Introduction
Human immunodeficiency virus type 1 (HIV-1)
preferentially destroys CD4+ T lymphocytes and leads to
disturbed T cell homeostasis, characterized by the depletion of
CD4+ T cells (1). The
sustained deletion and dysfunction of CD4+ T cells
caused by HIV-1 infection can result in opportunistic infections
and tumors, diagnosed as acquired immune deficiency syndrome
(1–3). The majority of attempts to manage
HIV-1 infection have focused on CD4+ T cell recovery,
whereas changes in CD8+ T cells have received less
attention. However, the overall course of HIV-1 infection is
largely shaped by CD8+ T cell responses. The CD4/CD8
ratio has been reported to be a useful marker for clinical outcome,
immune dysfunction and viral reservoir size in HIV-1-infected
patients (4,5). Cytotoxic T lymphocytes (CTLs), the
critical effector CD8+ T cells, are vital in host
defense against HIV-1 by impeding viral replication through
cytolytic and non-cytolytic pathways (6). The dynamics of effector
CD8+ T cell expansion in acute HIV-1 infection is
similar to that in other viral infections (7). However, CD8+ T cells in
patients with chronic HIV-1 infection exhibit characteristics of
exhaustion and immunosenescence (7,8).
Although may patients with HIV-1 maintain high circulating
CD8+ T cell counts, the suppression of HIV-1 replication
is attenuated by the disturbed differentiation and homeostasis of
the CD8 T cell compartment (9).
The molecular mechanisms controlling peripheral
CD8+ T cell differentiation in humans remain to be fully
elucidated. Several microRNAs are critical in the development of
hematopoietic cells (10,11). A study by Zhang and Bevan
demonstrated that CD8+ T cells with knockout of dicer
are defective in cell accumulation and survival (12). Compared with naïve CD8+
T cells, miRNA (miR)-21 and miR-155 were found to be upregulated in
effector CD8+ T cells (13). The sustained expression of miR-155
has been associated with effector and effector memory cells,
whereas lower expression levels of miR-155 have been associated
with central memory cells (14).
miR-155 has also been shown to be an important regulator for the
differentiation of T helper cells in HIV-1-infected individuals,
particularly Th17 and regulatory T cells (15,16).
However, the profile of miR-155 in HIV-1-infected individuals and
its association with CD8+ T cell differentiation remain
to be fully elucidated.
In the present study, the expression levels of
miR-155 were investigated in CD8+ T cells from patients
with HIV-1, with or without highly active antiretroviral therapy
(HAART), and correlation between the levels of miR-155 and
percentages of CD8+ T cell subsets was examined. It was
found that the expression of miR-155 in CD8+ T cells of
patients with HIV-1 was increased, particularly in the discord
controllers and HAART-naïve patients. The level of miR-155 in
CD8+ T cells was positively correlated with the
percentages of effector and effector memory subsets, and negatively
correlated with the percentages of naïve and central memory
subsets.
Subjects and methods
Subjects
Patients with HIV-1 were recruited from the First
Affiliated Hospital, School of Medicine, Zhejiang University
(Hangzhou, China) between June 2015 and December 2015. HIV-1
infection was diagnosed on the basis of positive results from
serological and HIV-1 RNA detection assays. The subjects were
excluded if they had received systemic antibiotics, vaccination or
any immunomodulatory drug in the previous 3 months. A total of 35
apparently healthy uninfected control subjects from the community
clinics were also recruited. The present study received approval
from the Ethics Review Boards of the First Affiliated Hospital,
School of Medicine, Zhejiang University (approval no. 2015–06103).
All subjects were volunteers and provided written informed consent
prior to involvement in the study.
Flow cytometry
The subsets of CD8+ T cells were analyzed
using four-color flow cytometry with a FACScan flow cytometer (BD
Biosciences, Franklin Lakes, NJ, USA) and a commercial flow
cytometry assay kit (BD Biosciences). Cell staining was performed
at room temperature using a cocktail of the following
fluorochrome-conjugated antibodies, at the manufacturer's
recommended dilution: Anti-CD3-PerCP-CY5.5 (cat. no: 561478),
anti-CD8-APC-Cy7 (cat. no: 561967), anti-CD45RA-PE (cat. no:
560975) and anti-CD62L-FITC (cat. no: 561914) (BD Biosciences).
Briefly, 50 µl whole blood was added to the antibody cocktail,
mixed and then incubated for 20 min in the dark at room
temperature. The red cells in whole blood were lysed with red cell
lysis buffer (Beyotime Institute of Biotechnology, Shanghai,
China), washed three times with 1 ml phosphate-buffered saline, and
subsequently detected using FACScan flow cytometry. The percentages
of
CD3+CD8+CD45RA+CD62L+,
CD3+CD8+CD45RA−CD62L−,
CD3+CD8+CD45RA+CD62L−
and
CD3+CD8+CD45RA−CD62L+T
cells were determined.
Detection of miR-155
Peripheral blood mononuclear cells (PBMCs) were
isolated from all patients from 5 ml venous whole blood samples,
using a density gradient centrifugation method with Ficoll-Paque
PLUS (GE Healthcare Life Sciences, Marlborough, MA, USA) at 2,000 ×
g for 10 min at room temperature. The CD8+ T lymphocytes
were purified from the PBMCs using MACS human CD8 microbeads for
positive selection (Miltenyi Biotec GmbH, Bergisch Gladbach,
Germany). The purity of the CD8+ T lymphocytes was
>90%. The total RNA was extracted from the CD8+ T
lymphocytes using TRIzol (Thermo Fisher Scientific, Inc., Waltham,
MA, USA). The quantitative analysis of miR-155 was performed using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis with a Bulge-Loop™ miRNA qRT-PCR Starter kit
(Guangzhou RiboBio, Co., Ltd., Guangzhou, China) and thehsa-miR-155
qRT-PCR primer set (Guangzhou RiboBio, Co., Ltd.). A U6 small
nuclear RNA primer set (Guangzhou RiboBio, Co., Ltd.) was used as
the internal control. The experiments were performed according to
the protocol provided in the manufacturer of the kit using a 10 µl
reaction system. Briefly, the miRNA RT reaction mix included 1 µl
RNA template (68 ng/µl), 1 µl miRNA RT primer, 2 µl 5X reverse
transcription buffer, 2 µl RTase mix and 4 µl RNase-free water. The
mix was incubated at 42°C for 60 min followed by incubation at 70°C
for10 min. The miRNA qPCR reaction system contained 10 µl
SYBR-Green Master mix, 0.8 µl miRNA forward primer, 0.8 µl miRNA
reverse primer and 2 µl RT product; the final volume was normalised
to 20 µl with DNase-free water. Real-time PCR was performed for 40
cycles of denaturation (95°C, 45 sec), annealing (62°C, 30 sec) and
extension (72°C, 30 sec). Double-stranded DNA was measured at 86°C
following each cycle. Each sample was repeated three times. The
relative expression levels of miRNA were calculated using the
2ΔΔCq method (17).
Statistical analysis
Statistical analyses were performed using SPSS for
Windows version 20.0 (IBM SPSS, Armonk, NY, USA). Student's t-test
was used to compare between two groups and one-way analysis of
variance was used when comparing more than three groups.
χ2 was used for categorical variables. The correlation
was tested using Spearman's correlation test. All tests were
two-tailed. P<0.05 was considered to indicate a statistically
significant difference.
Results
Clinical information of subjects
A total of 94 HIV-1-infected patients were recruited
to the present study, including 31 HAART-naïve patients and 63
patients receiving HAART. The patients receiving HAART were divided
into two groups according to the recovery of their CD4+
T counts and viral suppression: 34 typical controllers had a
CD4+ T count >500 cells/ml and a viral load <400
copies/ml; 29 discord controllers had a CD4+ T count
<500 cells/ml or a viral load >400 copies/ml. The three
groups of HIV-1-infectedpatientsand normal controls were all
appropriately age- and sex-matched. The mean durations of HIV-1
infection for the typical and discord controllers were 6.38±2.51
and 4.96±1.36 years, respectively, which were significantly longer,
compared with that for the HAART naïve patients (2.12±1.43 years;
P<0.05). The durations of HAART for the typical and discord
controllers were 4.17±1.13 and 3.84±1.62 years, respectively. The
typical controllers had a significantly lower viral load, and a
higher CD4+ T cell count and CD4/CD8 ratio, compared
with the other HIV-1 patients (P<0.05). The regimens for
patients receiving HAART were primarilyd 4T+3TC+NVP and AZT+3TC+NVP
(49 cases; 78%). The detailed participant data is presented in
Table I.
 | Table I.Clinical data from the study
participants. |
Table I.
Clinical data from the study
participants.
| Characteristic | Typical controller
(n=34) | Discord controller
(n=29) | HAART-naïve
(n=31) | Control (n=35) | P-value |
|---|
| Males, n (%) | 21 (62%) | 17 (59%) | 19 (61%) | 27 (77%) | 0.317 |
| Age (years) | 41.57±10.01 | 38.35±15.36 | 36.41±13.47 | 39.63±11.79 | 0.254 |
| Years with
HIV-1 | 6.38±2.51 | 4.96±1.36 | 2.12±1.43 | NA | 0.027 |
| Years on HAART | 4.17±1.13 | 3.84±1.62 | NA | NA | 0.184 |
| Viral load (log
10) | 1.631±0.75 | 3.91±0.49 | 4.32±1.54 | NA | <0.001 |
| CD4+
cells (/µl) | 655.45±263.81 | 353.68±187.95 | 431.77±284.56 | 798.32±261.19 | 0.001 |
| CD8+
cells (/µl) | 618.76±330.17 | 785.35±296.54 | 801.52±272.21 | 518.23±241.36 | 0.008 |
| CD4/CD8 ratio | 1.21±0.42 | 0. 54±0.22 | 0.57±0.30 | 1.63±0.41 | 0.017 |
| HAART regimen, n
(%) |
|
|
|
| 0.116 |
| d4T+3TC+NVP | 11 (32%) | 9 (31%) | NA | NA | – |
| AZT+3TC+NVP | 17 (50%) | 12 (41%) | NA | NA | – |
| Other | 6 (18%) | 8 (28%) | NA | NA | – |
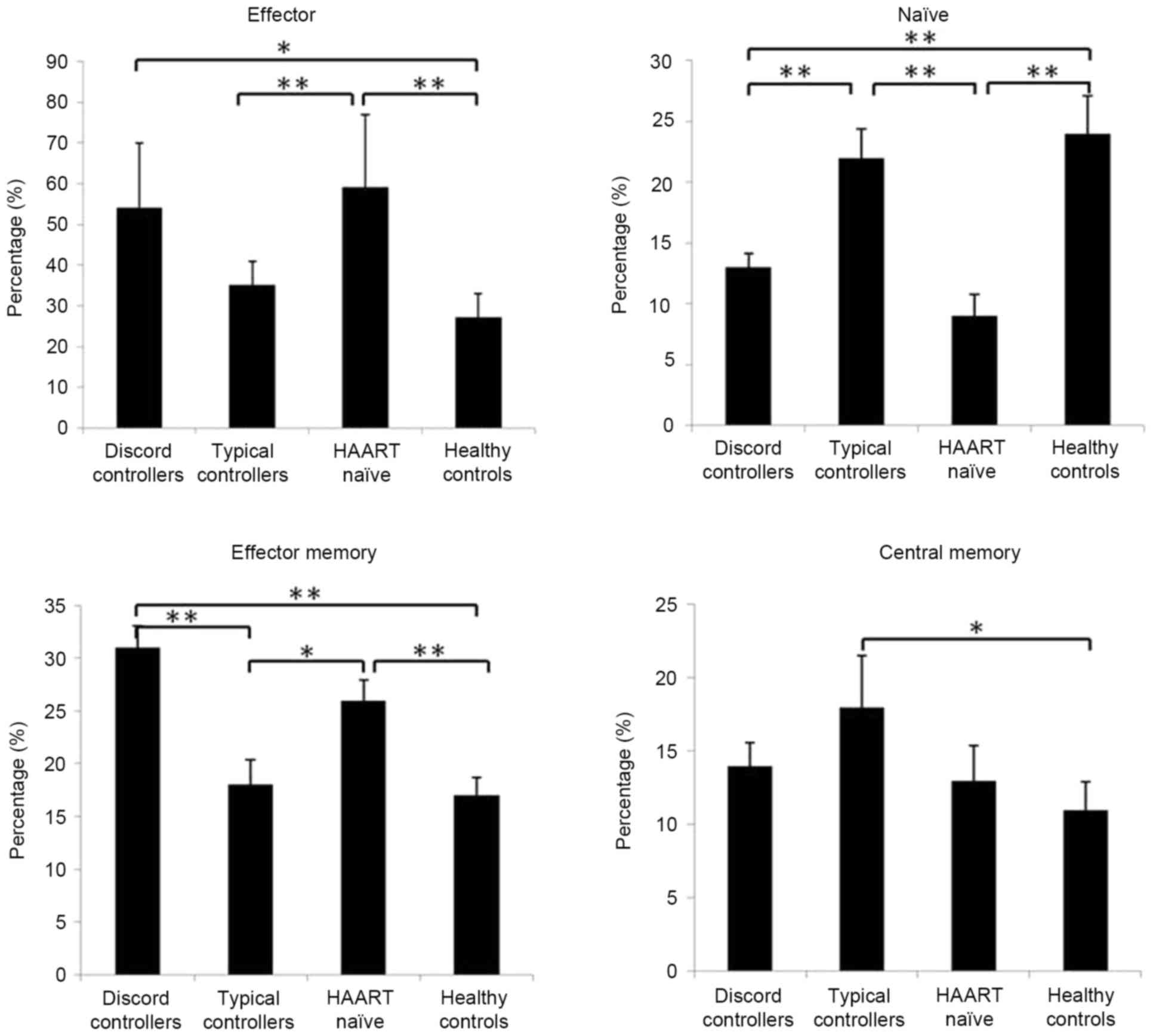
Subsets of circulating CD8+ T cells are
altered in patients with HIV-1. Based on the expression of CD45RA
and CD62L, human CD8+ T cells can be divided into four
subsets with distinct homing and functional properties: Naïve
(CD45RA+CD62L+), central memory
(CD45RA−CD62L+), effector memory
(CD45RA−CD62L−) and effector
(CD45RA+CD62L−) cells (18,19).
In the present study, the CD8+ T cell subsets were
determined in patients with HIV-1. Compared with the normal
controls and typical controllers, the discord controllers and HAART
naïve patients showed higher percentages of effector and effector
memory CD8+ T cells, and a lower percentage of naïve
CD8+ T cells (P<0.05; Fig. 1). The typical controllers had a
similar composition of CD8+ T cell subsets to the normal
controls, but had a higher percentage of central memory
CD8+ T cells (P<0.05; Fig. 1).
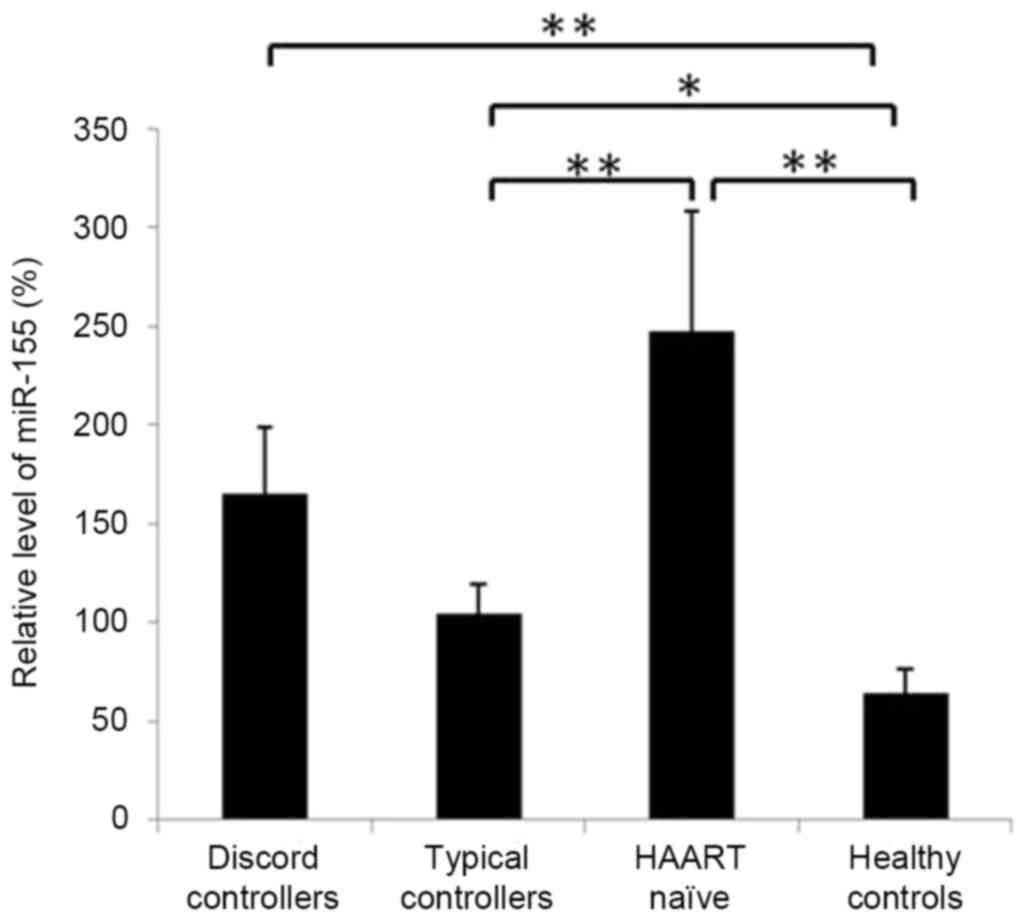
Expression of miR-155 is elevated in CD8+
T cells of patients with HIV-1. The present study compared the
expression levels ofmiR-155 in CD8+ T cells of typical
and discord controllers with HAART and HAART-naïve patients with
normal levels. It was found that the levels of miR-155 in
CD8+ T cells of all three groups of HIV-1 patients were
significantly higher, compared with that in the normal controls
(P<0.05; Fig. 2). Although
increased, the expression levels of miR-155 in the CD8+
T cells of typical controllers were almost normal, and were
significantly lower, compared with the levels in the discord
controllers and HAART-naïve patients, in which viremia was not
suppressed (P<0.01; Fig.
2).
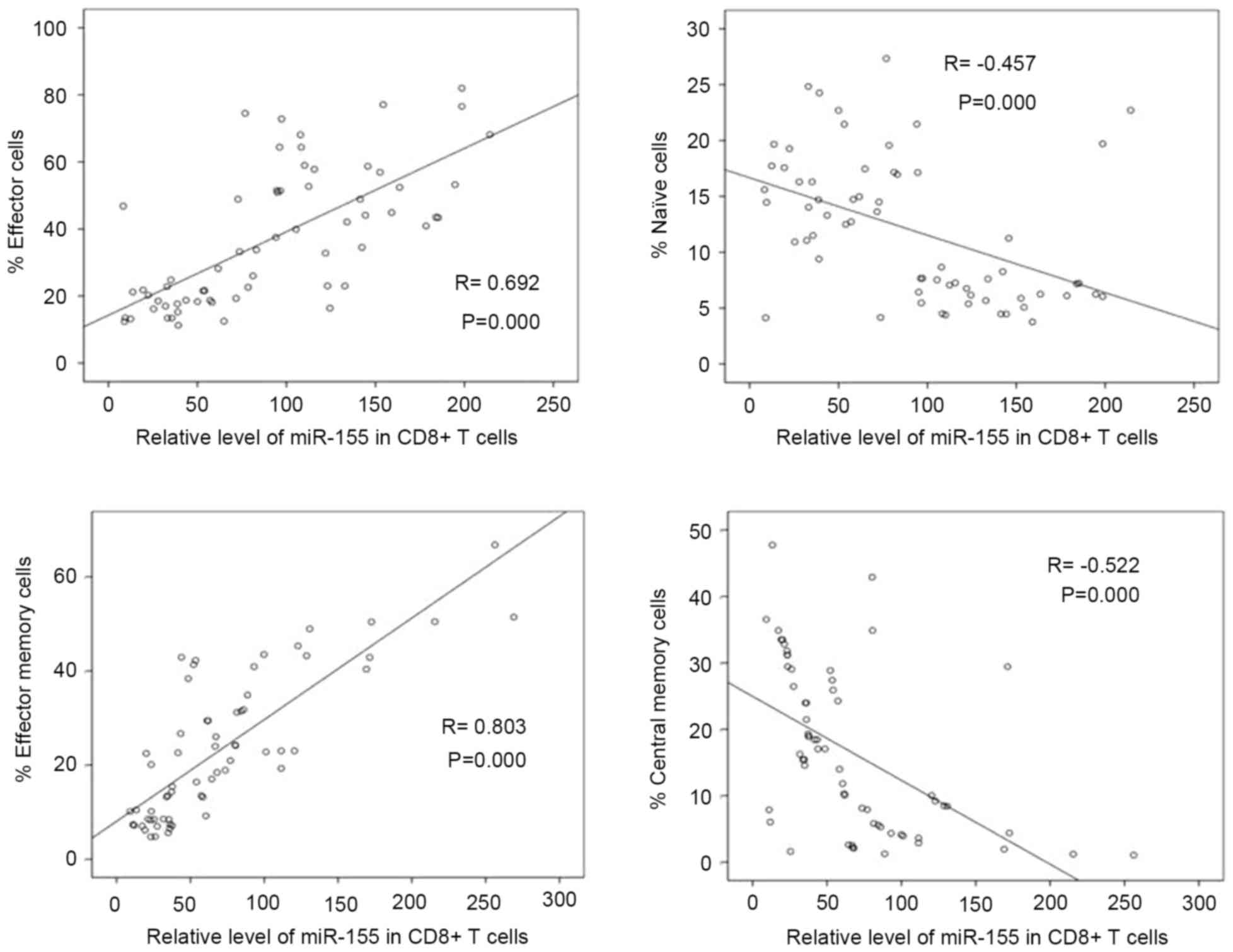
Correlation between levels of miR-155 and
percentages of CD8+ T cell subsets in patients with
HIV-1. To determine whether miR-155 is associated with
CD8+ T cell differentiation in HIV-1-infected
individuals, the present study analyzed the correlation between
levels of miR-155 and percentages of CD8+ T cell
subsets. In all patients with HIV-1, the expression of miR-155 in
CD8+ T cells was positively correlated with the
percentages of effector and effector memory CD8+ T cells
(r=0.692 and 0.803 respectively; P<0.01; Fig. 3), and was negatively correlated
with the percentage of naïve and central memory CD8+ T
cells (r=−0.457 and −0.522 respectively; P<0.01; Fig. 3).
Discussion
The cellular immune response is critical in
controlling the viral replication of HIV-1 (6–8). The
majority of circulating CD8+ T cells in healthy
individuals are naïve and central memory cells. When combined with
antigens, CD8+ T cells become activated and undergo a
program of clonal expansion (20).
The majority of effector cells die through apoptosis whereas
certain surviving CD8 T cells become long-lasting virus-specific
memory CD8+ T cells, forming a protective mechanism upon
antigen re-exposure (21,22). In the present study, higher
percentages of effector and effector memory cells, and a lower
percentage of naïve CD8+ T cells were found in discord
controllers and HAART-naïve patients, in which viral replication
was not suppressed. In typical controllers with higher
CD4+ T counts and suppressed viremia, the
CD8+ T cell subsets were restored almost to normal
levels. These results are consistent with previous studies, which
reported that the majority of circulating CD8+ T cells
in individuals with chronic HIV-1 infection were mature effector
and effector memory CD8+ T cells (23,24).
However, in a cross sectional study by Groves et al
(19), discord controllers and
typical controllers had higher numbers of naive CD8+ T
cells and reduced CD8+ T cell activation, compared with
the patients with rapidly progressing disease. Notably, Groves
et al defined discord controllers as patients with a viral
RNA load <2,000 copies/ml and <450 CD4+ T
cells/mm3, with the viral load of discord controllers
ranging between 100.3 and 1,043.0 copies/ml. In the present study,
the viral load of the discord controllers ranged between 1,491 and
23,386 copies/ml. The higher naïve CD8+ T cells and
lower CD8+ T cell activation may be associated with
lower virus replication, although no direct association between
CD8+ T cell subsets and HIV-1 RNA load was found. These
results suggested that a more preserved CD8+ T cell
compartment is associated with the control of plasma viremia.
The expansion of effector CD8+ T cells is
associated with certain microRNAs, which can control gene
expression at the post-transcriptional level. miR-155 is an
important microRNA, which regulates the immune response and is
important in controlling lymphocyte differentiation at multiple
levels (25). miR-155 is essential
for normal B cell differentiation and antibody production (26), and controls the differentiation of
CD4+ T cells into the Th1, Th2, and Th17 subsets of
helper T cells (15,27). In addition, miR-155 is essential
for efficient antigen presentation by dendritic cells (28). miR-155 is also reported to regulate
CD8+ T cell differentiation. Antigen-specific
CD8+ effector T cells express high levels of miR-155
(29). Naïve and central memory
cells express low levels of miR-155, and effector memory cells
express intermediate levels of miR-155 (13). However, the association between
miR-155 and CD8+ T cell differentiation in
HIV-1-infected individuals has not been reported. In the present
study, increased levels of miR-155 were found in CD8+ T
cells of HIV-1-infected patients, which was higher in the discord
controllers and HAART-naïve patients. The expression of miR-155 in
CD8+ T cells was positively correlated with the
percentages of effector and effector memory CD8+ T
cells, and negatively correlated with the percentages of naïve and
central memory CD8+ T cells. These results were
consistent with those of previous studies on other pathogen
infections. Lind et al (30) demonstrated that miR-155 was
essential for optimal CD8+ T cell responses against
influenza virus and Listeria infection, and was crucial for the
generation of CD8+ T cell memory against pathogens. Tsai
et al (14) showed that
miR-155 was an important regulator in effector and memory
virus-specific CD8+ T cell responses in murid herpes
virus 68-infected mice. Compared with the wild-type mice, chimeric
mice lacking miR-155 in CD8+ T cells showed a weaker
effector response and a skewing toward memory precursor cells with
significantly higher viral titers (14). However, the present study found no
correlation between the levels of miR-155 levels and virus
replication.
The expression of miR-155 has been reported to be
associated with HIV infection. miR-155 can affect disease
progression by regulating the transformation of naïve Tregs and
naïve CD4 subsets into activate subsets (31,32).
In addition, miR-155 exerts an anti-HIV-1 effect by targeting
several HIV-1 dependency factors involved in post-entry and
pre-integration events, for example, DC-SIGN and TRIM32, leading to
severely reduced HIV-1 infection (33). The present study suggested that
miR-155 may be an important regulator of the CTL response in
patients with HIV. However, how the expression of miR-155 is
altered by HIV-1 infection remains to be elucidated, as does the
causal association between the expression of miR-155 and virus
replication. The mechanisms underlying altered levels of miR-155 in
the CD8+ T compartment of HIV-1-infected patients
requires further investigation. A limitation of the present study
is that miR-155 was detected in total CD8+ T cells, but
not in subsets. Further investigations on the expression of miR-155
in subsets of CD8+ T cells in patients with HIV-1 may
provide additional information onmiR-155 and the differentiation of
CD8+ T cells.
Acknowledgements
This study was supported by the Zhejiang Provincial
Science and Technology Foundation (grant no. 2014C33252 &
2015C33183), the National Key Technologies R&D Program for the
12th Five-year Plan of China (grant no. 2012ZX10001-004) and the
National Natural Science Foundation of China (grant no.
81402726).
Glossary
Abbreviations
Abbreviations:
|
HIV
|
human immunodeficiency virus
|
|
CTLs
|
cytotoxic T lymphocytes
|
|
HAART
|
highly active antiretroviral
therapy
|
|
PBMCs
|
peripheral blood mononuclear cells
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
References
|
1
|
Morou A, Palmer BE and Kaufmann DE:
Distinctive features of CD4+ T cell dysfunction in
chronic viral infections. Curr Opin HIV AIDS. 9:446–451. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jin CZ, Zhao Y, Zhang FJ, Yao HP, Wu LJ,
Zhao HX, Wei HS and Wu NP: Different plasma levels of interleukins
and chemokines: Comparison between children and adults with AIDS in
China. Chin Med J (Engl). 122:530–535. 2009.PubMed/NCBI
|
|
3
|
Chitra Y, Urgen S, Dayananda I and
Brajachand SN: Effect of anti retroviral therapy (ART) on CD4 T
lymphocyte count and the spectrum of opportunistic infections in
HIV/AIDS in Manipur. J Commun Dis. 41:19–24. 2009.PubMed/NCBI
|
|
4
|
Lu W, Mehraj V, Vyboh K, Cao W, Li T and
Routy JP: CD4:CD8 ratio as a frontier marker for clinical outcome,
immune dysfunction and viral reservoir size in virologically
suppressed HIV-positive patients. J Int AIDS Soc. 18:200522015.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Serrano-Villar S, Sainz T, Lee SA, Hunt
PW, Sinclair E, Shacklett BL, Ferre AL, Hayes TL, Somsouk M, Hsue
PY, et al: HIV-Infected Individuals with low CD4/CD8 ratio despite
effective antiretroviral therapy exhibit altered T cell subsets,
heightened CD8+ T cell activation, and increased risk of
non-AIDS morbidity and mortality. PLoS Pathog. 10:e10040782014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Saeidi A, Buggert M, Che KF, Kong YY, Velu
V, Larsson M and Shankar EM: Regulation of CD8+ T-cell
cytotoxicity in HIV-1 infection. Cell Immunol. 298:126–133. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mudd JC and Lederman MM: CD8 T cell
persistence in treated HIV infection. Curr Opin HIV AIDS.
9:500–505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Demers KR, Reuter MA and Betts MR: CD8(+)
T-cell effector function and transcriptional regulation during HIV
pathogenesis. Immunol Rev. 254:190–206. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cao W, Mehraj V, Kaufmann DE, Li T and
Routy JP: Elevation and persistence of CD8 T-cells in HIV
infection: The Achilles heel in the ART era. J Int AIDS Soc.
19:206972016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Allantaz F, Cheng DT, Bergauer T,
Ravindran P, Rossier MF, Ebeling M, Badi L, Reis B, Bitter H,
D'Asaro M, et al: Expression profiling of human immune cell subsets
identifies miRNA-mRNA regulatory relationships correlated with cell
type specific expression. PLoS One. 7:e299792012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ooi AG, Sahoo D, Adorno M, Wang Y,
Weissman IL and Park CY: MicroRNA-125b expands hematopoietic stem
cells and enriches for the lymphoid-balanced and lymphoid-biased
subsets. Proc Natl Acad Sci USA. 107:21505–21510. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang N and Bevan MJ: Dicer controls
CD8+ T-cell activation, migration, and survival. Proc
Natl Acad Sci USA. 107:21629–21634. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Salaun B, Yamamoto T, Badran B,
Tsunetsugu-Yokota Y, Roux A, Baitsch L, Rouas R, Fayyad-Kazan H,
Baumgaertner P, Devevre E, et al: Differentiation associated
regulation of microRNA expression in vivo in human CD8+
T cell subsets. J Transl Med. 9:442011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tsai CY, Allie SR, Zhang W and Usherwood
EJ: MicroRNA miR-155 affects antiviral effector and effector Memory
CD8 T cell differentiation. J Virol. 87:2348–2351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Singh UP, Murphy AE, Enos RT, Shamran HA,
Singh NP, Guan H, Hegde VL, Fan D, Price RL, Taub DD, et al:
miR-155 deficiency protects mice from experimental colitis by
reducing T helper type 1/type 17 responses. Immunology.
143:478–489. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yao R, Ma YL, Liang W, Li HH, Ma ZJ, Yu X
and Liao YH: MicroRNA-155 modulates Treg and Th17 cells
differentiation and Th17 cell function by targeting SOCS1. PLoS
One. 7:e460822012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pender MP, Csurhes PA, Pfluger CM and
Burrows SR: Deficiency of CD8+ effector memory T cells
is an early and persistent feature of multiple sclerosis. Mult
Scler. 20:1825–1832. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Groves KC, Bibby DF, Clark DA, Isaksen A,
Deayton JR, Anderson J, Orkin C, Stagg AJ and McKnight A: Disease
progression in HIV-1-infected viremic controllers. J Acquir Immune
Defic Syndr. 61:407–416. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mohan T, Bhatnagar S, Gupta DL and Rao DN:
Current understanding of HIV-1 and T-cell adaptive immunity:
Progress to date. Microb Pathog. 73:60–69. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prlic M, Williams MA and Bevan MJ:
Requirements for CD8 T-cell priming, memory generation and
maintenance. Curr Opin Immunol. 19:315–319. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Joshi NS and Kaech SM: Effector CD8 T cell
development: A balancing act between memory cell potential and
terminal differentiation. J Immunol. 180:1309–1315. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Papagno L, Spina CA, Marchant A, Salio M,
Rufer N, Little S, Dong T, Chesney G, Waters A, Easterbrook P, et
al: Immune activation and CD8+ T-cell differentiation
towards senescence in HIV-1 infection. PLoS Biol. 2:E202004.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Appay V, Papagno L, Spina CA, Hansasuta P,
King A, Jones L, Ogg GS, Little S, McMichael AJ, Richman DD and
Rowland-Jones SL: Dynamics of T cell responses in HIV infection. J
Immunol. 168:3660–3666. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Seddiki N, Brezar V, Ruffin N, Lévy Y and
Swaminathan S: Role of miR-155 in the regulation of lymphocyte
immune function and disease. Immunology. 142:32–38. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Landgraf P, Rusu M, Sheridan R, Sewer A,
Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M,
et al: A mammalian microRNA expression atlas based on small RNA
library sequencing. Cell. 129:1401–1414. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Escobar T, Yu CR, Muljo SA and Egwuagu CE:
STAT3 activates miR-155 in Th17 cells and acts in concert to
promote experimental autoimmune uveitis. Invest Ophthalmol Vis Sci.
54:4017–4025. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Thai TH, Calado DP, Casola S, Ansel KM,
Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, et
al: Regulation of the germinal center response by microRNA-155.
Science. 316:604–608. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Gracias DT, Stelekati E, Hope JL,
Boesteanu AC, Doering TA, Norton J, Mueller YM, Fraietta JA, Wherry
EJ, Turner M and Katsikis PD: The microRNA miR-155 controls CD8 (+)
T cell responses by regulating interferon signaling. Nat Immunol.
14:593–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lind EF, Elford AR and Ohashi PS:
Micro-RNA 155 is required for optimal CD8+ T cell
responses to acute viral and intracellular bacterial challenges. J
Immunol. 190:1210–1216. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bignami F, Pilotti E, Bertoncelli L, Ronzi
P, Gulli M, Marmiroli N, Magnani G, Pinti M, Lopalco L, Mussini C,
et al: Stable changes in CD4+ T lymphocyte miRNA
expression after exposure to HIV-1. Blood. 119:6259–6267. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Seddiki N, Swaminathan S, Phetsouphanh C
and Kelleher AD: miR-155 is differentially expressed in Treg
subsets, which may explain expression level differences of miR-155
in HIV-1 infected patients. Blood. 119:6396–6397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Martinez-Nunez RT, Louafi F, Friedmann PS
and Sanchez-Elsner T: MicroRNA-155 modulates the pathogen binding
ability of dendritic cells (DCs) by down-regulation of DC-specific
intercellular adhesion molecule-3 grabbing non-integrin (DC-SIGN).
J Biol Chem. 284:16334–16342. 2009. View Article : Google Scholar : PubMed/NCBI
|