Introduction
The emergence of antibiotic resistance and the
potential for transfer of resistance genes has raised great public
concern (1). New Delhi
metallo-β-lactamase-1 (NDM-1), encoded by
blaNDM-1, was originally identified in 2009 in a
Klebsiella pneumoniae isolate from a Swedish patient
transferred from India (2).
NDM-1-producing bacteria, including clinical isolates of
Enterobacteriaceae and Acinetobacter spp., have since
been reported across the Indian subcontinent and worldwide
(2–9). NDM-1-producing bacteria are generally
resistant to almost all β-lactam antibiotics, including
carbapenems, which have brought great challenges to antibiotic
therapy. The size of plasmids harboring blaNDM-1
varies considerably, ranging from 50 to >400 kb. In addition,
the plasmids belong to different incompatibility (Inc) groups,
including IncA/C, IncFI/FII, IncL/M and a non-typed group (10,11).
Our previous study identified an IncA/C plasmid,
designated pHS36-NDM, that was identified in a carbapenem-resistant
Salmonella Stanley strain isolated from the stool sample of
an 11-month-old girl with community-acquired acute diarrhea, and
could be transferred from Salmonella to E. coli C600
and K. pneumoniae (12).
The plasmid was highly similar to NDM-1-carrying plasmids reported
in Sweden, Italy and Japan, however the source of
blaNDM-1 remains unclear due to the lack of
relationship with the patient's social and travel history. In order
to further understand the underlying mechanism of resistance,
spread and passage stability, the present study sequenced pHS36-NDM
and performed a comparison with reported NDM-1-harboring plasmids,
and analyzed the phenotype-related genetic characteristics of
pHS36-NDM.
Materials and methods
Plasmid extraction and sequencing
A carbapenem-resistant E. coli C600
transconjugant was created by performing transconjugation between
the Salmonella Stanley strain and E.coli C600 Rifr,
as previously described (12).
pHS36-NDM plasmid DNA was purified from the carbapenem-resistant
E. coli C600 transconjugant using a Qiagen Plasmid Midi kit
(Qiagen GmbH, Hilden, Germany) according to the manufacturer's
protocol. Pyrosequencing was performed using the GS-FLX Titanium
System (Roche Diagnostics, Basel, Switzerland), according to the
manufacturer's protocol, as previously described (13). Paired-end reads were collected at a
single site and assembled using Newbler software version 2.3 (Roche
Diagnostics). The assembly was further improved manually and with
the aid of custom Perl scripts.
Bioinformatics analysis of pHS36-NDM
DNA sequence
Open reading frames (ORFs) were predicted and
annotated using the RAST server (http://rast.nmpdr.org/). Each predicted protein was
compared with the National Centre for Biotechnology Information
(NCBI) protein database using the protein basic local alignment
sequence tool (BLASTP; National Institutes of Health, Bethesda, MD,
USA; http://blast.ncbi.nlm.nih.gov/Blast.cgi), with a
minimum cutoff of 30% in identity and >80% in length coverage.
Gene sequences were further compared and aligned with the GenBank
database using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Proteins
were assigned to clusters of orthologous groups (http://www.ncbi.nlm.nih.gov/COG/), and genes were
initially annotated using In Silico Molecular Cloning (IMC)
Genomics Edition (version 4.1.21D; In Silico Biology Inc.,
Yokohama, Japan). The circular representation of pHS36-NDM was
generated with IMC. Linear comparative representations were based
on results from Vector NTI 8 (Invitrogen; Thermo Fisher Scientific,
Inc., Waltham, MA, USA).
Genetic contexts of pHS36-NDM
To investigate the homology of pHS36-NDM with
previously reported NDM-1-containing plasmids, and the differences
between flanking regions of the blaNDM-1 gene,
pHS36-NDM was compared with the following 8
blaNDM-1-harboring plasmids: pMR0211 from
Providencia stuartii (GenBank accession no. JN687470.1),
pNDM-1_Dok01 from E. coli (GenBank accession no.
AP012208.1), pNDM-KN from K. pneumoniae (GenBank accession
no. NC_019153.1), pNDM-HN380 from K. pneumoniae (GenBank
accession no. JX104760.1), pNDM-BJ01 from A. lwoffii
(GenBank accession no. JQ001791.1), pKpANDM-1 from K.
pneumoniae (GenBank accession no. FN396876.1), pNDM-OM from
K. pneumoniae (GenBank accession no. JX988621.1) and
pGUE-NDM from E. coli (GenBank accession no. JQ364967.1).
Furthermore, these plasmids were used as references for annotating
pHS36-NDM.
Results
General features of plasmid
pHS36-NDM
Sequencing of pHS36-NDM revealed it to be 138,001 bp
in size with 52.0% guanine-cytosine content (Fig. 1). The plasmid contained 181
putative ORFs, of which 147 were on the same (plus) strand as the
replication initiator gene, repA, and 34 were on the minus
strand. BLASTN analysis revealed that the sequence of pHS36-NDM was
well conserved with E. coli pNDM102337 (99% identity;
GenBank accession no. JF714412.2), and K. pneumoniae
pNDM10469 (99% identity; GenBank accession no. JN861072.1),
indicating that pHS36-NDM may be frequently transmitted amongst
virulent Enterobacteriaceae. Several common functional
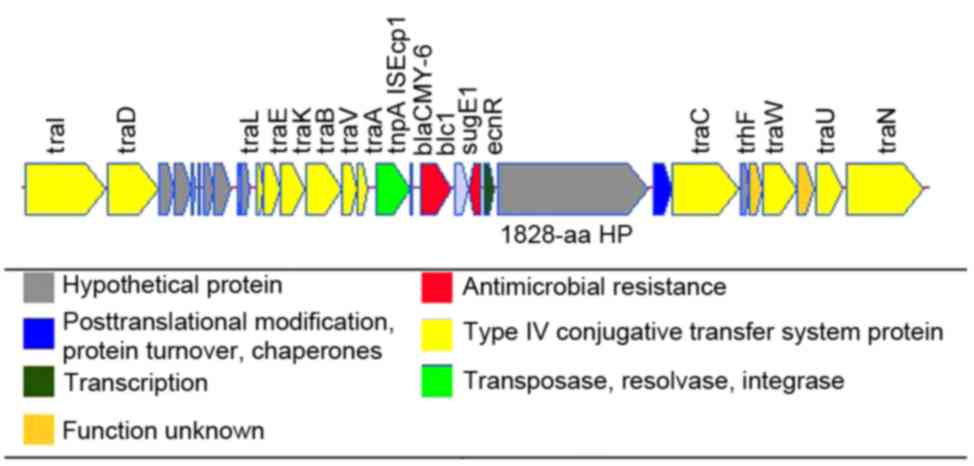
regions were predicted (Fig. 1),
including: A replication region containing repA and
plasmid-partitioning genes parA and parB, the
ISEcp1-blaCMY module region containing the
type IV secretion system (T4SS) conjugative transfer genes and
CMY-6 β-lactamase gene, blaCMY-6; and a
blaNDM-1-containing transposon region flanked by
a class 1 integron and insertion sequence common repeat 27
transposase (ISCR27).
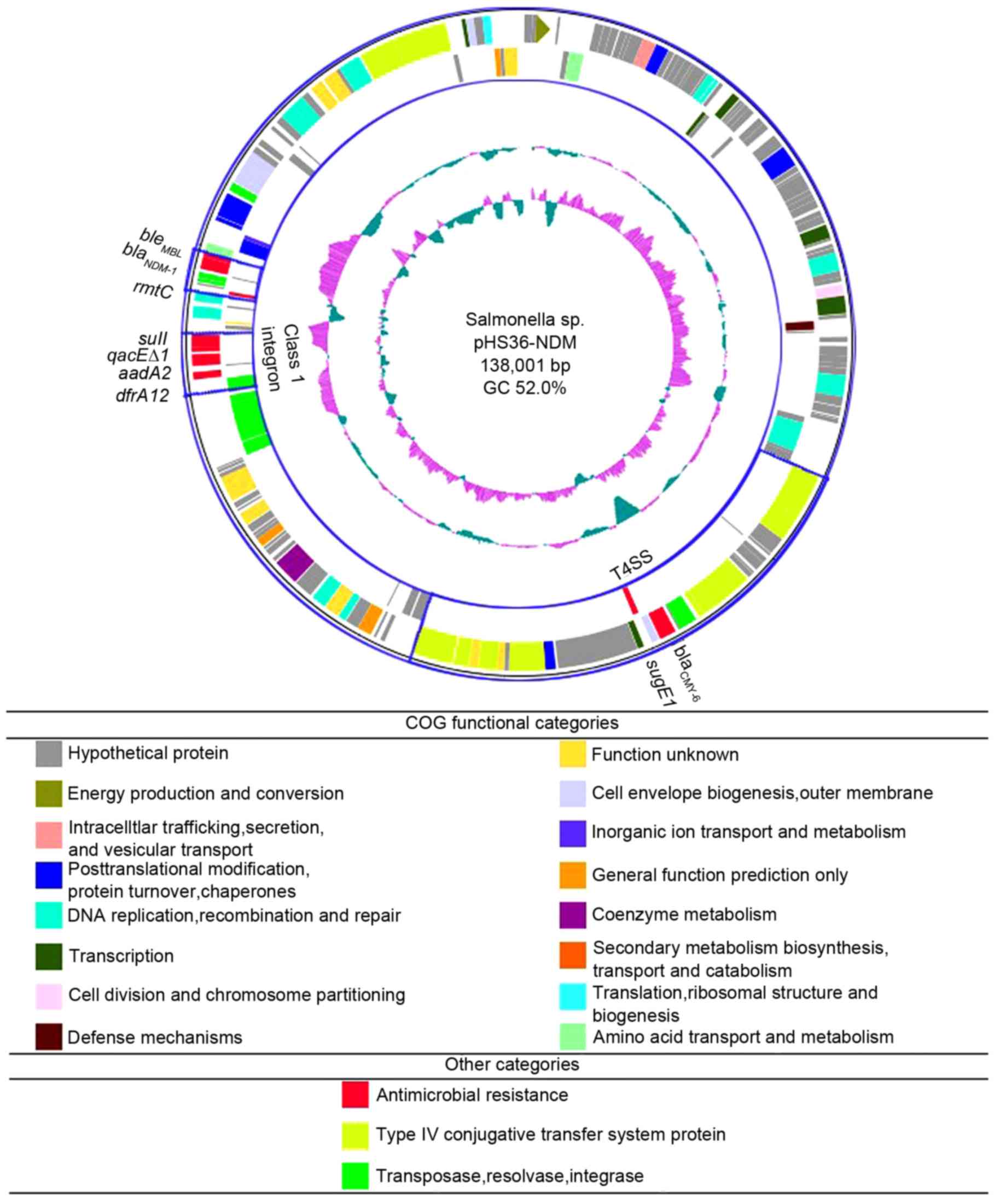 | Figure 1.Circular representation of pHS36-NDM.
The innermost circle represents GC skew; the second inner circle
represents GC content (blue, GC+; purple, GC-). The outermost
double circle depicts gene distribution relative to repA
(represented as an arrowhead) with colored boxes depicting
functional category and direction of transcription; the outer band
is the plus strand; inner band is the minus strand. The loci for a
T4SS operon and class 1 integron are indicated in blue boxes. NDM,
New Delhi metallo-β-lactamase-1; GC, guanine + cytosine;
repA, replication protein A; T4SS, type IV secretion system;
blaCMY6, CMY-6 β-lactamase gene; sugE1,
quaternary ammonium compound resistance gene; dfrA12,
trimethoprim resistance gene; aadA2, aminoglycoside
resistance gene; qacEΔ1, truncated quaternary ammonium
compound resistance gene; sul1, sulfonamide resistance gene;
rmtC, rRNA methyltransferase; blaNDM-1,
New Delhi metallo-β-lactamase-1; bleMBL,
bleomycin resistance gene. |
The T4SS cluster and stable plasmid
inheritance protein A (stbA) are responsible for horizontal
transfer and passage stability
The transfer region of pHS36-NDM structurally
belonged to a T4SS, comprising 15 transfer (tra) genes
(traI, traD, traL, traE, traK, traB, traV, traA, traC, traW,
traU, traN, traF, traH and traG), which were responsible
for conjugation. The transfer region is conserved amongst
pNDM-1_Dok01, pNDM102337, pNDM10469 and is conserved in gene order
(14). The stbA gene
identified in pHS36-NDM is necessary for the low copy number
plasmid to be passed to the daughter cells (15,16).
Two multidrug resistance genes islands
are responsible for the in vitro drug resistance phenotype
One or more copies of the
ISEcp1-blaCMY transposition unit genes have been
reported within T4SSs in IncA/C plasmids (17,18).
In pHS36-NDM, the ISEcp1-blaCMY transposition
unit contained two resistance genes, blaCMY-6 and
a quaternary ammonium compound resistance gene, sugE
(Fig. 2), flanked by ISEcp1
transposases. Furthermore, blaCMY-6 and
sugE of pHS36-NDM were embedded in the T4SS cluster at
60,922 bp, flanked by traA and a hypothetical protein 1,828
amino acids in size. This hypothetical protein had 100% identity
with that found in pNDM102337, pNDM10469 and pNDM-1_Dok01, and high
similarity with pMR0211; pMR0211 has 4 extra amino acids at its
5′terminus. In addition, a short sequence was identified at the
3′terminus of pH S36-NDM blaCMY-6 (ATT TCC CTA),
which is almost identical to the 5′terminus (ATT TCC TTA), located
adjacent to traA.
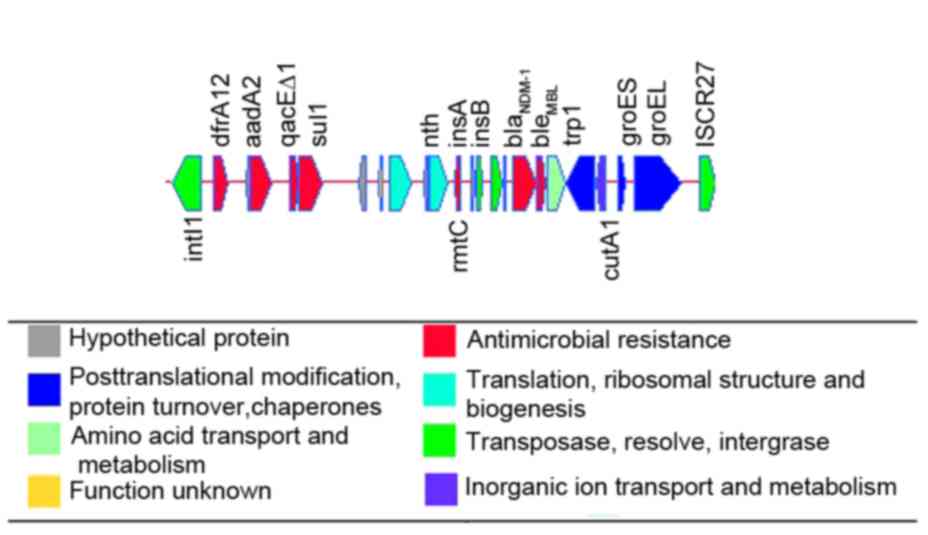
The second island was the intI-ISCR27 transposition
unit (Fig. 3). Seven drug
resistance genes were clustered together in a 14,802 bp accessory
region bordered by intI1 [99,852-100,865 (−)] and
ISCR27 [114,073-114,653 bp (+)]. The accessory region is
14,802 bp long and contains 24 genes, including genes conferring
resistance to trimethoprim (dfrA12), aminoglycosides
(aadA2 and rmtC), quaternary ammonium compounds
(qacE∆1), sulfonamides (sul1), β-lactams including
carbapenems (blaNDM-1) and bleomycin
(bleMBL). One part of this accessory region, the
class 1 integron, was composed of intl1 and the antibiotic
resistance markers dfrA12, aadA2, qacE∆1 and
sul1. Class 1 integrons have been found in several
Gram-negative bacteria, such as pNDM-1_Dok01 (3).
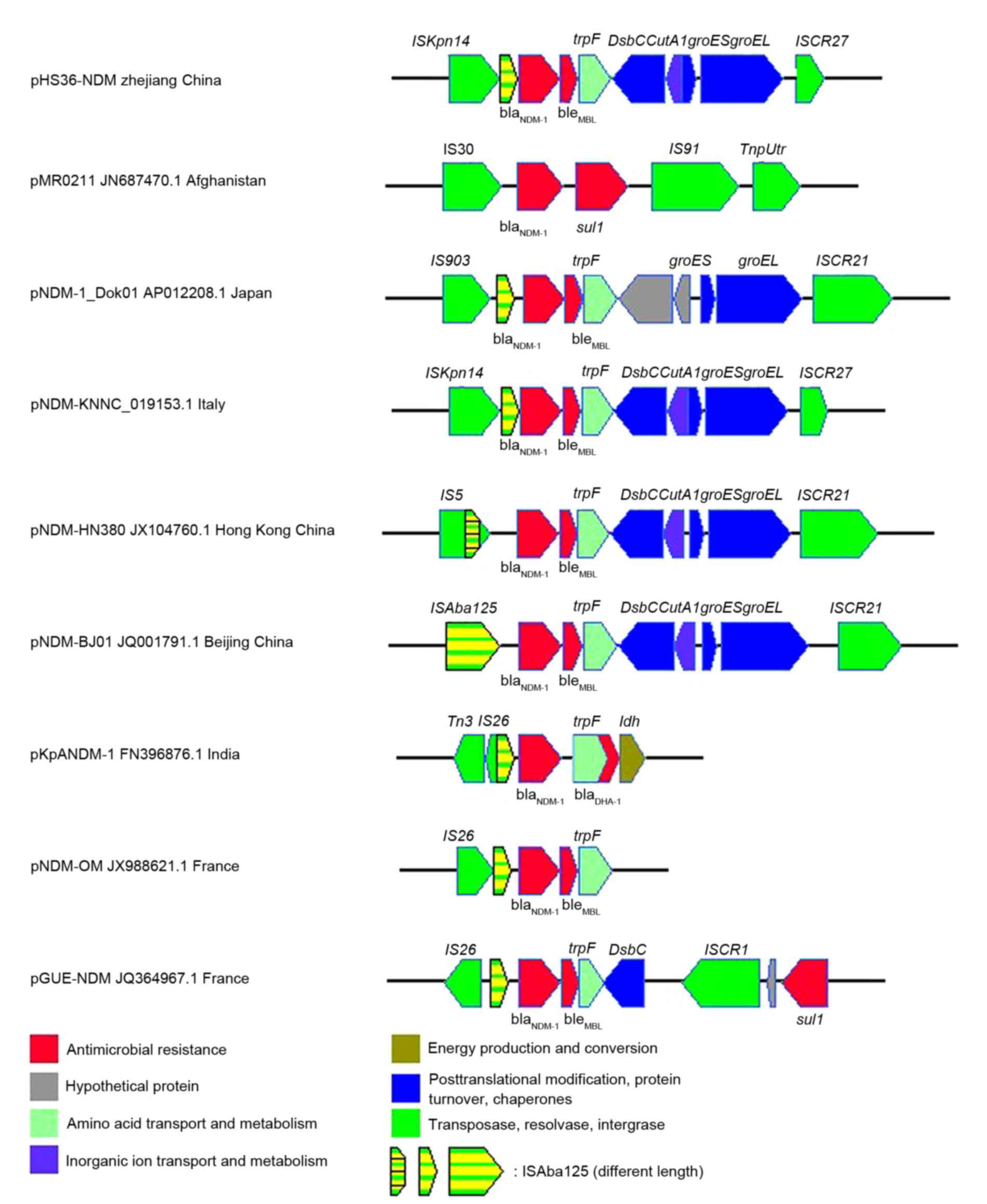
Sequence analysis of elements flanking
blaNDM-1
The pHS36-NDM blaNDM-1 gene was
localized in a 7 kb region that was flanked by ISCR27 and the
insertion sequence ISKpn14 (Fig.
4). Immediately downstream of the blaNDM-1
gene, a bleMBL gene was identified, encoding a
putative protein conferring resistance to bleomycin.
bleMBL was followed by an N-(5′-phosphoribosyl)
anthranilate isomerase (trpF) gene, found previously in
other blaNDM-1-bearing plasmids (Fig. 4). Furthermore, genes encoding a
bifunctional protein-disulfide isomerase/oxidoreductase
(dsbC) and CutA1 periplasmic divalent cation tolerance
protein (cutA1) were commonly identified in other
NDM-harboring plasmids (Fig. 4).
Finally, chaperonins groES and groEL, which are
involved in general stress responses, together with ISCR27,
were also present in the regions flanking
blaNDM-1 (Fig.
4). Although in many plasmids, the heat shock chaperone
groEL-groES cluster is adjacent to an rhs gene, which
belongs to the retrotransposon hot spot family that are known as
hotspots for integration (19), no
such rhs gene insertion was found in this region of
pHS36-NDM.
Notably, a truncated ISAba125 insertion
sequence adjacent to the ISKpn14 insertion sequence was
identified immediately upstream of the blaNDM-1
gene (Fig. 4). This truncated
ISAba125 contained a 235-nucleotide promoter sequence, which
drives blaNDM-1 gene expression. A truncated
ISAba125 of varying sizes containing this specific promoter
was identified in almost all blaNDM-1-containing
plasmids (Fig. 4). Furthermore,
the bleMBL gene, followed by trpF, was
observed to be consistently adjacent to the 3′end of
blaNDM-1. These results suggest that the
blaNDM-1 gene may originally have been linked to
ISAba125.
Discussion
Salmonella is an important foodborne pathogen
that it frequently causes infection and worldwide outbreaks
(20). Although Salmonella
is increasingly resistant to cephalosporins and quinolones,
resistance to carbapenems is rare. The present study revealed the
complete sequence of plasmid pHS36-NDM, which harbors a
blaNDM-1 gene that was observed to confer
carbapenem resistance to a Salmonella Stanley isolated from
a child in China.
pHS36-NDM presented a well-conserved plasmid
structure with E. coli pNDM102337, K. pneumoniae
pNDM10469, E. coli pNDM10505 (GenBank accession no.
JF503991.1) and K. pneumoniae pNDM-KN, indicating that the
plasmid may frequently be transmitted amongst virulent
Enterobacteriaceae. The presence of T4SS genes and
stbA indicated the capability of horizontal transfer and
passage stability of the pHS36-NDM plasmid. Previous studies have
demonstrated that the T4SS is a double walled transmembrane
structure, which macromolecular nanomachines utilize for the
transport of proteins or DNA across the bacterial cell envelope of
Gram-negative bacteria (21,22).
The transfer of pHS36-NDM to daughter cells may be dependent on the
presence of the stbA gene (16). Furthermore, the plasmid harbored
several mobile genetic elements including ISEcp1, insertion
sequence IS4321, transposon Tn1696, a class 1
integron, ISKpn14, ISAb125 and ISCR27,
which increased the plasticity of the plasmid. This feature of
pHS36-NDM may contribute to its marked genetic stability in the
donor and the transconjugant strains, and to the high conjugation
frequency that has previously been reported between different types
of bacteria (12). Further
analysis indicated that, in the
ISEcp1-blaCMY transposition unit, the
blaCMY-6 3′ATTTCCTTA tandem repeat sequence and
the 5′downstream sequence of traA are conserved across all
IncA/C NDM-harboring plasmids investigated, such as pNDM-1_Dok01,
pNDM102337 and pNDM10469, indicating that the short flanking repeat
sequence is probably a transposon insertion site conserved in the
IncA/C plasmid backbone.
Our previous study demonstrated that the
Salmonella strain containing pHS36-NDM was resistant to all
β-lactam antibiotics, including cephalosporins and carbapenems, but
susceptible to chloramphenicol, ciprofloxacin, tetracycline,
fosfomycin, and azithromycin (12). These antibiotic susceptibility
results were consistent with the resistance genes identified in the
two transposition units, ISEcp1-blaCMY and
intI1-ISCR27. pHS36-NDM was highly conserved with
pNDM-KN from K. pneumoniae in Italy, pNDM10469 from K.
pneumoniae and pNDM102337 from E. coli in Japan,
however, the affected patient and her family had not traveled to
any countries within the year, including 14 countries with a high
prevalence of NDM-1 producers. Furthermore, the patient had been
living in a small rural village in southern China without a special
diet (12). Although it is
important to investigate the origin and transmission of
blaNDM-1, there was no evidence to suggest that
pHS36-NDM originated from Italy or Japan. Comparing pHS36-NDM with
other blaNDM-1-bearing plasmids reported in
China, including pKPN5047 (GenBank accession no. KC311431.1) and
pNDM-HN380 from K. pneumoniae, pNDM-BJ01 from A.
lwoffii and pYE315203 (GenBank accession no. JX254913.2) from
C. freundii, revealed that these plasmids belonged to a
different Inc type and shared low homology in the surrounding
structure of blaNDM-1. These results implied that
it is unlikely that the gene cluster in pHS36-NDM originated from
other previously discovered blaNDM-1-carrying
plasmids in China. It is more likely that this occurred due to
independent evolutionary events, resulting from the abuse of
antibiotics on a global scale. Toleman et al (23) proposed two possible routes of
blaNDM-1 construction, which involve a deletion
event and a rolling-circle replication event. According to the
present study, it is postulated that blaNDM-1 and
the surrounding region in pHS36-NDM may have been formed via
multiple recombination events by genetic elements of different
origins.
Acknowledgements
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos.
81120108024 and 81102509), the National Key Programs for Infectious
Diseases of China (grant no. 2013ZX10004216-001-004) and Shanghai
Municipal Science and Technology (grant no. 12JC1401700).
References
|
1
|
Levy SB and Marshall B: Antibacterial
resistance worldwide: Causes, challenges and responses. Nat Med. 10
Suppl 12:S122–S129. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yong D, Toleman MA, Giske CG, Cho HS,
Sundman K, Lee K and Walsh TR: Characterization of a new
metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin
esterase gene carried on a unique genetic structure in Klebsiella
pneumoniae sequence type 14 from India. Antimicrob Agents
Chemother. 53:5046–5054. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sekizuka T, Matsui M, Yamane K, Takeuchi
F, Ohnishi M, Hishinuma A, Arakawa Y and Kuroda M: Complete
sequencing of the bla(NDM-1)-positive IncA/C plasmid from
Escherichia coli ST38 isolate suggests a possible origin from plant
pathogens. PLoS One. 6:e253342011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carattoli A, Villa L, Poirel L, Bonnin RA
and Nordmann P: Evolution of IncA/C blaCMY-2-carrying plasmids by
acquisition of the blaNDM-1 carbapenemase gene. Antimicrob Agents
Chemother. 56:783–786. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lowe CF, Kus JV, Salt N, Callery S, Louie
L, Khan MA, Vearncombe M and Simor AE: Nosocomial transmission of
New Delhi metallo-β-lactamase-1-producing Klebsiella pneumoniae in
Toronto, Canada. Infect Control Hosp Epidemiol. 34:49–55. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ho PL, Li Z, Lo WU, Cheung YY, Lin CH,
Sham PC, Cheng VC, Ng TK, Que TL and Chow KH: Identification and
characterization of a novel incompatibility group X3 plasmid
carrying bla NDM-1 in Enterobacteriaceae isolates with
epidemiological links to multiple geographical areas in China.
Emerg Microbes Infect. 1:e392012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu H, Hu Y, Pan Y, Liang H, Wang H, Wang
X, Hao Q, Yang X, Yang X, Xiao X, et al: Novel plasmid and its
variant harboring both a bla(NDM-1) gene and type IV secretion
system in clinical isolates of Acinetobacter lwoffii. Antimicrob
Agents Chemother. 56:1698–1702. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bonnin RA, Nordmann P, Carattoli A and
Poirel L: Comparative genomics of IncL/M-type plasmids: Evolution
by acquisition of resistance genes and insertion sequences.
Antimicrob Agents Chemother. 57:674–676. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bonnin RA, Poirel L, Carattoli A and
Nordmann P: Characterization of an IncFII plasmid encoding NDM-1
from Escherichia coli ST131. PLoS One. 7:e347522012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kumarasamy KK, Toleman MA, Walsh TR,
Bagaria J, Butt F, Balakrishnan R, Chaudhary U, Doumith M, Giske
CG, Irfan S, et al: Emergence of a new antibiotic resistance
mechanism in India, Pakistan, and the UK: A molecular, biological,
and epidemiological study. Lancet Infect Dis. 10:597–602. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ho PL, Lo WU, Yeung MK, Lin CH, Chow KH,
Ang I, Tong AH, Bao JY, Lok S and Lo JY: Complete sequencing of
pNDM-HK encoding NDM-1 carbapenemase from a multidrug-resistant
Escherichia coli strain isolated in Hong Kong. PLoS One.
6:e179892011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang J and Wang M, Ding H, Ye M, Hu F,
Guo Q, Xu X and Wang M: New Delhi metallo-β-lactamase-1 in
carbapenem-resistant Salmonella strain, China. Emerg Infect Dis.
19:2049–2051. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang Y, Luan G, Xu Y, Wang Y, Shen M,
Zhang C, Zheng W, Huang J, Yang J, Jia X and Ling B:
Characterization of carbapenem-resistant Acinetobacter baumannii
isolates in a Chinese teaching hospital. Front Microbiol.
6:9102015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fronzes R, Schäfer E, Wang L, Saibil HR,
Orlova EV and Waksman G: Structure of a type IV secretion system
core complex. Science. 323:266–268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nazemi A, Mirinargasi M, Merikhi N and
Sharifi SH: Distribution of pathogenic genes aatA, aap, aggR, among
Uropathogenic Escherichia coli (UPEC) and their linkage with StbA
gene. Indian J Microbiol. 51:355–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guynet C, Cuevas A, Moncalián G and de la
Cruz F: The stb operon balances the requirements for vegetative
stability and conjugative transfer of plasmid R388. PLoS Genet.
7:e10020732011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Del Castillo CS, Hikima J, Jang HB, Nho
SW, Jung TS, Wongtavatchai J, Kondo H, Hirono I, Takeyama H and
Aoki T: Comparative sequence analysis of a multidrug-resistant
plasmid from Aeromonas hydrophila. Antimicrob Agents Chemother.
57:120–129. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Call DR, Singer RS, Meng D, Broschat SL,
Orfe LH, Anderson JM, Herndon DR, Kappmeyer LS, Daniels JB and
Besser TE: blaCMY-2-positive IncA/C plasmids from Escherichia coli
and Salmonella enterica are a distinct component of a larger
lineage of plasmids. Antimicrob Agents Chemother. 54:590–596. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Rye HS, Roseman AM, Chen S, Furtak K,
Fenton WA, Saibil HR and Horwich AL: GroEL-GroES cycling: ATP and
nonnative polypeptide direct alternation of folding-active rings.
Cell. 97:325–338. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Villa L, Guerra B, Schmoger S, Fischer J,
Helmuth R, Zong Z, García-Fernández A and Carattoli A: IncA/C
plasmid carrying bla(NDM-1), bla(CMY-16), and fosA3 in a salmonella
enterica serovar Corvallis strain isolated from a migratory wild
bird in Germany. Antimicrob Agents Chemother. 59:6597–6600. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Babic A, Berkmen MB, Lee CA and Grossman
AD: Efficient gene transfer in bacterial cell chains. MBio.
2:pii:e00027–e11. 2011. View Article : Google Scholar
|
|
22
|
Wozniak RA and Waldor MK: Integrative and
conjugative elements: Mosaic mobile genetic elements enabling
dynamic lateral gene flow. Nat Rev Microbiol. 8:552–563. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Toleman MA, Spencer J, Jones L and Walsh
TR: blaNDM-1 is a chimera likely constructed in Acinetobacter
baumannii. Antimicrob Agents Chemother. 56:2773–2776. 2012.
View Article : Google Scholar : PubMed/NCBI
|