of mutation
sites in different populations or evaluating the segregation of
mutations are currently available.
As mentioned above, the disease develops when one of
the two tumor suppressor genes, TSC1 or TSC2, is
mutated. This is the result of close cooperation of encoded
proteins within the cell. Hamartin and tuberin form an
intracellular complex responsible for the inhibition of Ras
homologue enriched in brain (Rheb). Subsequently, Rheb activates
the mammalian target of rapamycin (mTor) kinase, which is
responsible for the regulation of protein translation. Thus, when
the TS complex has not been formed or is non-functional, Rheb
inhibition is inefficient and mTor promotes cell cycle progression,
leading to uncontrolled proliferation and, possibly, tumor
development.
The aim of the present study was to compare the
frequency of TSC1 and TSC2 mutations, as well as the
types of specific mutations presenting in different populations.
Therefore, a meta-analysis of five large-scale sequencing studies
(1–5) was performed, including TS patients
from four distinct populations: American, British, Polish and
Taiwanese.
Materials and methods
Taiwanese population
Eighty-four unrelated patients with a familial or
sporadic form of TS were included in the study by Hung et
al(5). Denaturing
high-performance liquid chromatography (DHPLC) and direct
sequencing were used for mutation detection. Mutations were
identified in a total of 64 patients. The diagnostic criteria used
were according to Roach et al(6).
American and Polish population
Two hundred and twenty-four unrelated patients with
a familial or sporadic form of TS were included in the study by
Dabora et al(4). DHPLC,
long-range polymerase chain reaction (PCR) and quantitative PCR
were used for mutation detection. Mutations were identified in a
total of 186 patients. The diagnostic criteria used were according
to Roach et al(6).
American population
One hundred and twenty-six unrelated patients with a
familial or sporadic form of TS were included in the study by Niida
et al(3). Single-stranded
conformational polymorphism (SSCP) method followed by direct
sequencing was applied for mutation detection. The diagnostic
criteria used were according to Gomez (7) and Roach et al(8).
British population
One hundred and fifty unrelated patients with a
familial or sporadic form of TS were included in the study by Jones
et al(1,2). SSCP, heteroduplex analysis,
pulsed-field gel electrophoresis, Southern blot analysis and long
PCR were applied for mutation detection. The diagnostic criteria
used were according to Roach et al(6).
Cohort of patients
Large rearrangements and polymorphisms were excluded
from this meta-analysis. Eventually, a group of 381 patients was
obtained, in whom small mutations in TSC1 or TSC2
were detected. The cohort of patients included 136 American, 98
British, 83 Polish and 64 Taiwanese patients. Due to insufficient
data, one of the patients from the American population (with a
TSC2 mutation) was not included in this meta-analysis.
Results
Frequency of TSC1 and TSC2 mutations in
the different populations
One of the most obvious results of this
meta-analysis was the fact that the frequency of TSC1 and
TSC2 mutations was different in the analyzed populations.
There were significant differences in the prevalence of TSC1
and TSC2 mutations between Polish/Taiwanese populations and
American/British populations (Table
I). Approximately 9.6 and 12.7% of Polish and Taiwanese
patients, respectively, had TSC1 mutations, while their
American and British counterparts exhibited an approximately 2-fold
higher ratio of mutated hamartin (26.5 and 22.4%, respectively).
Statistical analysis using the Chi-square test showed that there
were significant differences between these populations
(P=0.011660).
 | Table IFrequency of TSC1 and
TSC2 mutations in the analyzed populations. |
Table I
Frequency of TSC1 and
TSC2 mutations in the analyzed populations.
| Frequency of
mutations, n (% of total) |
|---|
|
|
|---|
| Population | TSC1 | TSC2 | Total |
|---|
| American | 36 (26.5) | 100 (73.5) | 136 |
| British | 22 (22.4) | 76 (77.6) | 98 |
| Polish | 8 (9.6) | 75 (90.4) | 83 |
| Taiwanese | 9 (12.7) | 55 (87.3) | 64 |
| Total | 74 (19.5) | 306 (80.5) | 380 |
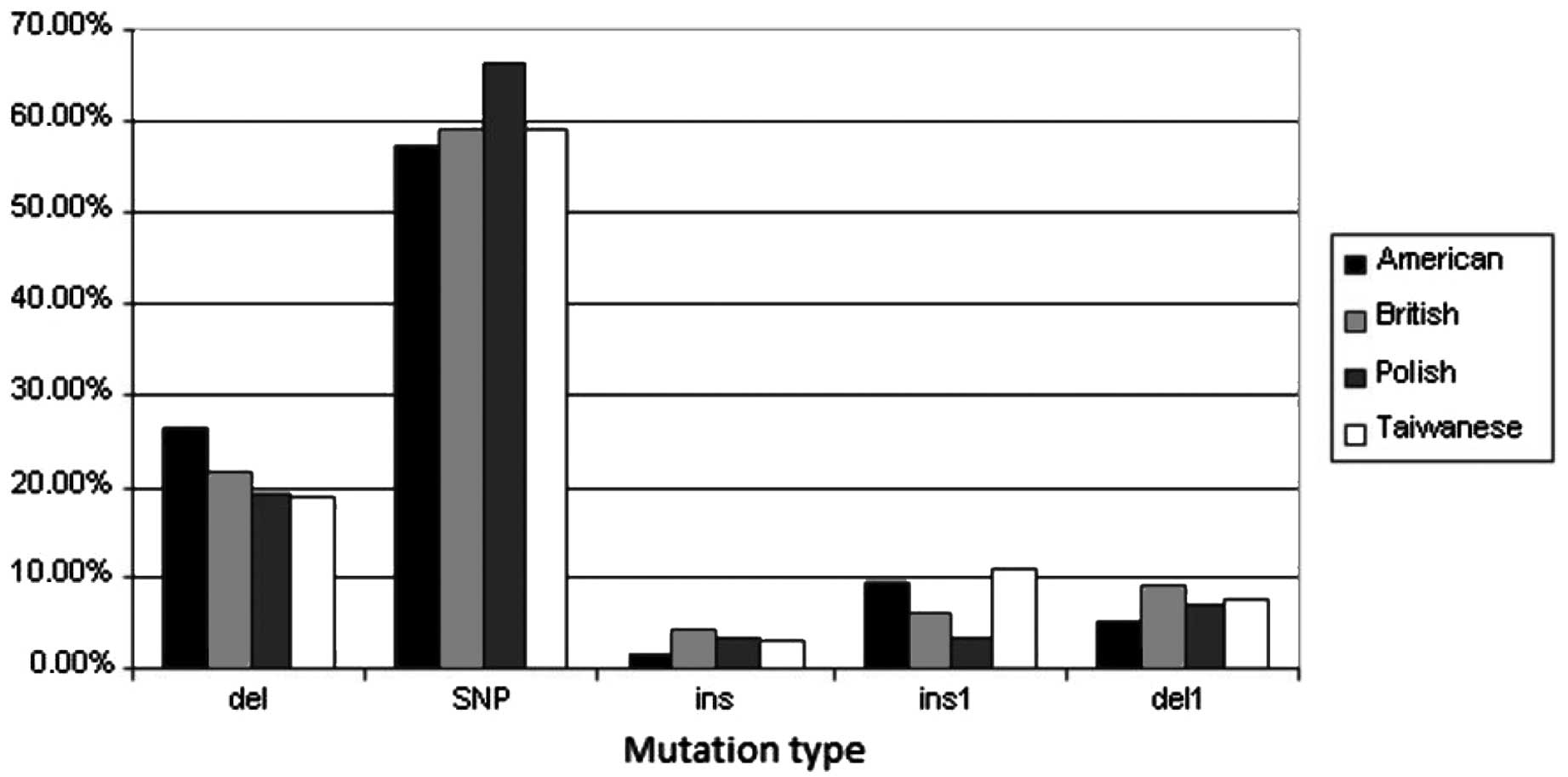
Types of mutations
There were no significant differences in the types
of mutations occurring in the affected genes in the populations
analyzed (Chi-square test, P=0.692681). The majority of the
mutations were single nucleotide changes. The second most important
type of mutations were deletions of >1 nucleotide. The third
type were insertions of single nucleotides (in the American and
Taiwanese populations) or deletions of single nucleotides (in the
Polish and British populations). There were also single deletions
(in the American and Taiwanese populations) or single insertions
(in the British population) and both single or multiple insertions.
In all the patients analyzed, insertions of >1 nucleotide were
the least frequent. The results are shown in Table II.
 | Table IITypes of mutations found in the cohort
of patients analyzed.a |
Table II
Types of mutations found in the cohort
of patients analyzed.a
| Types of mutations, n
(% of total) |
|---|
|
|
|---|
| Population | Deletions >1 | SNP | Insertions >1 | Single
insertions | Single deletions | Total |
|---|
| American | 36 (26.47) | 78 (57.35) | 2 (1.47) | 13 (9.56) | 7 (5.15) | 136 |
| British | 21 (21.43) | 58 (58.19) | 4 (4.08) | 6 (6.12) | 9 (9.18) | 98 |
| Polish | 16 (19.28) | 55 (66.27) | 3 (3.61) | 3 (3.61) | 6 (7.23) | 83 |
| Taiwanese | 12 (18.75) | 38 (59.38) | 2 (3.13) | 7 (10.94) | 5 (7.81) | 64 |
| Total | 85 | 229 | 11 | 29 | 27 | 381 |
However results of this study may not completely
reflect the actual distribution of mutations, since the original
studies used in this meta-analysis employed different methods for
the detection of mutations. Additionally, comparative studies have
shown that DHPLC is a more sensitive method compared to SSCP
(9,10).
Analysis of sequences flanking the
mutations
The application analyzing the complexity of
nucleotide sequences, Linguistic Complexity (available at:
http://csweb.haifa.ac.il/library/#complex.html), was
used to analyze the degree of complexity of the sequences flanking
the mutations. Fifteen nucleotides located directly upstream and
downstream of the mutation were analyzed. The level of sequence
complexity in sequences flanking the mutation in the Polish
population was found to be statistically higher compared to the
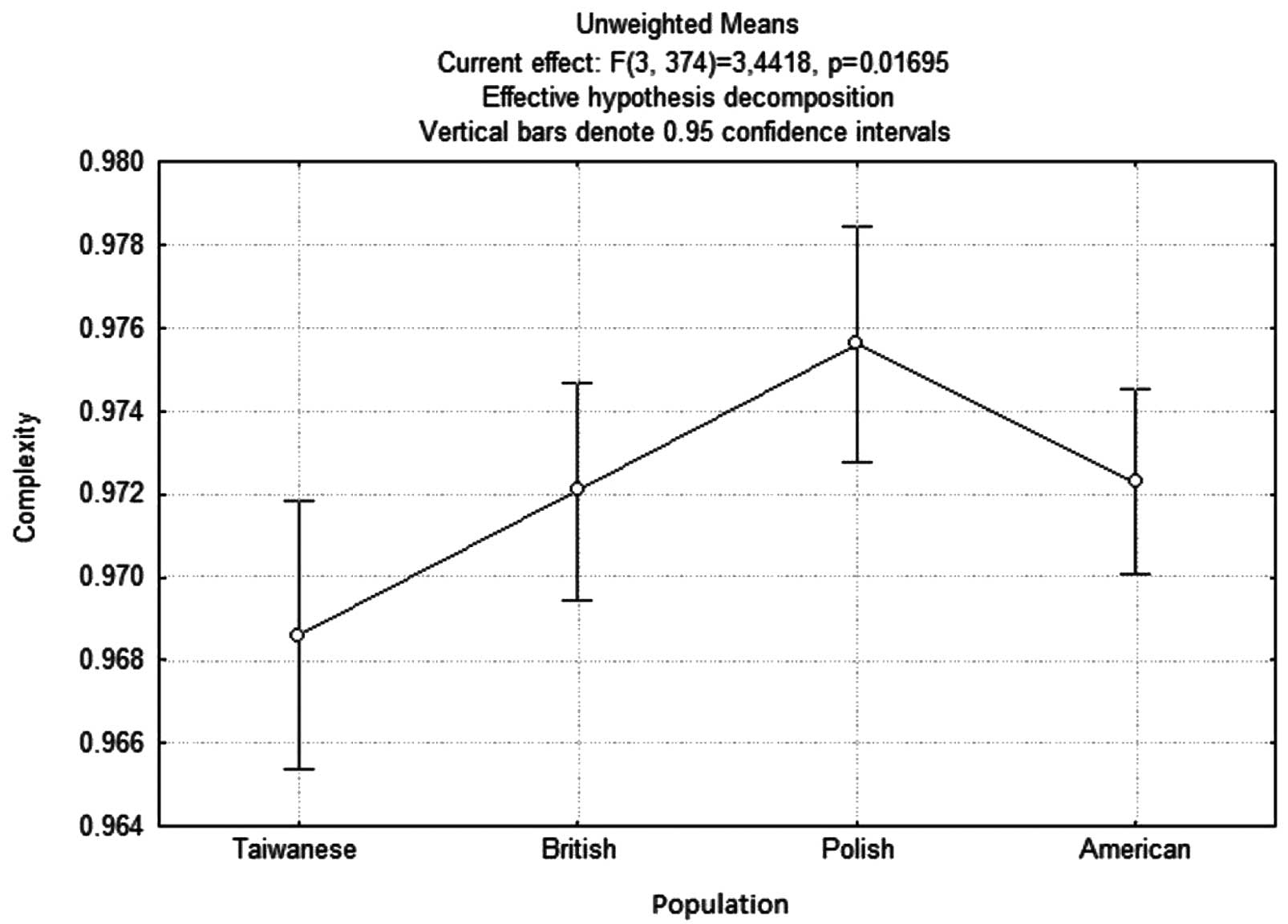
remaining populations (Fig. 1).
One-way Anova analysis was used to show that the differences were
statistically significant (P=0.01695).
Frequency of mutation types in the
different populations
No statistically significant differences were
observed in the frequency of mutation types in individual
populations (Fig. 2). The most
frequent type of mutations were deletions spanning >1
nucleotide.
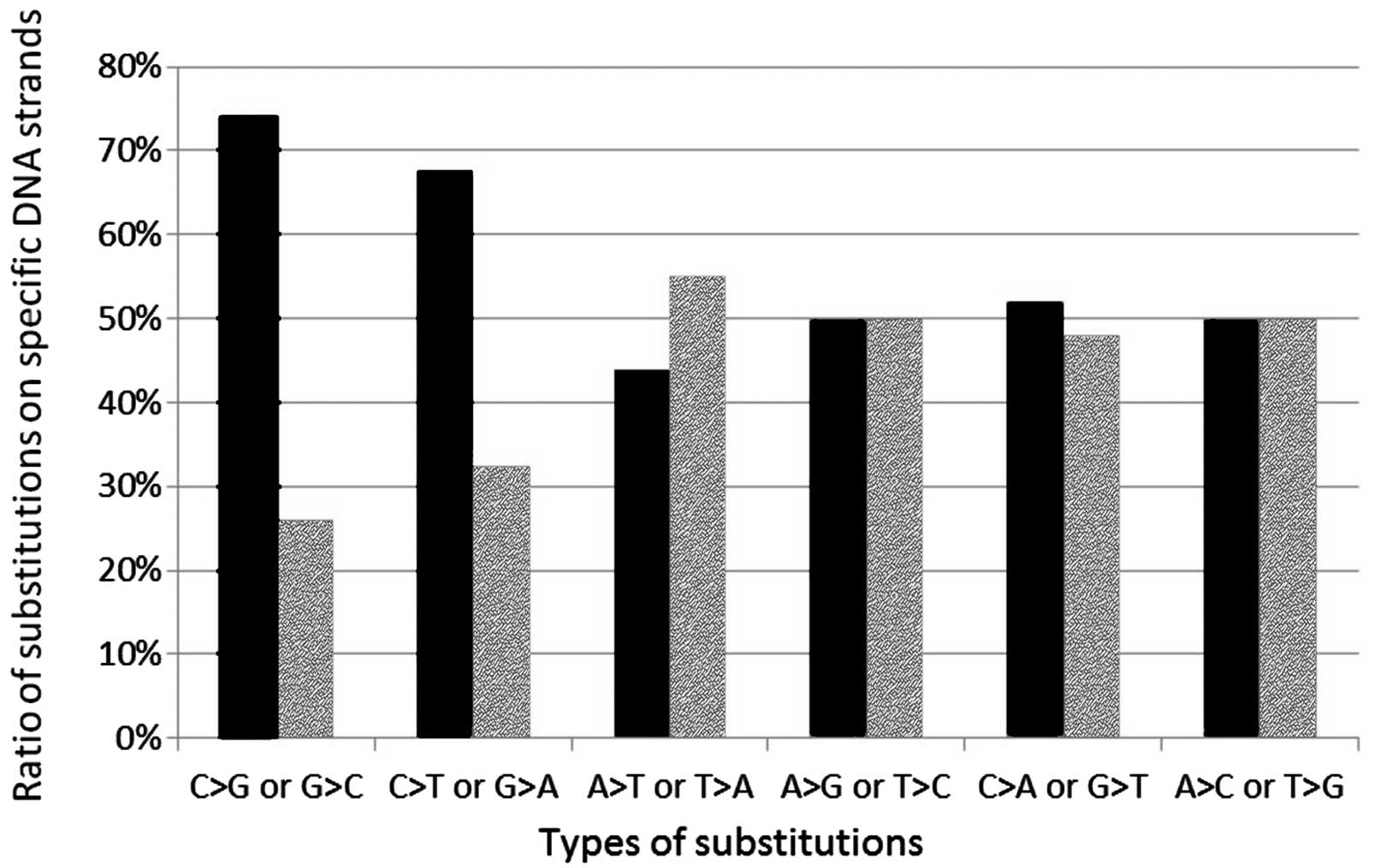
Frequency of pair substitutions
The frequency of pair substitutions was analyzed in
the four populations. The ratio of corresponding mutations (e.g.,
C>G and G>C) is usually approximately 1:1 because if
susceptibility of the two DNA strands to a given type of mutation
is the same, there should be an equal number of C>G mutations on
both strands. However, a C>G mutation on the coding strand is
detected as G>C on a transcribed strand. Notably, in the case of
two types of substitutions (C>G/G>C and C>T/G>A), the
ratio of the corresponding mutations was approximately 3:1 and 2:1,
respectively. The difference was statistically significant
(P<10−6 for C>T/G>A and P=0.037 for
C>G/G>C using the Chi-square test; Fig. 3).
Discussion
The potential differences in the distribution of
mutations between distinct populations including Polish and
Taiwanese were investigated in the present study.
Notably, statistically significant differences were
found between the frequency of TSC1 and TSC2
mutations between Polish/Taiwanese populations and American/British
populations. The TSC1 mutation is associated with a milder
clinical presentation of the disease. Thus, it is suggested that
the higher incidence of TSC2 mutations in Polish and
Taiwanese patients may be explained by a more pronounced clinical
picture. Additionally, the higher ratio of TSC1 mutations in
British and American patients might be due to the highly efficient
patient organizations in these countries, working actively to
increase disease awareness among the population. According to
Dabora et al(4), patients
with sporadic TSC1 mutations usually have milder disease
compared to patients with TSC2 mutations. Particularly, they
present with less frequent seizures and moderate to severe mental
retardation, fewer subependymal nodules and cortical tubers, less
severe kidney involvement, no retinal hamartomas, and less severe
facial angiofibroma.
In the present study, the level of sequence
complexity in sequences flanking the mutation in the Polish
population was found to be significantly higher compared to the
remaining populations analyzed. This was an interesting finding,
since mutations tend to generally appear in the sequences of lower
complexity. This finding suggests that there is a factor that
contributes to the occurrence of mutations in the Polish
population, where sequences are more complex. The identification of
this mutagenic factor, the activity of which is particularly
notable in more complex sequences, and the prevention of its
activity could lead to the reduction of TS morbidity in Poland, and
would also provide a better understanding of this disease.
Pleasance et al(11) reported the effect of substances
present in cigarette smoke on gene modifications in small cell lung
cancer cases. According to results of this study, G>T
transversions caused by polycyclic aromatic hydrocarbons occur more
often in the loci of methylated CpG dinucleotides in TP53, and
guanine, which is subjected to transversion into cytosine, is more
often preceded by adenine (11).
Additional studies describe the formation of C>T and CC>TT
mutations in skin cancer cases under the influence of ultraviolet
(UV) radiation. Radiation leads to the formation of covalent bonds
between adjacent pyrimidines, and subsequent mutations usually
appear in dipirimidine sequences (12,13).
It has also been noted that the same mutations, susceptible to UV
radiation, are more frequent in CpG dinucleotides (13,14).
Notably, the ratio of corresponding mutations was
not found to be equal (1:1), but significantly different in the
case of C>G/G>C and C>T/G>A pairs. Substitutional
strand asymmetry results from transcription and replication, with
both of these processes involving annealing of the two DNA strands.
Substitutional asymmetry resulting from replication has been
described in bacteria and human mitochondrial DNA (15,16),
while substitutional asymmetry resulting from transcription has
been described in mammals (17).
In the present study, an increased frequency of C>G/G>C and
C>T/G>A mutations in the coding strand was found in the
analyzed populations. Compared to 4,590 genes evaluated by Mugal
et al(18), where the
relative difference between mutation frequencies on both strands
(C>T/G>A) was rarely shown to be >35%, TSC1 and
TSC2 appear to be particularly prone to strand bias, as the
relative difference reaches 35% (18). However, additional studies and the
analysis of larger population groups are needed for further
investigation of this phenomenon.
References
|
1
|
Jones AC, Daniells CE, Snell RG, Tachataki
M, Idziaszczyk SA, Krawczak M, Sampson JR and Cheadle JP: Molecular
genetic and phenotypic analysis reveals differences between TSC1
and TSC2 associated familial and sporadic tuberous sclerosis. Hum
Mol Genet. 6:2155–2161. 1997. View Article : Google Scholar
|
|
2
|
Jones AC, Shyamsundar MM, Thomas MW,
Maynard J, Idziaszczyk S, Tomkins S, Sampson JR and Cheadle JP:
Comprehensive mutation analysis of TSC1 and TSC2 - and phenotypic
correlations in 150 families with tuberous sclerosis. Am J Hum
Genet. 64:1305–1315. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Niida Y, Lawrence-Smith N, Banwell A,
Hammer E, Lewis J, Beauchamp RL, Sims K, Ramesh V and Ozelius L:
Analysis of both TSC1 and TSC2 for germline mutations in 126
unrelated patients with tuberous sclerosis. Hum Mutat. 14:412–422.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dabora SL, Jozwiak S, Franz DN, Roberts
PS, Nieto A, Chung J, Choy YS, Reeve MP, Thiele E, Egelhoff JC,
Kasprzyk-Obara J, Domanska-Pakiela D and Kwiatkowski DJ: Mutational
analysis in a cohort of 224 tuberous sclerosis patients indicates
increased severity of TSC2, compared with TSC1, disease in multiple
organs. Am J Hum Genet. 68:64–80. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hung CC, Su YN, Chien SC, Liou HH, Chen
CC, Chen PC, Hsieh CJ, Chen CP, Lee WT, Lin WL and Lee CN:
Molecular and clinical analyses of 84 patients with tuberous
sclerosis complex. BMC Med Genet. 7:722006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Roach ES, Gomez MR and Northrup H:
Tuberous sclerosis complex consensus conference: revised clinical
diagnostic criteria. J Child Neurol. 13:624–628. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gomez MR: Phenotypes of the tuberous
sclerosis complex with a revision of diagnostic criteria. Ann N Y
Acad Sci. 615:1–7. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roach ES, Smith M, Huttenlocher P, Bhat M,
Alcorn D and Hawley L: Diagnostic criteria: tuberous sclerosis
complex. Report of the Diagnostic Criteria Committee of the
National Tuberous Sclerosis Association. J Child Neurol. 7:221–224.
1992. View Article : Google Scholar
|
|
9
|
Eng C, Brody LC, Wagner TM, Devilee P,
Vijg J, Szabo C, Tavtigian SV, Nathanson KL, Ostrander E and Frank
TS; Steering Committee of the Breast Cancer Information Core (BIC)
Consortium. Interpreting epidemiological research: blinded
comparison of methods used to estimate the prevalence of inherited
mutations in BRCA1. J Med Genet. 38:824–833. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bunn CF, Lintott CJ, Scott RS and George
PM: Comparison of SSCP and DHPLC for the detection of LDLR
mutations in a New Zealand cohort. Hum Mutat. 19:3112002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pleasance ED, Stephens PJ, O’Meara S,
McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman
C, Varela I, Nik-Zainal S, Davies HR, Ordoñez GR, Mudie LJ, Latimer
C, Edkins S, Stebbings L, Chen L, Jia M, Leroy C, Marshall J,
Menzies A, Butler A, Teague JW, Mangion J, Sun YA, McLaughlin SF,
Peckham HE, Tsung EF, Costa GL, Lee CC, Minna JD, Gazdar A, Birney
E, Rhodes MD, McKernan KJ, Stratton MR, Futreal PA and Campbell PJ:
A small-cell lung cancer genome with complex signatures of tobacco
exposure. Nature. 463:184–190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Daya-Grosjean L and Sarasin A: The role of
UV induced lesions in skin carcinogenesis: an overview of oncogene
and tumor suppressor gene modifications in xeroderma pigmentosum
skin tumors. Mutat Res. 571:43–56. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pleasance ED, Cheetham RK, Stephens PJ,
McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR,
Bignell GR, Ye K, Alipaz J, Bauer MJ, Beare D, Butler A, Carter RJ,
Chen L, Cox AJ, Edkins S, Kokko-Gonzales PI, Gormley NA, Grocock
RJ, Haudenschild CD, Hims MM, James T, Jia M, Kingsbury Z, Leroy C,
Marshall J, Menzies A, Mudie LJ, Ning Z, Royce T, Schulz-Trieglaff
OB, Spiridou A, Stebbings LA, Szajkowski L, Teague J, Williamson D,
Chin L, Ross MT, Campbell PJ, Bentley DR, Futreal PA and Stratton
MR: A comprehensive catalogue of somatic mutations from a human
cancer genome. Nature. 463:191–196. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pfeifer GP, You YH and Besaratinia A:
Mutations induced by ultraviolet light. Mutat Res. 571:19–31. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lobry JR: Asymmetric substitution patterns
in the two DNA strands of bacteria. Mol Biol Evol. 13:660–665.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tanaka M and Ozawa T: Strand asymmetry in
human mitochondrial DNA mutations. Genomics. 22:327–335. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Green P, Ewing B, Miller W, Thomas PJ and
Green ED; NISC Comparative Sequencing Program.
Transcription-associated mutational asymmetry in mammalian
evolution. Nat Genet. 33:514–517. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mugal CF, von Grünberg HH and Peifer M:
Transcription-induced mutational strand bias and its effect on
substitution rates in human genes. Mol Biol Evol. 26:131–142. 2009.
View Article : Google Scholar : PubMed/NCBI
|