Introduction
Hepatocellular carcinoma (HCC), the predominant
subtype primary liver cancer, is the fifth most common malignancy
in men and the seventh in women worldwide (1). Every year, there are ~750,000 new
cases and 700,000 deaths due to HCC around the world (2). Research in epidemiology has indicated
that many risk factors contribute to the HCC occurrence and
development, including hepatitis B virus or hepatitis C virus
infection, dietary aflatoxin B1 contamination, chronic alcohol
abuse and tobacco consumption, lack of dietary antioxidants,
arsenic exposure, obesity and non-alcoholic fatty liver disease
(3). Other risks for HCC include
geographic region, socio-economic status, gender, ethnicity and
environmental exposures (4).
Despite development in the therapy of HCC, the prognosis of HCC
patients remains unsatisfactory (5). This is primarily due to the fact that
only a small number of patients can receive surgery due to the
limitations such as the stage of the HCC, the number and the size
of the nodules, and the liver function (6). Additionally, the high rate of
recurrence and metastasis after surgery also results in poor
prognosis (7). Therefore, it is of
great significance to explore the molecular mechanism underlying
HCC progression, and investigate novel therapeutic strategies for
the treatments of HCC.
MicroRNAs (miRs) are a large family of single
strand, non-coding, and small RNAs consisting of 19–25 nucleotides
(8). As a new type of gene
expression regulators, miRNAs may regulate gene expression through
base pairing with the 3′ untranslated regions (3′UTRs) of their
target genes and thus participate in a variety of biological and
pathological processes, including cell proliferation, cycle,
apoptosis, differentiation, metabolism, invasion and metastasis
(9–11). Since the initial observation,
>1,000 miRNAs have been validated, and are estimated to modulate
the expression of >60% of protein-coding genes (12). Accumulated studies have
demonstrated that a larger number of miRNAs were dysregulated in
human cancers with differential regulation (‘up’ or ‘down’)
observed in neoplastic compared with normal cells (13). For example, miR-200b (14) and miR-379-5p (15) were downregulated, whereas miR-155
(16) and miR-130b (17) were upregulated in HCC. The
involvement of miRNAs in cancer carcinogenesis and progression is
well identified, as miRNAs can function as oncogenes or tumor
suppressor genes depending on the roles of their target genes
(18). Therefore, further
exploration of the abnormal expressed miRNAs and their direct
target genes would provide efficacious therapeutic targets for
patients with HCC.
In the present study, the authors focused on the
expression and roles of miR-296-5p in HCC. Results demonstrated
that miR-296-5p was significantly downregulated in HCC, and reduced
miR-296-5p expression levels were correlated with tumor size, TNM
stage and metastasis. miR-296-5p overexpression suppressed cell
proliferation, migration and invasion of HCC cells. Moreover, AKT2
was validated as the direct downstream and functional target gene
of miR-296-5p in HCC. miR-296-5p acted as a tumor suppressor in HCC
via direct targeting AKT2.
Materials and methods
Ethics statement and HCC samples
The current study was approved by the ethics
committee of Weifang Medical University (Weifang, China). Written
informed consent approving the use of their tissue samples for
research purposes was also obtained from all the HCC patients. A
total of 79 HCC tissues and adjacent non-tumor liver tissues were
obtained from patients underwent surgical resection of primary HCC
at the Associated Hospital of Weifang Medical University (Weifang,
China). All the tissue samples were histopathologically confirmed
and stored in liquid nitrogen until use.
Cell lines, cell culture and
transfection
Human HCC cell lines (HepG2, BEL-7402, SNU-182,
SMMC-7721, Huh7), the human normal liver cell line L02 and HEK293T
cell line were obtained from American Type Culture Collection
(ATCC; Manassas, VA, USA). All cell lines were cultured according
to ATCC's recommendations.
miR-296-5p mimics, miRNA negative control (NC), and
the small interfering (si)RNAs targeted for AKT2 (AKT2 siRNA) and
NC (NC siRNA) were synthesized by Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). For transfection, cells were seeded in six-well
plates at a density of 8×105 cells per well and cultured
overnight. miRNA or siRNA was transfected into cells by using
Lipofectamine 2000 (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), according to manufacturer's instructions. At 48
h following transfection, the transfection efficiency of miRNA and
siRNA was determined by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR).
RNA isolation and RT-qPCR
Total RNA was isolated from the cell lines or tissue
samples using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), following to the manufacturer's protocol. Total RNA was
reversed transcription into cDNA by using M-MLV Reverse
Transcription system (Promega Corporation, Madison, WI, USA).
Expression levels of miR-296-5p were determined by SYBR premix Ex
Taq™ kit (Takara Biotechnology Co., Ltd., Dalian, China) and
internal control of U6 RNA. AKT2 mRNA expression was quantified
using SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) with GADPH as an internal control. All RT-qPCR
was performed on an ABI PRISM® 7700 Sequence Detection System
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Relative
expression of miRNA and mRNA was evaluated as folder changes using
2−ΔΔCq method (19).
Cell Counting Kit 8 (CCK8) assay
Cell proliferation was determined with CCK8 (Dojindo
Molecular Technologies, Inc., Kumamoto, Japan) assay. In briefly,
transfected cells were harvested at 24 h post-transfection, and
seeded at a density of 3,000 cells/well in 96-well plates. Cells
were incubated for different periods (0, 24, 48, 72 and 96 h) in a
humidified atmosphere containing 5% CO2 at 37°C.
Subsequently, 10 µl CCK8 solution was added in each well and
incubated for additional 2 h. The absorbance was detected at 450 nm
using a microplate reader (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). Each experiment was repeated at least three times.
Transwell migration and invasion
assay
Transwell migration and invasion assays were
performed by using 24-well Transwell chambers (EMD Millipore,
Billerica, MA, USA) with an 8 mm polycarbonate membrane. For the
Transwell invasion assay, the polycarbonate membranes were coated
with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA). Briefly,
5×104 transfected cells in 200 µl fetal bovine serum (FBS; Gibco;
Thermo Fisher Scientific, Inc.)-free culture medium were placed
into the upper chamber, and the lower chamber was filled with 500
µl culture medium containing 20% FBS. After incubation for 48 h in
a humidified atmosphere containing 5% CO2 at 37°C, the
cells on the upper chamber of the polycarbonate membranes were
wiped off by cotton bud. The migrated and invaded cells were fixed
in 4% paraformaldehyde (Beyotime Institute of Biotechnology,
Haimen, China), stained with 0.5% crystal violet (Beyotime
Institute of Biotechnology), and washed three times with
phosphate-buffered saline (PBS; Gibco; Thermo Fisher Scientific,
Inc.). Finally, cells in five microscopic fields were counted and
photographed with a light microscope (Olympus Corporation, Tokyo,
Japan).
Western blot analysis
Cellular proteins were isolated by using
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology). Proteins were resolved on 10% SDS-PAGE and then
transferred onto polyvinylidene fluoride (EMD Millipore) membranes.
Subsequently, the membranes were blocked with 5% skim milk in TBS
containing 0.1% Tween 20 (TBST) at room temperature for 1 h, and
incubated with primary antibodies, mouse anti-human monoclonal AKT2
antibody (1:1,000 dilution; cat. no. sc-5270; Santa Cruz
Biotechnology Inc., Dallas, TX, USA) and anti-human monoclonal
GADPH antibody (1:1,000 dilution; cat. no. sc-365062; Santa Cruz
Biotechnology, Inc.), at 4°C overnight. On the following day, the
membranes were washed in TBST three times, and treated with
corresponding goat anti-mouse horseradish peroxidase-conjugated
secondary antibody (1:1,000 dilution; cat. no. sc-2005; Santa Cruz
Biotechnology, Inc.) diluted in TBST at room temperature for 1 h.
After washing in TBST, the signals were detected with enhanced
chemiluminescence system Western Blotting Detection Reagents (GE
Healthcare Life Sciences, Chalfont, UK) and photographed by
FluorChem imaging system (version 4.1.0; Alpha Innotech, San
Leandro, CA, USA).
Luciferase reporter assay
For the luciferase reporter assay, luciferase
reporter vectors (pMIR-AKT2-3′UTR Wt 1, pMIR-AKT2-3′UTR Wt 2,
pMIR-AKT2-3′UTR Mut 1 and pMIR-AKT2-3′UTR Mut 2) were synthesized
by Shanghai GenePharma Co., Ltd. (Shanghai, China). HEK293T cells
were seeded in 24-well plates at a density of 1.5×105 cells per
well and cotransfected with pMIR-AKT2-3′UTR Wt (1 and 2) or
pMIR-AKT2-3′UTR Mut (1 and 2) and miR-296-5p mimics or NC using
Lipofectamine 2000. At 48 h post-transfection, cells were harvested
and luciferase activities were measured by using Dual-Luciferase
Reporter Assay System (Promega Corporation), following to the
manufacturer's protocol. Firefly luciferase activities were
normalized to Renilla luciferase activities. Each sample was
performed in triplicate.
Statistical analysis
Data were presented as mean ± standard deviation,
and compared using Student's t-test in SPSS 16.0 statistical
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
miR-296-5p was downregulated in HCC
tissues and cell lines
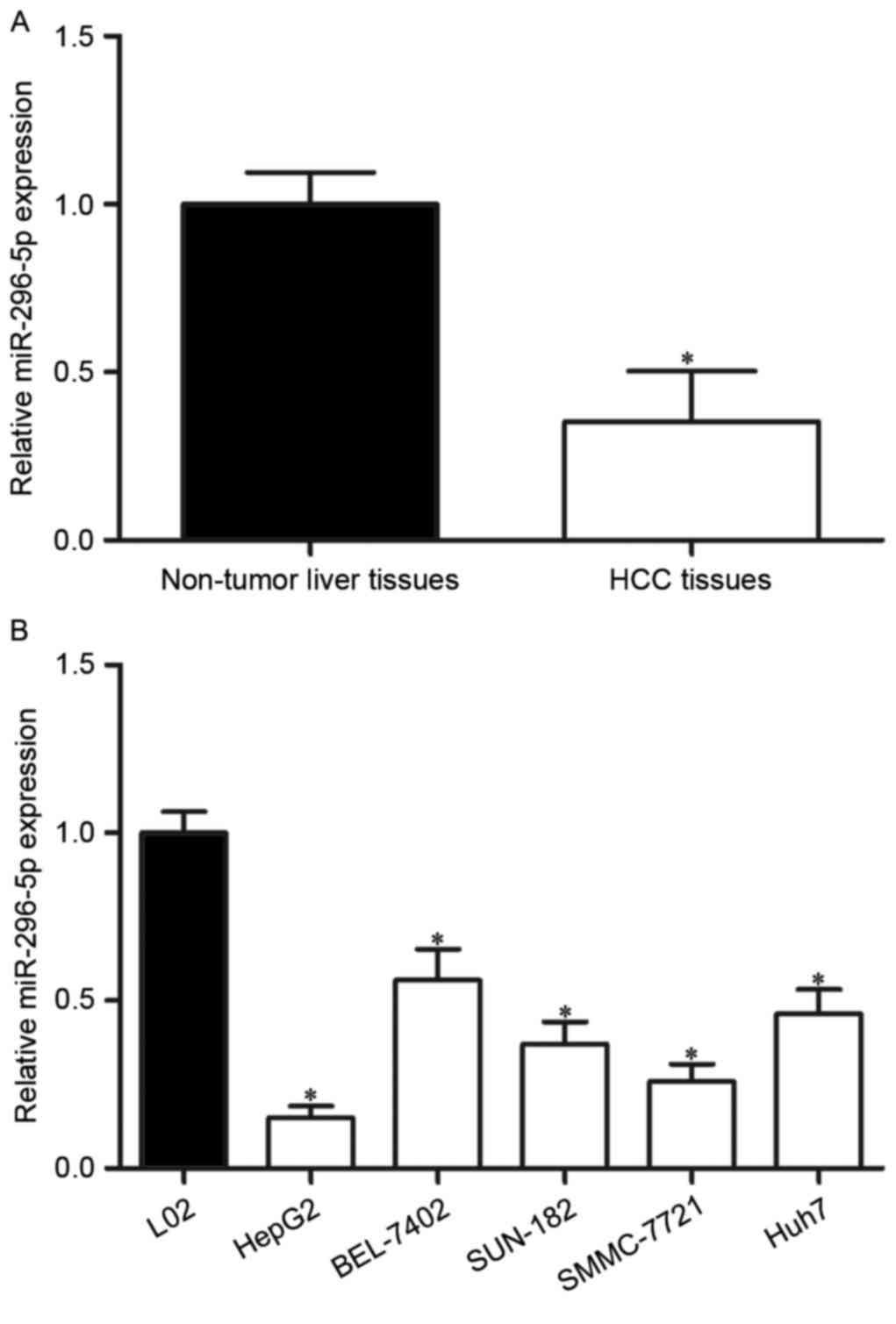
To explore the roles of miR-296-5p in HCC, its
expression in HCC tissues and adjacent non-tumor liver tissues was
measured by using RT-qPCR. The results demonstrated that miR-296-5p
was obvious downregulated in HCC tissues compared with that in
adjacent non-tumor liver tissues (Fig.
1A; P<0.05).
To validate whether the abnormal expression of
miR-296-5p was also the case in HCC cell lines, its expression in
five HCC cell lines was then detected (HepG2, BEL-7402, SNU-182,
SMMC-7721 and Huh7), as well as the human normal liver cell line
L02. The results confirmed that miR-296-5p expression levels were
reduced in each HCC cell lines than in L02 (Fig. 1B; P<0.05).
The relationship between miR-296-5p
expression and clinicopathological characteristics in HCC
The clinicopathological characteristics of
miR-296-5p expression in patients with HCC was investigated. As
presented in Table I, miR-296-5p
expression levels were significantly correlated with tumor size
(P=0.005), TNM stage (P=0.013) and metastasis (P=0.033), but not
with other clinicopathological characteristics, including age
(P>0.05), gender (P>0.05), tumor number (P>0.05) and
differentiation (P>0.05), in patients with HCC.
 | Table I.Correlations between miR-296-5p
expression and clinicopathological characteristics in
hepatocellular carcinoma. |
Table I.
Correlations between miR-296-5p
expression and clinicopathological characteristics in
hepatocellular carcinoma.
|
|
| miR-296-5p
expression |
|
|---|
|
|
|
|
|
|---|
| Clinical
features | Case number | Low | High | P-value |
|---|
| Age |
|
|
| 0.666 |
| <50
years | 44 | 23 | 21 |
|
| ≥50
years | 35 | 20 | 15 |
|
| Gender |
|
|
| 0.849 |
| Male | 54 | 29 | 25 |
|
|
Female | 25 | 14 | 11 |
|
| Tumor size |
|
|
| 0.005 |
| <5
cm | 39 | 15 | 24 |
|
| ≥5
cm | 40 | 28 | 12 |
|
| Tumor number |
|
|
| 0.789 |
|
Single | 47 | 25 | 22 |
|
|
Multiple | 32 | 18 | 14 |
|
| TNM Stage |
|
|
| 0.013 |
|
I–II | 30 | 11 | 19 |
|
|
III–IV | 49 | 32 | 17 |
|
| Metastasis |
|
|
| 0.033 |
| No | 59 | 28 | 31 |
|
|
Yes | 20 | 15 | 5 |
|
| Differentiated |
|
|
| 0.975 |
|
High | 55 | 30 | 25 |
|
|
Low | 24 | 13 | 11 |
|
miR-296-5p inhibited the
proliferation, migration and invasion of HCC cells in vitro
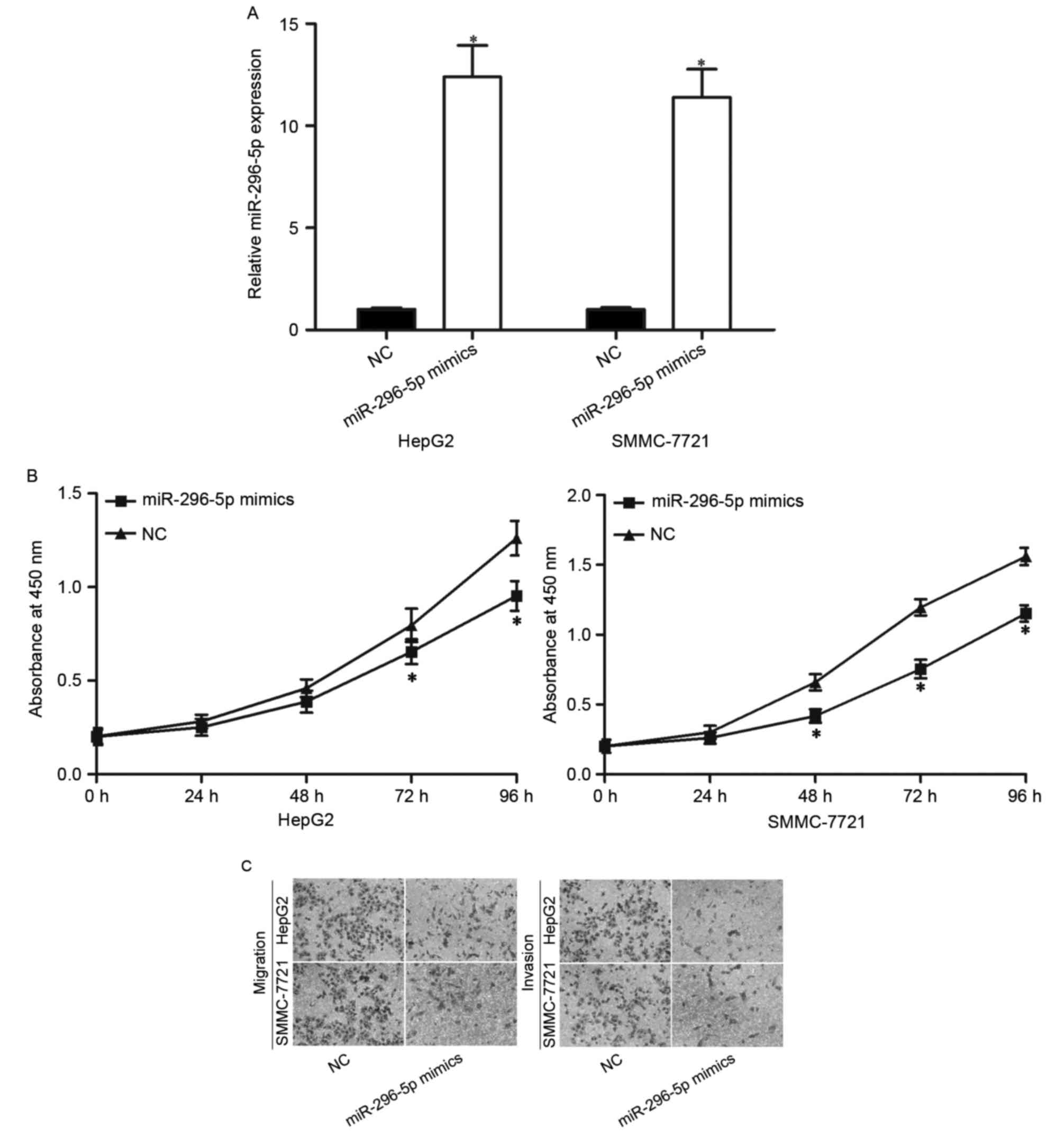
To determine the functions of miR-296-5p on HCC
progression, the authors transfected HepG2 and SMMC-7721 cells with
miR-296-5p mimics or NC. Following transfection, RT-qPCR reported
that miR-296-5p was remarkably increased in both HepG2 and
SMMC-7721 cells transfected with miR-296-5p mimics (Fig. 2A; P<0.05).
Considering that miR-296-5p expression was
correlated with tumor size, the authors explored the effect of
miR-296-5p on HCC proliferation using CCK8 assay. As demonstrated
in Fig. 2B, restoration of
miR-296-5p expression resulted in the inhibition of HepG2 and
SMMC-7721 cells proliferation (P<0.05). Furthermore, given the
association between miR-296-5p expression and HCC metastasis,
Transwell migration and invasion assays were performed to evaluate
the effects of miR-296-5p on HCC cells metastasis. It was observed
that enforced miR-296-5p expression decreased the migration and
invasion abilities in HepG2 and SMMC-7721 cells (Fig. 2C; P<0.05). These results
demonstrated that miR-296-5p acted as a tumor suppressor in HCC,
through inhibiting cellular growth and metastasis in
vitro.
AKT2 was a direct target gene of
miR-296-5p
To further explore the molecular mechanism of
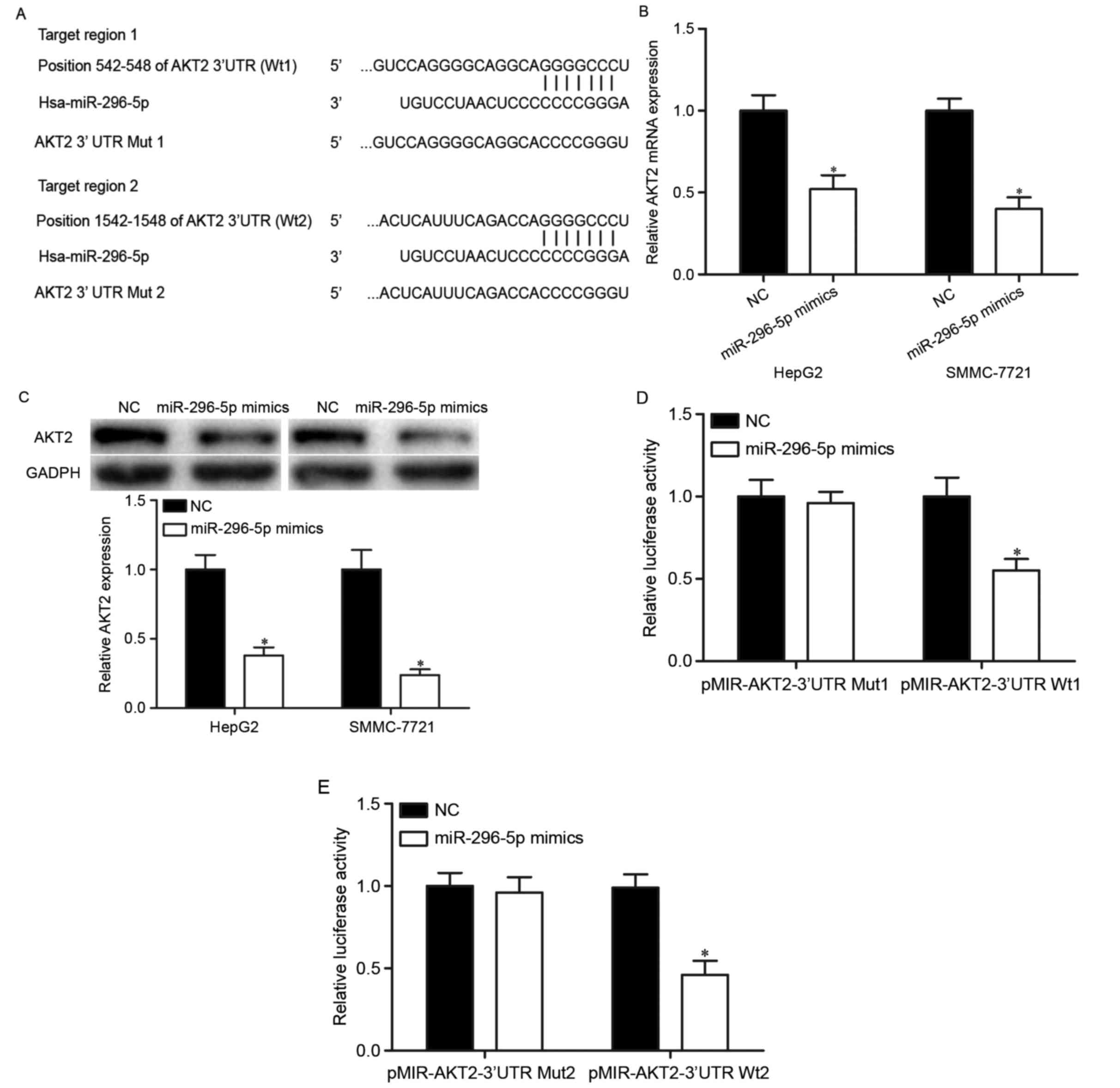
miR-296-5p mediated suppressive roles in HCC, the authors analyzed
the direct target genes of miR-296-5p. Firstly, TargetScan
(http://www.targetscan.org) and miRanda
(http://www.microrna.org) were used to predicate
the potential target genes of miR-296-5p. The analyses suggested
that 3′UTR of AKT2 contained the highly conserved putative
miR-296-5p binding sites (Fig.
3A). Subsequently, RT-qPCR and western blot analysis were
performed. The results indicated that upregulation of miR-296-5p
suppressed AKT2 expression in HepG2 and SMMC-7721 cells at both
mRNA (Fig. 3B; P<0.05) and
protein (Fig. 3C; P<0.05)
levels. Finally, luciferase reporter assay was adopted to validate
direct targeting of AKT2 by miR-296-5p. As demonstrated in Fig. 3D and E, the luciferase activities
were significantly reduced in HEK293T cells co-transfected with
pMIR-AKT2-3′UTR Wt (1 and 2) and miR-296-5p mimics. However,
miR-296-5p did not significantly alleviate the luciferase
activities in HEK293T cells transfected with pMIR-AKT2-3′UTR Mut (1
and 2). Taken together, these results strongly demonstrated that
AKT2 was a direct downstream target of miR-296-5p in HCC.
AKT2 was a downstream mediator of the
biological roles of miR-296-5p in HCC
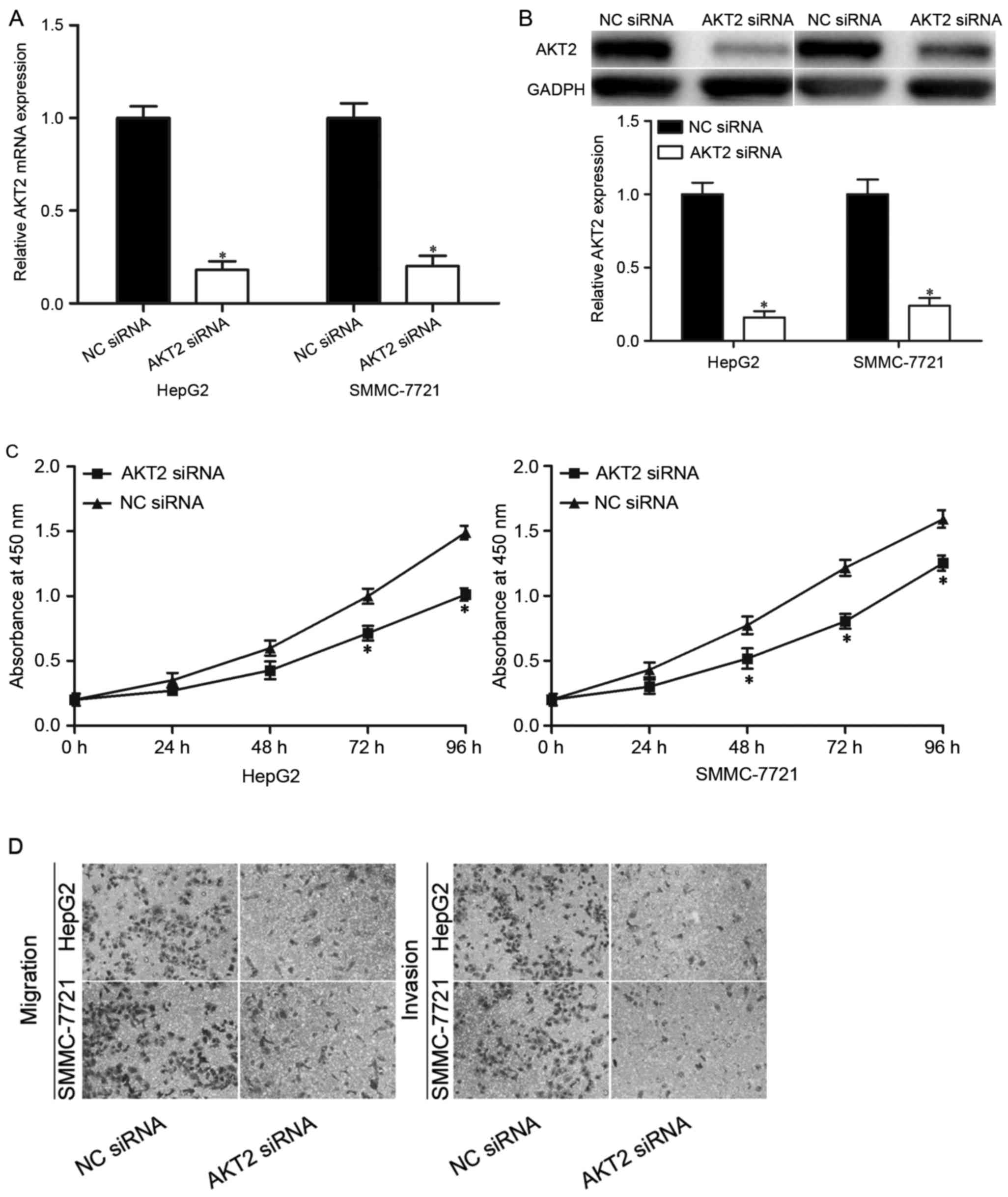
To investigate whether AKT2 mediated the biological
roles of miR-296-5p in HCC, AKT2 siRNA was adopted to knockdown
AKT2 expression in HepG2 and SMMC-7721 cells. Following
transfection, RT-qPCR and western blot analysis revealed that AKT2
siRNA could reduced AKT2 mRNA (Fig.
4A; P<0.05) and protein (Fig.
4B; P<0.05) expression levels in HepG2 and SMMC-7721 cells
compared with NC siRNA groups.
The CCK8 assay presented that the proliferation
ability of the HepG2 and SMMC-7721 cells transfected with AKT2
siRNA was markedly lower than that of the NC siRNA groups (Fig. 4C; P<0.05). Moreover, Transwell
migration and invasion assays indicated that silencing of AKT2 led
to a significant reduction of migration and invasion capacities in
HepG2 and SMMC-7721 cells (Fig.
4D; P<0.05). These results were consistent with the findings
that miR-296-5p overexpression can inhibit HCC cells proliferation,
migration and invasion in vitro, which provided further
evidences that AKT2 was a downstream mediator of the biological
roles of miR-296-5p in HCC.
Discussion
Increasing studies demonstrated that the abnormal
expression of miRNAs may be involved in HCC occurrence and
development, and some of these miRNAs may act as tumor suppressors
or oncogenes (20–22). Therefore, a better understanding of
the specific molecular events involved by miRNAs correlated with
carcinogenesis and progression of HCC is significant in exploring
new therapeutic strategies to improve the prognosis of patients
with this disease. In the present study, for the first time, the
authors revealed the expression and roles of miR-296-5p in HCC.
miR-296-5p was obvious downregulated in HCC tissues and cell lines
compared with adjacent non-tumor liver tissues and human normal
liver cell line, respectively. Reduced miR-296-5p levels were
correlated with tumor size, TNM stage and metastasis in HCC. The
gain-of-function demonstrated that miR-296-5p suppressed cell
proliferation, migration and invasion of HCC. Mechanistically, AKT2
was identified as a direct target gene of miR-296-5p via
bioinformatics analysis, RT-qPCR, western blotting and luciferase
reporter assay. Furthermore, the knockdown of AKT2 may phenocopy
the suppressive roles of miR-296-5p overexpression in HCC. These
results indicated that miR-296-5p may be a novel therapeutic target
for preventing HCC from rapidly growth and metastasis.
It has been reported that miR-296-5p was
dysregulated in diverse cancer types. For example, in breast
cancer, miR-296-5p was downregulated in tumor tissues, and low
expression levels of miR-296-5p predicted shorter disease-free
survival independently of classic clinicopathological parameters.
In addition, reduced miR-296-5p was associated with an earlier
spread of breast cancer in the overall series and with distant
metastases in the subset (23).
The downregulation of miR-296-5p was also identified in non-small
cell lung cancer (24) and
prostate cancer (25). However,
miR-296-5p expression was upregulated in esophageal cancer tissues,
and low expression of miR-296-5p was able to distinguish long-term
survivors with node-positive disease from those dying within 20
months by predicting survival (26). Gastric cancer also expressed high
levels of miR-296-5p (27). The
present study indicated that miR-296-5p was downregulated in HCC,
and correlated with pathological factors in HCC. Altogether,
miR-296-5p expression has tissue specificity, and may be a
prognostic marker for HCC.
The functions of miR-296-5p were mainly reported in
human cancer research. In non-small cell lung caner, miR-296-5p
targeted PLK1 to inhibit cell viability (24). In prostate cancer, miR-296-5p
overexpression decreased cell growth and invasion capacity by
negative regulation of HMGA1 (28). Lee et al (25) also reported that upregulation of
miR-296-5p suppressed cell proliferation and anchorage-independent
growth of prostate cancer cell lines through targeting Pin1. Savi
et al (23) revealed that
restoration of miR-296-5p into tumors of a breast cancer xenograft
model significantly decreased tumor growth via directly targeting
SCRIB. These findings verified that miR-296-5p may act as a tumor
suppressor in tumorigenesis and tumor development. Oppositely, it
was demonstrated to act as an oncogene (26,27).
In esophageal cancer, miR-296-5p underexpression repressed cell
proliferation in vitro and in vivo via targeting
cyclin D1 and p27. Furthermore, downregulation of miR-296-5p could
confer sensitivity of both P-glycoprotein-related and
P-glycoprotein-nonrelated drugs on esophageal cancer cells, and may
promote ADR-induced apoptosis, accompanied by increased
accumulation and decreased releasing amount of ADR (26). Li and his colleagues (27) reported that enforced miR-296-5p
expression promoted cell proliferation in gastric cancer through
downregulation of CDX1. These conflicting findings revealed that
the roles of miR-296-5p have tissue specificity, and could be
explained by the ‘imperfect complementarity’ of the interactions
between miR-296-5p and their direct target genes.
miRs serve significant roles in carcinogenesis and
progression of cancer by targeting key regulator. In addition, miRs
can act as either oncogenes or tumor suppressors, primarily
depending on the functions of their target genes in cancer.
Therefore, in the present work, the mechanism by which miR-296-5p
inhibited HCC cells proliferation, migration and invasion was
explored. In the current study, AKT2 was identified as a novel
direct downstream and functional target of miR-296-5p in HCC. AKT
is a key element in PI3K/AKT pathway. The PI3K/AKT pathway is
associated with aggressive phenotypes and poor outcomes in many
human cancers (29). In addition,
the PI3K/AKT pathway involves in a great deal of cellular processes
such as cell proliferation, apoptosis, migration, invasion and
metabolism (30,31). A previous study verified that AKT2
was upregulated in HCC tissues, and expression levels of AKT2 were
associated with histopathological differentiation, portal invasion
and number of tumor nodules in HCC (32). Multivariate analysis also revealed
that AKT2 was an independent prognostic marker for patients with
HCC (32). A recent studies also
indicated that AKT2 may be regulated by multiple miRs in HCC. For
example, miR-302b targeted AKT2 to inhibit HCC cells invasion and
metastasis (33). miR-137
suppressed HCC cells growth and metastasis through downregulation
of AKT2 (34). Taken together,
these data provided solid evidence to support that the miRs/AKT2
axis is a potential therapeutic target in HCC.
In conclusion, the current study indicated that
miR-296-5p acted as a tumor suppressor in HCC, through inhibiting
cell proliferation, migration and invasion.
The miR-296-5p/AKT2 axis serves important roles in
HCC carcinogenesis and progression, and miR-296-5p/AKT2 based
targeted therapy hampers HCC tumor growth and metastasis.
Restoration of miR-296-5p may represent a new therapeutic strategy
for patients with HCC. However, the molecular mechanism of low
expression of miR-296-5p in HCC requires further research.
References
|
1
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Borel F, Konstantinova P and Jansen PL:
Diagnostic and therapeutic potential of miRNA signatures in
patients with hepatocellular carcinoma. J Hepatol. 56:1371–1383.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yu MC and Yuan JM: Environmental factors
and risk for hepatocellular carcinoma. Gastroenterology. 127 5
Suppl 1:S72–S78. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
El-Serag HB and Rudolph KL: Hepatocellular
carcinoma: Epidemiology and molecular carcinogenesis.
Gastroenterology. 132:2557–2576. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bi Q, Tang S, Xia L, Du R, Fan R, Gao L,
Jin J, Liang S, Chen Z, Xu G, et al: Ectopic expression of MiR-125a
inhibits the proliferation and metastasis of hepatocellular
carcinoma by targeting MMP11 and VEGF. PLoS One. 7:e401692012.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Karaman B, Battal B, Sari S and Verim S:
Hepatocellular carcinoma review: Current treatment and
evidence-based medicine. World J Gastroenterol. 20:18059–18060.
2014.PubMed/NCBI
|
|
7
|
Jonas S, Bechstein WO, Steinmüller T,
Herrmann M, Radke C, Berg T, Settmacher U and Neuhaus P: Vascular
invasion and histopathologic grading determine outcome after liver
transplantation for hepatocellular carcinoma in cirrhosis.
Hepatology. 33:1080–1086. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kuhn AR, Schlauch K, Lao R, Halayko AJ,
Gerthoffer WT and Singer CA: MicroRNA expression in human airway
smooth muscle cells: Role of miR-25 in regulation of airway smooth
muscle phenotype. Am J Respir Cell Mol Biol. 42:506–513. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hwang HW and Mendell JT: MicroRNAs in cell
proliferation, cell death, and tumorigenesis. Br J Cancer. 96
Suppl:R40–R44. 2007.PubMed/NCBI
|
|
11
|
Kato M and Slack FJ: microRNAs: Small
molecules with big roles - C. elegans to human cancer. Biol Cell.
100:71–81. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li XY, Feng XZ, Tang JZ, Dong K, Wang JF,
Meng CC, Wang J, Mo YW and Sun ZW: MicroRNA-200b inhibits the
proliferation of hepatocellular carcinoma by targeting DNA
methyltransferase 3a. Mol Med Rep. 13:3929–3935. 2016.PubMed/NCBI
|
|
15
|
Chen JS, Li HS, Huang JQ, Dong SH, Huang
ZJ, Yi W, Zhan GF, Feng JT, Sun JC and Huang XH: MicroRNA-379-5p
inhibits tumor invasion and metastasis by targeting FAK/AKT
signaling in hepatocellular carcinoma. Cancer Lett. 375:73–83.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guan C, Yang F, He X, Li T, Yang Q, He H
and Xu M: Clinical significance of microRNA-155 expression in
hepatocellular carcinoma. Oncol Lett. 11:1574–1580. 2016.PubMed/NCBI
|
|
17
|
Chang RM, Xu JF, Fang F, Yang H and Yang
LY: MicroRNA-130b promotes proliferation and EMT-induced metastasis
via PTEN/p-AKT/HIF-1α signaling. Tumour Biol. 37:10609–10619. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dong Y, Zou J, Su S, Huang H, Deng Y, Wang
B and Li W: MicroRNA-218 and microRNA-520a inhibit cell
proliferation by downregulating E2F2 in hepatocellular carcinoma.
Mol Med Rep. 12:1016–1022. 2015.PubMed/NCBI
|
|
21
|
Chen X, Bo L, Zhao X and Chen Q:
MicroRNA-133a inhibits cell proliferation, colony formation
ability, migration and invasion by targeting matrix
metallopeptidase 9 in hepatocellular carcinoma. Mol Med Rep.
11:3900–3907. 2015.PubMed/NCBI
|
|
22
|
Li G, Yang F, Xu H, Yue Z, Fang X and Liu
J: MicroRNA-708 is downregulated in hepatocellular carcinoma and
suppresses tumor invasion and migration. Biomed Pharmacother.
73:154–159. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Savi F, Forno I, Faversani A, Luciani A,
Caldiera S, Gatti S, Foa P, Ricca D, Bulfamante G, Vaira V and
Bosari S: miR-296/Scribble axis is deregulated in human breast
cancer and miR-296 restoration reduces tumour growth in vivo. Clin
Sci (Lond). 127:233–242. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Xu C, Li S, Chen T, Hu H, Ding C, Xu Z,
Chen J, Liu Z, Lei Z, Zhang HT, et al: miR-296-5p suppresses cell
viability by directly targeting PLK1 in non-small cell lung cancer.
Oncol Rep. 35:497–503. 2016.PubMed/NCBI
|
|
25
|
Lee KH, Lin FC, Hsu TI, Lin JT, Guo JH,
Tsai CH, Lee YC, Lee YC, Chen CL, Hsiao M and Lu PJ:
MicroRNA-296-5p (miR-296-5p) functions as a tumor suppressor in
prostate cancer by directly targeting Pin1. Biochim Biophys Acta.
1843:2055–2066. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Hong L, Han Y, Zhang H, Li M, Gong T, Sun
L, Wu K, Zhao Q and Fan D: The prognostic and chemotherapeutic
value of miR-296 in esophageal squamous cell carcinoma. Ann Surg.
251:1056–1063. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li T, Lu YY, Zhao XD, Guo HQ, Liu CH, Li
H, Zhou L, Han YN, Wu KC, Nie YZ, et al: MicroRNA-296-5p increases
proliferation in gastric cancer through repression of
Caudal-related homeobox 1. Oncogene. 33:783–793. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei JJ, Wu X, Peng Y, Shi G, Basturk O,
Yang X, Daniels G, Osman I, Ouyang J, Hernando E, et al: Regulation
of HMGA1 expression by microRNA-296 affects prostate cancer growth
and invasion. Clin Cancer Res. 17:1297–1305. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Manning BD and Cantley LC: AKT/PKB
signaling: Navigating downstream. Cell. 129:1261–1274. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Agarwal E, Brattain MG and Chowdhury S:
Cell survival and metastasis regulation by Akt signaling in
colorectal cancer. Cell Signal. 25:1711–1719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kang B, Hao C, Wang H, Zhang J, Xing R,
Shao J, Li W, Xu N, Lu Y and Liu S: Evaluation of
hepatic-metastasis risk of colorectal cancer upon the protein
signature of PI3K/AKT pathway. J Proteome Res. 7:3507–3515. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu X, Sakon M, Nagano H, Hiraoka N,
Yamamoto H, Hayashi N, Dono K, Nakamori S, Umeshita K, Ito Y, et
al: Akt2 expression correlates with prognosis of human
hepatocellular carcinoma. Oncol Rep. 11:25–32. 2004.PubMed/NCBI
|
|
33
|
Wang L, Yao J, Sun H, Sun R, Chang S, Yang
Y, Song T and Huang C: miR-302b suppresses cell invasion and
metastasis by directly targeting AKT2 in human hepatocellular
carcinoma cells. Tumour Biol. 37:847–855. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu LL, Lu SX, Li M, Li LZ, Fu J, Hu W,
Yang YZ, Luo RZ, Zhang CZ and Yun JP: FoxD3-regulated microRNA-137
suppresses tumour growth and metastasis in human hepatocellular
carcinoma by targeting AKT2. Oncotarget. 5:5113–5124. 2014.
View Article : Google Scholar : PubMed/NCBI
|