Introduction
Diabetic kidney disease (DKD) is a common
complication of diabetes, which is characterized by albuminuria, an
impaired glomerular filtration rate (GFR) or a combination of the
two (1,2). DKD accounts for ~50% of all cases of
end-stage renal disease (ESRD) in the United States and the US ESRD
program is a large medical expense/economic burden and costs a
great amount of money to run. However, a number of genes closely
associated with DKD development have yet to be identified despite
many years of intensive study (3).
Therefore, the identification of genes associated with DKD
development is urgently required, as well as the subsequent
elucidation of its molecular mechanism.
Some advancements have been made in the elucidation
of the pathological mechanism involved in the development of DKD.
The loss of podocytes is an early feature of DKD (4). The levels of almost all
podocyte-specific genes including genes for congenital nephrotic
syndrome of the finish type (NPHS1), glomerular podocin
(NPHS2), the Wilm's tumor gene (WT1) and vascular
endothelial growth factor (VEGF) are all severely reduced in
DKD (3). Some other studies have
also demonstrated that NPHS1 (5,6),
NPHS2 (7), bone
morphogenetic protein 7 (8),
WT1 (4) and VEGF
(9,10) are decreased in DKD. In addition,
tubulointerstitial fibrosis is a prominent feature of progressive
DKD and is likely to be one of the final common pathways leading to
the development of ESRD, with patients subsequently requiring
dialysis or transplantation (11,12).
A previous study revealed that using
angiotensin-converting-enzyme-inhibitors and angiotensin II
receptor antagonists in patients with diabetes mellitus can
respectively improve mortality and delay the progression of DKD
(13). In addition, a human
genetic study highlighted that the complement system potentially
serves a role in low-grade inflammation and the development of DKD
(14). Therefore, the aim of the
present study was to identify the key genes associated with the
development of DKD and elucidate its underlying mechanism.
In the present study, the microarray data of
GSE30528 was downloaded the from Gene Expression Omnibus database
(GEO; www.ncbi.nlm.nih.gov/geo/). The gene expression
profiles in DKD were analyzed and functional analysis was performed
for differentially expressed genes (DEGs) in DKD glomerular and
tubular kidney biopsy tissues in comparison with normal tissues. In
addition, the protein-protein interaction (PPI) network was also
constructed. These results were used to discover the key genes
associated with DKD development and to clarify the underlying
mechanism.
Materials and methods
Affymetrix microarray data
The array data for GSE30528 was downloaded from the
GEO database, which was first recorded by Woroniecka et al
(3) and was based on the GPL571
platform (Affymetrix Human Genome U133A 2.0 Array; Affymetrix,
Inc.; Thermo Fisher Scientific, Inc., Waltham, MA, USA). A total of
44 samples were used to develop the original array data, and of
these 9 DKD [age, 64±13.56 years; 5 females, 4 males; body mass
index (BMI), 32.74±7.9 kg/m2] and 13 healthy,
disease-free control samples (age, 51.38±12.01 years; 5 females, 8
males; BMI, 29.59±9.08 kg/m2) were selected for analysis
in the present study.
Data processing and DEG analysis
The raw expression data was preprocessed using the
robust multiarray average algorithm (15) and the Affy package in Bioconductor
(bioconductor.org/packages/release/bioc/html/affy.html);
the expression levels of the probes were then obtained. If several
probes mapped to one gene symbol, then the mean value was set as
the final expression value of this gene. The DEGs in DKD glomerular
and tubular kidney biopsy tissues where then compared with normal
tissues using the limma package (16). |logFC| >1 and P<0.05 were
considered as the cutoff criterion.
Gene Ontology (GO) and pathway
enrichment analysis
GO is used for the unification of biology,
collecting defined, structured and controlled vocabulary for gene
annotation, which mainly includes the following 3 categories:
Molecular function, biological process and cellular component
(17). The Kyoto Encyclopedia of
Genes and Genomes (KEGG) is a database for the classification of
relevant gene sets into their respective pathways (18).
In order to analyze the DEGs on a functional level,
GO annotation and KEGG pathway enrichment analyses were performed
for DEGs using GO Function version 1.24.0 (19) software in Bioconductor version 3.5
(www.bioconductor.org/packages/release/bioc/html/GOFunction.html),
and gene annotation information was obtained from the org. Hs. eg.
db and GO. db package. P<0.05 and gene counts >2 were set as
the cut off values.
PPI network analysis
The Search Tool for the Retrieval of Interacting
Genes (STRING) database provides the experimental and predicted
interaction information of proteins (20). Protein pair interactions in STRING
were presented with a combined score. The DEGs were mapped into
PPIs and a combined score of >0.7 was identified as the cutoff
standard for the important protein pairs. The PPI network was
constructed using Cytoscape software version 2.8.2 (www.cytoscape.org/) (21).
Module analysis
ClusterONE version 1.0 (www.paccanarolab.org/cluster-one/) in the Cytoscape
software package was used to analyze the PPI network modules with a
minimum size of 3 and a minimum density of 0.5. Modules with
P<0.01 were set as significant clustering modules.
Results
Data processing and DEGs analysis
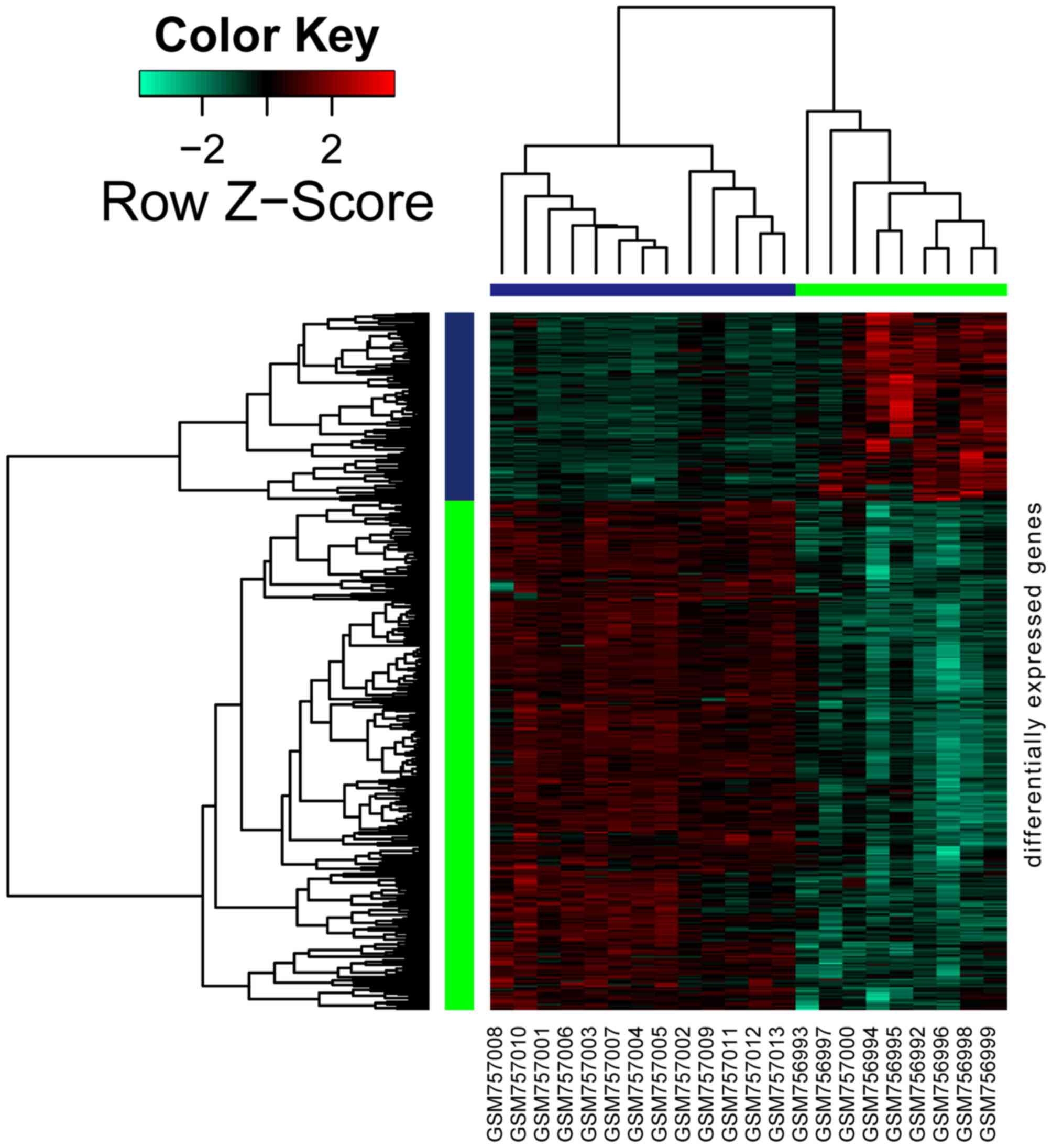
As shown in Fig. 1,
a total of 426 genes were differentially expressed in DKD samples
when compared with normal samples, amongst which 115 genes were
upregulated and 311 were downregulated.
GO and pathway enrichment
analysis
GO and KEGG pathway analyses were performed for
upregulated and downregulated DEGs. The top 5 GO terms are shown in
Table I. The overrepresented GO
terms of upregulated DEGs were primarily associated with
extracellular region, antigen binding, extracellular space, the
defense response, the immune response and peptidase regulator
activity (Table IA). The
downregulated DEGs were mainly involved in cardiovascular system
development, circulatory system development, actin cytoskeleton,
cell junction, cytoskeletal protein binding and integrin binding
(Table IB).
 | Table I.Gene Ontology analysis for
differentially expressed genes. |
Table I.
Gene Ontology analysis for
differentially expressed genes.
| A, Upregulated |
|---|
|
|---|
| Term | Description | Counts (n) |
P-valuea |
|---|
| GO-BP terms |
|
GO:0006952 | Defense
response | 44 | <0.0005 |
|
GO:0006955 | Immune
response | 41 |
2.22×10−16 |
|
GO:0002376 | Immune system
process | 50 |
3.00×10−15 |
|
GO:0001775 | Cell
activation | 31 |
1.37×10−14 |
|
GO:0045321 | Leukocyte
activation | 26 |
1.76×10−13 |
| GO-CC terms |
|
GO:0005576 | Extracellular
region | 63 |
2.06×10−12 |
|
GO:0005615 | Extracellular
space | 32 |
4.37×10−12 |
|
GO:0031982 | Vesicle | 53 |
9.90×10−11 |
|
GO:0031988 | Membrane-bounded
vesicle | 52 |
1.13×10−10 |
|
GO:0044421 | Extracellular
region part | 53 |
3.50×10−10 |
| GO-MF terms |
|
GO:0003823 | Antigen
binding |
8 |
2.97×10−07 |
|
GO:0061134 | Peptidase regulator
activity | 10 |
6.67×10−07 |
|
GO:0005539 | Glycosaminoglycan
binding |
9 |
2.84×10−07 |
|
GO:0004866 | Endopeptidase
inhibitor activity |
8 |
|
|
8092×10−06 |
|
GO:0061135 | Endopeptidase
regulator activity |
8 |
1.11×10−05 |
|
| B,
Downregulated |
|
| Term | Description | Counts (n) |
P-valuea |
|
| GO-BP terms |
|
GO:0072358 | Cardiovascular
system development | 50 |
3.77×10−15 |
|
GO:0072359 | Circulatory system
development | 50 |
3.77×10−15 |
|
GO:0009653 | Anatomical
structure morphogenesis | 91 |
4.66×10−15 |
|
GO:0048731 | System
development | 121 |
3.40×10−14 |
|
GO:0032502 | Developmental
process | 147 |
5.66×10−14 |
| GO-CC terms |
|
GO:0015629 | Actin
cytoskeleton | 31 |
1.79×10−12 |
|
GO:0030054 | Cell junction | 47 |
6.04×10−10 |
|
GO:0070161 | Anchoring
junction | 28 |
1.82×10−09 |
|
GO:0005912 | Adherens
junction | 27 |
3.39×10−09 |
|
GO:0044421 | Extracellular
region part | 98 |
3.00×10−08 |
| GO-MF terms |
|
GO:0008092 | Cytoskeletal
protein binding | 40 |
9.40×10−11 |
|
GO:0005178 | Integrin
binding | 12 |
1.23×10−07 |
|
GO:0032403 | Protein complex
binding | 35 |
4.24×10−07 |
|
GO:0050839 | Cell adhesion
molecule binding | 14 |
8.04×10−07 |
|
GO:0003779 | Actin binding | 21 |
1.40×10−06 |
The upregulated DEGs were mainly enriched in 17 KEGG
pathways, including primary immunodeficiency, extracellular
matrix-receptor interactions, rheumatoid arthritis and systemic
lupus erythematosus (Table IIA).
In addition, major histocompatibility complex, class II, DP α1
(HLA-DPA1) was the common gene in the rheumatoid arthritis
and systemic lupus erythematosus pathways. The downregulated DEGs
were mainly enriched in 16 KEGG pathways, such as tight junction
and adherens junction.
 | Table II.Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis of the top 10 DEGs. |
Table II.
Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis of the top 10 DEGs.
| A, Upregulated |
|---|
|
|---|
| Term | Description | Counts (n) | P-value |
|---|
| 5150 | Staphylococcus
aureus infection | 5 |
1.89×10−04c |
| 5020 | Prion diseases | 4 |
3.58×10−04c |
| 5340 | Primary
immunodeficiency | 4 |
3.58×10−04c |
| 4512 | Extracellular
matrix-receptor interaction | 5 |
1.42×10−03b |
| 5323 | Rheumatoid
arthritis | 5 |
1.92×10−03b |
| 5142 | Chagas disease
(American trypanosomiasis) | 5 |
3.45×10−03b |
| 4610 | Complement and
coagulation cascades | 4 |
4.61×10−03b |
| 4974 | Protein digestion
and absorption | 4 |
8.12×10−03b |
| 5322 | Systemic lupus
erythematosus | 5 |
1.06×10−02a |
| 4640 | Hematopoietic cell
lineage | 4 |
1.08×10−02a |
|
| B,
Downregulated |
|
| Term | Description | Counts (n) | P-value |
|
| 4520 | Adherens
junction | 8 |
3.85×10−05d |
| 4510 | Focal adhesion | 13 |
4.51×10−05d |
| 4810 | Regulation of actin
cytoskeleton | 13 |
8.65×10−05d |
| 4530 | Tight junction | 9 |
5.06×10−04c |
| 5410 | Hypertrophic
cardiomyopathy | 7 |
6.17×10−04c |
| 5414 | Dilated
cardiomyopathy | 7 |
1.00×10−03b |
| 5412 | Arrhythmogenic
right ventricular cardiomyopathy | 6 |
1.87×10−03b |
| 4610 | Complement and
coagulation cascades | 5 |
7.26×10−03b |
| 4360 | Axon guidance | 7 |
7.65×10−03b |
| 5200 | Pathways in
cancer | 12 |
1.21×10−02a |
PPI network analysis
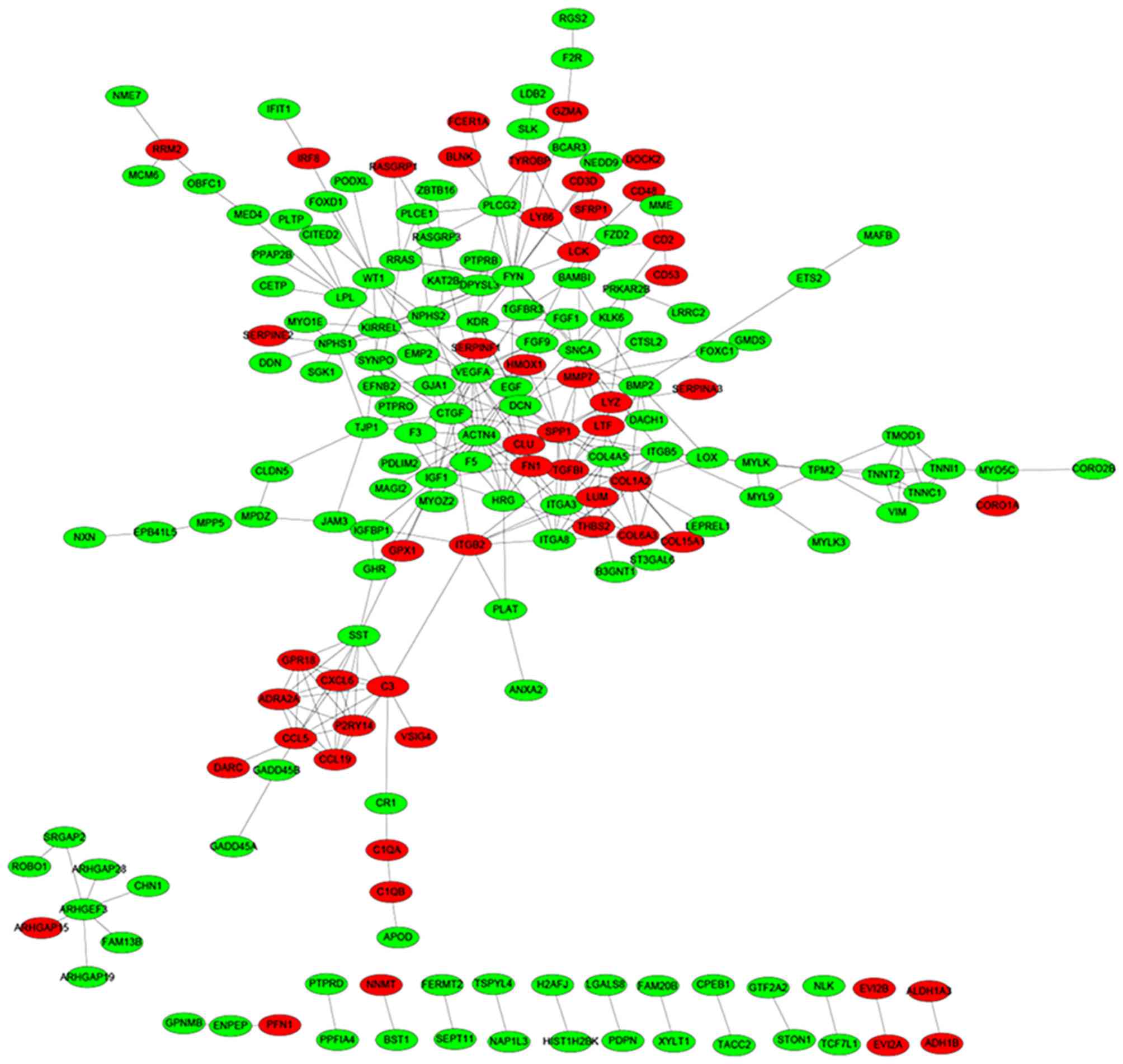
Based on the STRING database, a total of 335 protein
pairs with a combined score of >0.7 were obtained. As presented
in Fig. 2, the PPI network was
constructed with 335 edges and 184 nodes. The nodes of VEGFA
(degree score, 19), α-actinin-4 (ACTN4; degree score, 17),
proto-oncogene, Src family tyrosine kinase (FYN; degree score, 17),
collagen, type 1, α2 (COL1A2; degree score, 15) and insulin-like
growth factor 1 (IGF1; degree score, 15) were hub proteins in the
network.
Modules analysis
Two significant clustering modules were obtained
using ClusterONE software (Fig.
3). A total of 8 and 6 nodes were enriched in modules A and B,
respectively. As shown in Table
III, nodes in module A (density, 1.0; quality, 0.800;
P=1.606×10−4) were mainly enriched in GO: G-protein
coupled receptor protein signaling pathway, cell surface receptor
linked signal transduction, the immune response, the chemokine
signaling pathway and the cytokine-cytokine receptor interaction
pathway. Nodes in module B (density, 1.0; quality, 0.789; P=0.001)
were mainly enriched in GO: regulation of ATPase activity,
regulation of system process and regulation of hydrolase
activity.
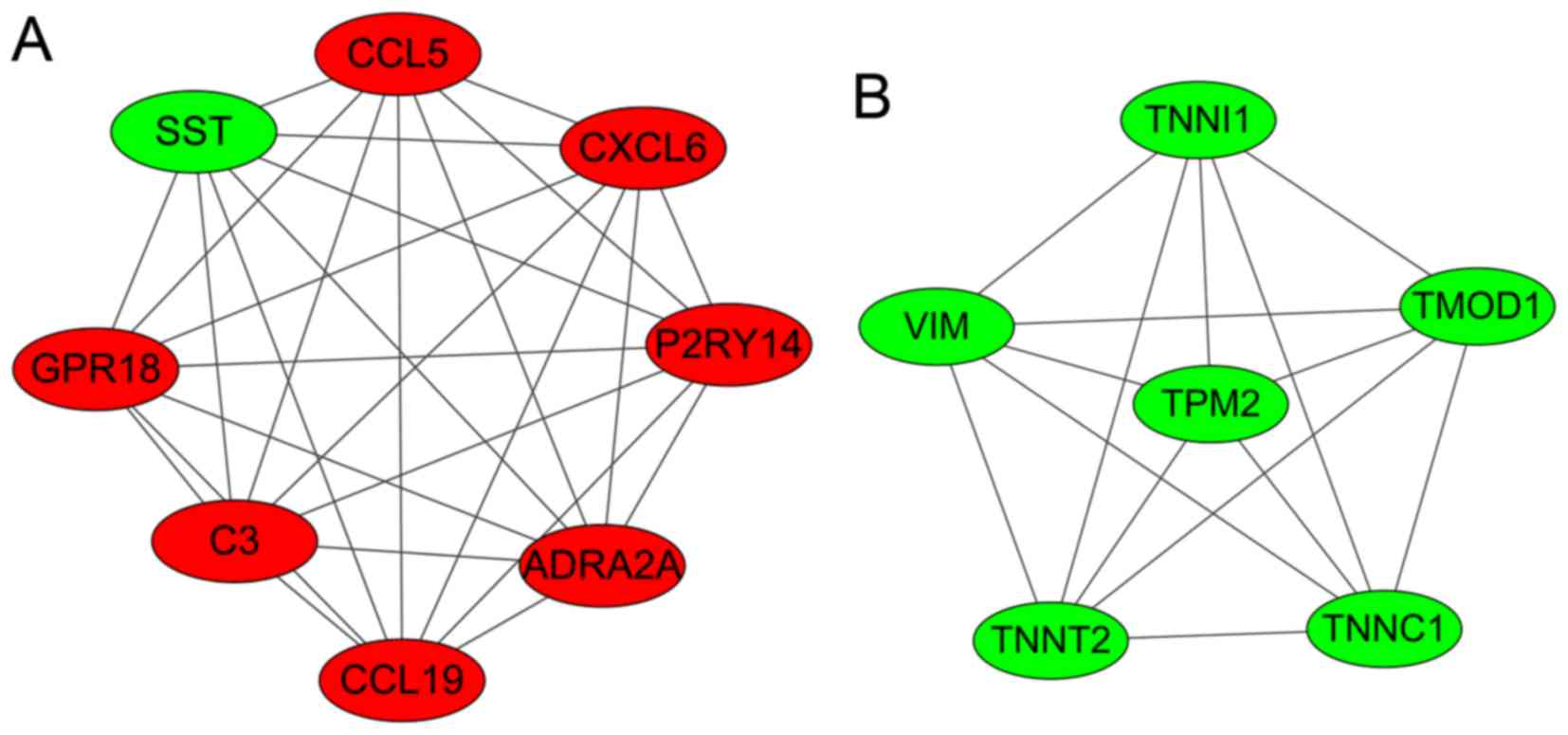 | Figure 3.Two significant clustering modules
(modules A and B) that were identified in the PPI networks. The red
nodes represent the upregulated genes and the green nodes represent
the downregulated genes. SST, somatostatin precursor; CCL5,
chemokine C-C motif ligand 5; CXCL6, chemokine C-X-C motif ligand
6; P2RY14, P2Y purinoceptor 14; ADRA2A, adrenoceptor α2A; C3,
complement component 3; GPR18, G-protein-coupled receptor 18; VIM,
vimentin; TNNI1, troponin I1; TMOD1, tropomodulin 1; TNNC1,
troponin C1; TNNT2, troponin T2; TPM2, tropomyosin 2β. |
 | Table III.Functional enrichment analysis of
protein-protein interaction network clustering modules. |
Table III.
Functional enrichment analysis of
protein-protein interaction network clustering modules.
| Term | Description | Counts (n) | P-value |
|---|
| Module A |
|
GO_BP:0007186 | G-protein coupled
receptor protein signaling pathway | 8 |
2.67×10−08a |
|
GO_BP:0007166 | Cell surface
receptor linked signal transduction | 8 |
9.06×10−07a |
|
GO_BP:0006955 | Immune
response | 5 |
2.08×10−04b |
| KEGG_
hsa04062 | Chemokine signaling
pathway | 3 |
1.25×10−02c |
| KEGG_
hsa04060 | Cytokine-cytokine
receptor interaction | 3 |
2.38×10−02c |
| Module B |
|
GO_BP:0043462 | Regulation of
ATPase activity | 3 |
1.31×10−05d |
|
GO_BP:0044057 | Regulation of
system process | 3 |
4.97×10−03e |
|
GO_BP:0051336 | Regulation of
hydrolase activity | 3 |
5.89×10−03f |
| KEGG_
hsa04260 | Cardiac muscle
contraction | 2 |
2.32×10−04b |
| KEGG_
hsa05410 | Hypertrophic
cardiomyopathy | 3 |
2.76×10−04b |
| KEGG_
hsa05414 | Dilated
cardiomyopathy | 2 |
3.24×10−04b |
Discussion
In the present study, using the gene expression
patterns downloaded from the GEO database, 426 DEGs in DKD
glomerular and tubular kidney biopsy tissues were obtained and
compared with matched normal tissues, identifying 115 upregulated
genes and 311 downregulated DEGs. The results demonstrated that
HLA-DPA1 was the common gene enriched in the rheumatoid
arthritis and systemic lupus erythematosus pathways, and the immune
response was a significant GO term enriched in module A. In
addition, VEGFA, ACTN4, FYN, COL1A2 and
IGF1 had higher degrees and were established as hub nodes in
the PPI network; they may therefore contribute to the progression
of DKD.
A previous study suggested that cells in the immune
system may be involved in the progression of DKD (22). Immune cells take part in vascular
injury under DKD-associated conditions (23). Other previous studies have also
indicated that the immune system is associated with DKD development
(24–26). Primary immunodeficiency (27), rheumatoid arthritis (28) and systemic lupus erythematosus
(29) are associated with the
immune system. In the present study, primary immunodeficiency,
rheumatoid arthritis and systemic lupus erythematosus were 3
significantly enriched pathways, and the immune response was a GO
term enriched in module A. Thus, the results of the present study
are in agreement with previous findings, and therefore indicate
that immune mechanisms may serve a role in DKD development.
The work of Woroniecka et al (3) suggested that HLA-DPA1 was a
differentially expressed transcript in the tubulointerstitium of
patients with DKD when compared with normal samples. Previous
studies revealed that HLA-DPA1, which is the closest
centromeric gene expressed to HLA-DOα, may contribute to the
differences in the associated risks of diabetes (30,31),
including DKD, which is a complication of diabetes. In the present
study, HLA-DPA1 was the common gene enriched in the
rheumatoid arthritis and systemic lupus erythematosus pathways.
Therefore, these results are in line with previous findings and
suggest that HLA-DPA1 may contribute to DKD development.
In addition, VEGFA, ACTN4, FYN, COL1A2 and IGF1 were
identified as hub proteins in the PPI network. VEGFA is an
important angiogenic growth factor that regulates endothelial
cells' permeability and vasculogenesis (32). It is also important for the
differentiation, proliferation, survival and migration of
endothelial cells within the glomerulus (33). Previous studies have suggested that
VEGFA may serve a significant role in retaining glomerular
endothelial cell function as a reduction in VEGFA levels induced
abnormal remodeling of glomerular capillaries (34,35).
VEGF may also serve a role in the pathogenesis of DKD (36) and the dysregulation of VEGFA may
serve a pathogenic role in inducing glomerular injury (37). In DKD, VEGFA has reduced mRNA
expression and may be a potential factor that can lead to the
development of DKD by inducing microvascular rarefaction and
tubular atrophy (9). In addition,
neoangiogenesis, which is caused by overexpression of VEGFA, may
stimulate the development of DKD and therefore blocking VEGFA or
its signaling may ameliorate DKD (38). These findings indicate that VEGFA
may serve a role in DKD progression.
ACTNs are actin-binding proteins that are critical
in cell adhesion and in the organization of the cytoskeleton
(39). Increasing evidence has
revealed that in diabetes, there are cytoskeletal changes in
podocytes. For instance, advanced glycosylation end products and
high glucose can decrease the expression of ACTN4 (40), and a reduced expression of ACTN4
may lead to proteinuria (a symptom of DKD) (41). In addition, FYN is a
tyrosine-specific phospho-transferase that belongs to the Src
family of tyrosine protein kinases (42). FYN phosphorylation is transiently
stimulated by high glucose levels (43). Src/FYN kinase inhibitors disrupt
signaling molecules in the VEGF signal transduction pathway
(44), and as mentioned above,
VEGF may be associated with DKD; thus, FYN may in turn be involved
in DKD. In addition, the accumulation of extracellular matrix
proteins such as COL1A2 is a key feature of DKD (45). A previous report demonstrated that
some key microRNAs (miR) act as effectors of transforming growth
factor (TGF)-β and the actions of high glucose in DKD (46). In mesangial cells and the kidney,
experimental diabetes was associated with the increased expression
of COL1A2, and miR-192 was increased by TGF-β treatment (45). IGF1 as a growth factor receptor has
been associated with type 1 DKD (47). In addition, IGF-1 has the capacity
to mediate the histological changes characteristic of DKD (48). These previous studies all indicate
that these proteins are associated with DKD. Therefore, the results
of the present study are in agreement with these findings and
provide further evidence that VEGFA, ACTN4, FYN, COL1A2 and IGF1
may serve important roles in DKD development directly or
indirectly.
In conclusion, the results of the present study
indicated that in addition to VEGFA, ACTN4,
FYN, COL1A2, IGF1 and HLA-DPA1, immune
mechanisms may also serve an important role in the development of
DKD. These genes may serve as target genes for the treatment of DKD
in future clinical practice. However, this conclusion has no
experimental verification; therefore, further evaluation of the
potential applications in clinical practice is required.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81070578 and
81270809).
References
|
1
|
Association AD: Standards of medical care
in diabetes-2010. Diabetes care. 33 Suppl:S11–S61. 2010. View Article : Google Scholar :
|
|
2
|
Levin A and Rocco M: KDOQI clinical
practice guidelines and clinical practice recommendations for
diabetes and chronic kidney disease. Am J Kidney Dis. 49(2 Suppl
2): S12–S154. 2007.
|
|
3
|
Woroniecka KI, Park AS, Mohtat D, Thomas
DB, Pullman JM and Susztak K: Transcriptome analysis of human
diabetic kidney disease. Diabetes. 60:2354–2369. 2011. View Article : Google Scholar :
|
|
4
|
Susztak K, Raff AC, Schiffer M and
Böttinger EP: Glucose-induced reactive oxygen species cause
apoptosis of podocytes and podocyte depletion at the onset of
diabetic nephropathy. Diabetes. 55:225–233. 2006. View Article : Google Scholar
|
|
5
|
Langham RG, Kelly DJ, Cox AJ, Thomson NM,
Holthöfer H, Zaoui P, Pinel N, Cordonnier DJ and Gilbert RE:
Proteinuria and the expression of the podocyte slit diaphragm
protein, nephrin, in diabetic nephropathy: Effects of angiotensin
converting enzyme inhibition. Diabetologia. 45:1572–1576. 2002.
View Article : Google Scholar
|
|
6
|
Doublier S, Salvidio G, Lupia E,
Ruotsalainen V, Verzola D, Deferrari G and Camussi G: Nephrin
expression is reduced in human diabetic nephropathy: Evidence for a
distinct role for glycated albumin and angiotensin II. Diabetes.
52:1023–1030. 2003. View Article : Google Scholar
|
|
7
|
Koop K, Eikmans M, Baelde HJ, Kawachi H,
de Heer E, Paul LC and Bruijn JA: Expression of podocyte-associated
molecules in acquired human kidney diseases. J Am Soc Nephrol.
14:2063–2071. 2003. View Article : Google Scholar
|
|
8
|
Turk T, Leeuwis JW, Gray J, Torti SV,
Lyons KM, Nguyen TQ and Goldschmeding R: BMP signaling and podocyte
markers are decreased in human diabetic nephropathy in association
with CTGF overexpression. J Histochem Cytochem. 57:623–631. 2009.
View Article : Google Scholar :
|
|
9
|
Lindenmeyer MT, Kretzler M, Boucherot A,
Berra S, Yasuda Y, Henger A, Eichinger F, Gaiser S, Schmid H,
Rastaldi MP, et al: Interstitial vascular rarefaction and reduced
VEGF-A expression in human diabetic nephropathy. J Am Soc Nephrol.
18:1765–1776. 2007. View Article : Google Scholar
|
|
10
|
Hohenstein B, Hausknecht B, Boehmer K,
Riess R, Brekken RA and Hugo CP: Local VEGF activity but not VEGF
expression is tightly regulated during diabetic nephropathy in man.
Kidney Int. 69:1654–1661. 2006. View Article : Google Scholar
|
|
11
|
Nath KA: The tubulointerstitium in
progressive renal disease. Kidney Int. 54:992–994. 1998. View Article : Google Scholar
|
|
12
|
Gilbert RE and Cooper ME: The
tubulointerstitium in progressive diabetic kidney disease: More
than an aftermath of glomerular injury? Kidney Int. 56:1627–1637.
1999. View Article : Google Scholar
|
|
13
|
Strippoli GF, Craig M, Deeks JJ, Schena FP
and Craig JC: Effects of angiotensin converting enzyme inhibitors
and angiotensin II receptor antagonists on mortality and renal
outcomes in diabetic nephropathy: Systematic review. BMJ.
329:8282004. View Article : Google Scholar :
|
|
14
|
Hansen TK, Forsblom C, Saraheimo M, Thorn
L, Wadén J, Høyem P, Østergaard J, Flyvbjerg A and Groop PH;
FinnDiane Study Group, : Association between mannose-binding
lectin, high-sensitivity C-reactive protein and the progression of
diabetic nephropathy in type 1 diabetes. Diabetologia.
53:1517–1524. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Altermann E and Klaenhammer TR:
PathwayVoyager: Pathway mapping using the Kyoto Encyclopedia of
Genes and Genomes (KEGG) database. BMC Genomics. 6:602005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Zhou X, Zhu J, Gu Y, Zhao W, Zou J
and Guo Z: GO-function: Deriving biologically relevant functions
from statistically significant functions. Brief Bioinform.
13:216–227. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39:D561–D568. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kohl M, Wiese S and Warscheid B:
Cytoscape: Software for visualization and analysis of biological
networks. Methods Mol Biol. 696:291–303. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ichinose K, Kawasaki E and Eguchi K:
Recent advancement of understanding pathogenesis of type 1 diabetes
and potential relevance to diabetic nephropathy. Am J Nephrol.
27:554–564. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Galkina E and Ley K: Leukocyte recruitment
and vascular injury in diabetic nephropathy. J Am Soc Nephrol.
17:368–377. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chow F, Ozols E, Nikolic-Paterson DJ,
Atkins RC and Tesch GH: Macrophages in mouse type 2 diabetic
nephropathy: Correlation with diabetic state and progressive renal
injury. Kidney Int. 65:116–128. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tuttle KR: Linking metabolism and
immunology: Diabetic nephropathy is an inflammatory disease. J Am
Soc Nephrol. 16:1537–1538. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mora C and Navarro JF: Inflammation and
diabetic nephropathy. Curr Diab Rep. 6:463–468. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Johnston SL, Virgo PF and Unsworth DJ:
Type 1 diabetes mellitus masking primary antibody deficiency. J
Clin Pathol. 53:236–237. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Prevoo ML, Van't Hof MA, Kuper HH, Van
Leeuwen MA, Van de Putte LB and Van Riel PL: Modified disease
activity scores that include twenty-eight-joint counts, development
and validation in a prospective longitudinal study of patients with
rheumatoid arthritis. Arthritis Rheum. 38:44–48. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Feng PH: Systemic lupus erythematosus: the
face of Asia. Ann N Y Acad Sci. 1108:114–120. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Varney MD, Valdes AM, Carlson JA, Noble
JA, Tait BD, Bonella P, Lavant E, Fear AL, Louey A, Moonsamy P, et
al: HLA DPA1, DPB1 alleles and haplotypes contribute to the risk
associated with type 1 diabetes: Analysis of the type 1 diabetes
genetics consortium families. Diabetes. 59:2055–2062. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Santin I, Castellanos-Rubio A, Aransay AM,
Gutierrez G, Gaztambide S, Rica I, Vicario JL, Noble JA, Castaño L
and Bilbao JR: Exploring the diabetogenicity of the HLA-B18-DR3
CEH: Independent association with T1D genetic risk close to
HLA-DOA. Genes Immun. 10:596–600. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dvorak HF, Brown LF, Detmar M and Dvorak
AM: Vascular permeability factor/vascular endothelial growth
factor, microvascular hyperpermeability, and angiogenesis. Am J
Pathol. 146:1029–1039. 1995.PubMed/NCBI
|
|
33
|
Schrijvers BF, Flyvbjerg A and De Vriese
AS: The role of vascular endothelial growth factor (VEGF) in renal
pathophysiology. Kidney Int. 65:2003–2017. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baelde HJ, Eikmans M, Doran PP, Lappin DW,
de Heer E and Bruijn JA: Gene expression profiling in glomeruli
from human kidneys with diabetic nephropathy. Am J Kidney Dis.
43:636–650. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bortoloso E, Del Prete D, Vestra M Dalla,
Gambaro G, Saller A, Antonucci F, Baggio B, Anglani F and Fioretto
P: Quantitave and qualitative changes in vascular endothelial
growth factor gene expression in glomeruli of patients with type 2
diabetes. Eur J Endocrinol. 150:799–807. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cha DR, Kim NH, Yoon JW, Jo SK, Cho WY,
Kim HK and Won NH: Role of vascular endothelial growth factor in
diabetic nephropathy. Kidney Int Suppl. 77:S104–S112. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eremina V, Sood M, Haigh J, Nagy A, Lajoie
G, Ferrara N, Gerber HP, Kikkawa Y, Miner JH and Quaggin SE:
Glomerular-specific alterations of VEGF-A expression lead to
distinct congenital and acquired renal diseases. J Clin Invest.
111:707–716. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Caldwell RB, Bartoli M, Behzadian MA,
El-Remessy AE, Al-Shabrawey M, Platt DH and Caldwell RW: Vascular
endothelial growth factor and diabetic retinopathy:
Pathophysiological mechanisms and treatment perspectives. Diabetes
Metab Res Rev. 19:442–455. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nikolopoulos SN, Spengler BA, Kisselbach
K, Evans AE, Biedler JL and Ross RA: The human non-muscle
alpha-actinin protein encoded by the ACTN4 gene suppresses
tumorigenicity of human neuroblastoma cells. Oncogene. 19:380–386.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
HA TS: High glucose and advanced
glycosylated end-products affect the expression of alpha-actinin-4
in glomerular epithelial cells. Nephrology (Carlton). 11:435–441.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Jefferson JA, Shankland SJ and Pichler RH:
Proteinuria in diabetic kidney disease: A mechanistic viewpoint.
Kidney Int. 74:22–36. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Resh MD: Fyn, a Src family tyrosine
kinase. Int J Biochem Cell Biol. 30:1159–1162. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Taniguchi K, Xia L, Goldberg HJ, Lee KW,
Shah A, Stavar L, Masson EA, Momen A, Shikatani EA, John R, et al:
Inhibition of Src kinase blocks high glucose-induced EGFR
transactivation and collagen synthesis in mesangial cells and
prevents diabetic nephropathy in Mice. Diabetes. 62:3874–3886.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
van Nieuw Amerongen GP and van Hinsbergh
VW: Targets for pharmacological intervention of endothelial
hyperpermeability and barrier function. Vascular pharmacol.
39:257–272. 2002. View Article : Google Scholar
|
|
45
|
Kato M, Zhang J, Wang M, Lanting L, Yuan
H, Rossi JJ and Natarajan R: MicroRNA-192 in diabetic kidney
glomeruli and its function in TGF-beta-induced collagen expression
via inhibition of E-box repressors. Proc Natl Acad Sci USA. 104:pp.
3432–3437. 2007; View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kato M, Arce L and Natarajan R: MicroRNAs
and their role in progressive kidney diseases. Clin J Am Soc
Nephrol. 4:1255–1266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Ewens KG, George RA, Sharma K, Ziyadeh FN
and Spielman RS: Assessment of 115 candidate genes for diabetic
nephropathy by transmission/disequilibrium test. Diabetes.
54:3305–3318. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Schreiber B, Hughes M and Groggel G:
Insulin-like growth factor-1 stimulates production of mesangial
cell matrix components. Clin Nephrol. 43:368–374. 1995.PubMed/NCBI
|