Introduction
Rheumatoid arthritis (RA) is one of the most common
articular diseases and is characterized by synovial hyperplasia,
which impairs quality of life. The estimated prevalence of RA is
0.5 to 1% worldwide (1,2). The pathological features of RA
include overgrowth of synoviocytes, chronic inflammation,
destruction of cartilage and bone and terminal-phase tissue
fibrosis (3,4). Biological agents, such as inhibitors
of tumor necrosis factor and interleukin-6, have recently been
developed to be used in RA treatments and have exhibited beneficial
therapeutic effects (5). However,
these therapies are expensive and may not be efficacious in a
significant number of RA patients.
Posttranslational modifications are crucial in the
regulation of protein functions by altering protein structure and
interactions (6). In RA, recent
studies have focused on the impact of citrullination (7–9).
Citrullination results in the loss of a positive charge and altered
biochemical features (10,11). Citrulline residues are synthesized
by the deamination of arginine residues using the
Ca2+-dependent enzyme, peptiydylarginine deiminase
(PADI). The PADI family consists of five members, PADI 1 to 4 and
6, which exhibit tissue specific expression (12). Patients with RA have increased
levels of citrullinated proteins, including vimentin, fibrinogen,
collagen and auto-antibodies against citrullinated protein
antibodies (ACPAs) are thought to be synthesized (13). Serum from the majority of patients
with RA contains auto-antibodies against a number of proteins and
ACPAs. ACPA is reported to have high sensitivity (60%) and
specificity (90%) as a clinical diagnostic biomarker for RA
(14–16).
PADI2 and PADI4 are expressed in the synovial tissue
of patients with RA and other types of arthritis (17–19).
PADI4 mediated citrullination has been implicated in a number of
inflammatory autoimmune diseases, including RA, lupus, colitis and
multiple sclerosis (20,21). In addition, PADI4 is a
RA-susceptibility gene, which was identified in genome-wide
association studies in patients with RA. The PADI4 haplotype is
associated with RA, and stabilization of PADI4 mRNA leads to
increased expression (22,23). PADI4 is a unique enzyme in the PADI
family due to its subcellular localization; other PADI family
members usually localize in the cytoplasm, whereas PADI4 is also
present in the nucleus (24). In
addition, PADI4 competes with arginine N-methyltransferase and
catalyzes citrullination of arginine residues in histones.
Furthermore, PADI4 promotes decondensation of chromatin structure
(25,26) and activates the transcription of a
number of genes and neutrophil extracellular traps (27,28).
Citrullination by PADI4 has been demonstrated to regulate protein
localization (29).
In addition to citrullination, protein
ubiquitination is important in the onset, pathogenesis and the
associated symptoms of RA. Endoplasmic reticulum (ER) stress
signals are associated with inflammation (30). ER stress signals, which are
triggered by an accumulation of unfolded or misfolded proteins in
the ER, induce a protective response known as the unfolded protein
response in order to maintain cellular homeostasis (31,32).
Accumulated misfolded proteins are degraded through the
ubiquitin-proteasome system in the cytoplasm. Secreted and membrane
proteins are synthesized in the ER, and pro-inflammatory cytokines
and extracellular matrix proteins have been revealed to be
associated with ER stress in patients with RA (33–35).
The E3 ubiquitin ligase, synoviolin (SYVN1), has
been identified in the synovial tissue of patients with RA. A
previous study demonstrated that the expression levels of SYVN1
increased in the RA synovium when compared with that in
osteoarthritis (36). SYVN1, a
mammalian homologue of Hrd1p/Der3p, serves important roles in the
ER-associated protein degradation (ERAD) pathway. SYVN1 deficient
mice exhibited resistance to collagen-induced arthritis, by
contrast, overexpression of SYVN1 induced arthropathy (36). In addition, SYVN1 has been revealed
to control the tumor suppressor p53 as a substrate and negatively
regulate its biological functions in transcription, the cell cycle
and apoptosis (37,38). As the expression of SYVN1 is higher
in patients that do not respond to treatment with infriximab, SYVN1
may have applications as a biomarker to facilitate the selection of
suitable treatments (39,40). Thus, these previous studies
indicated that SYVN1 may have crucial roles in synovial cell
hyperplasia in patients with RA. Therefore, the overexpression of
SYVN1 in patients with RA may result in a hyper-ERAD state, and in
turn, RA may be a disease of the ERAD system (41).
SYVN1 is implicated in a number of diseases in
addition to RA. It has been demonstrated that SYVN1 is associated
with liver and lung fibrosis (42–44)
and obesity in mouse models (45).
Analysis of post-neonatal SYVN1-knockout mice has indicated that
SYVN1 is associated with energy metabolism and mitochondrial
biosynthesis through peroxisome proliferator-activated receptor
gamma coactivator 1β (PGC-1β) degradation (45). Previous studies have identified a
number of small molecules that inhibit SYVN1 auto-ubiquitination
activity, suppress synoviocyte proliferation in patients with RA
and reduce the occurrence of arthritis in an RA model mouse
(46). In addition, these
inhibitors have been demonstrated to block the expression of
fibrosis-associated factors (42,44)
and reduce body weight and white adipose tissue (45). Thus, SYVN1 may represent a
potential therapeutic target.
Citrullinated proteins exhibit altered conformations
due to the increased hydrophobicity of citrulline residues, which
may be recognized as autoimmune antigens (11). Thus, it may be hypothesized that
accumulation of citrullinated proteins is associated with the
ubiquitin-proteasome system, a pathway that is induced by unfolded
proteins. Therefore, the present study investigated the
interactions between ubiquitination and citrullination.
Materials and methods
Plasmids
The sequence of full-length human PADI4 was
amplified from the cDNA of HL-60 cells obtained from American Type
Culture Collection (ATCC; cat. no. CCL240; Manassas, VA, USA) using
the following primers: Forward 5′-CGCGAATTCATGGCCCAGGGGACATTGAT-3
and reverse 5′-GCGGTCGACTCAGGGCACCATGTTCCACC-3. Mutant PADI4
(p.C645S), which is characterized by a Cys to Ser substitution at
amino acid (aa) 645 in the calcium binding site (26), was amplified using the following
primers: forward 5′-GAGGTGCACTCCGGCACCAACGT-3 and reverse
5′-ACGTTGGTGCCGGAGTGCACCTC-3, from the wild-type PADI4. The target
fragments were amplified using TaKaRa Ex Taq® DNA
Polymerase (Takara Biotechnology Co., Ltd., Dalian, China) under
the following conditions: 30 cycles at 94°C for 30 sec, at 55°C for
30 sec and at 72°C for 1 min, using the GeneAmp PCR System 9700
(Thermo Fisher Scientific, Inc., Waltham, MA, USA), and were then
cloned into the FLAG-tagged pcDNA3 vector (Invitrogen; Thermo
Fisher Scientific, Inc.).
Immunoprecipitation assay
Immunoprecipitation assays were performed on HEK 293
cells, as previously described (45). Briefly, HEK293 cells
(4×106), obtained from ATCC (cat. no. CRL-1573), were
transfected with FLAG (control cells) or SYVN1-FLAG constructs with
HA-PADI4 (experimental cells). Cells were lysed in lysis buffer,
containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1 mM EDTA, 1%
NP-40, 1 mM dithiothreitol (DTT), 10 µM MG-132, and protease
inhibitors, for 30 min at 4°C. Whole cell extracts (WCE) were
incubated with 2 µg of anti-FLAG (M2) antibodies in 1 ml of buffer
A (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1%
NP40, 5% glycerol, 10 µM MG-132, 1 µg/ml aprotinin and 1 µg/ml
leupeptin) for 4 h at 4°C. Proteins precipitated with anti-FLAG
antibody were subjected to 7.5% SDS-PAGE and detected using western
blot analysis with anti-HA or anti-FLAG antibodies.
In vivo ubiquitination assay
HEK293 cells (4×106) were transfected
with HA-ubiquitin, FLAG-PADI4 and SYVN1-FLAG expression plasmids.
Following incubation for 24 h, cells were treated with 10 µM MG-132
for 2 h. Cell extracts were prepared as described in the
immunoprecipitation assay methodology and separated by 7.5%
SDS-PAGE. Ubiquitinated proteins in WCE were detected using western
blot analysis with anti-HA, anti-SYVN1 and anti-PADI4 antibodies,
as described in the western blot analysis sub-section. Anti-SYVN1
antibodies detect endogenous proteins, however, anti-PADI4
antibodies do not.
For auto-ubiquitination of SYVN1 in vivo,
HEK293 cells were transfected with HA-ubiquitin, FLAG-PADI4 and
His-SYVN1. Following treatment with 10 µM MG-132 for 2 h, cells
(4×106) were harvested and lysed in the lysis buffer.
His-tagged SYVN1 was purified by incubation with Ni-Sepharose
chromatography resin (GE Healthcare Life Sciences) for 5 h at 4°C
and elution using buffer B (20 mM Tris-HCl pH 7.5, 0.5 M NaCl, 1 mM
EDTA, 1% Triton X-100, 1 mM DTT, 10 µM MG-132 and protease
inhibitors). Proteins were subjected to 7.5% SDS-PAGE and detected
using western blot analysis with anti-HA, anti-His and anti-FLAG
antibodies, as described in the western blot analysis
sub-section.
Pull-down assay
Pull-down assay was performed as previously
described (45). Glutathione
S-transferase (GST)-fused SYVN1 deletion mutant proteins were
expressed in Escherichia coli (E. coli) and purified
using glutathione sepharose beads. HA-tagged PADI4 was expressed
using the T7 in vitro transcription/translation system
(Promega Corporation, Madison, WI, USA) for 1 h at 30°C. GST-SYVN1
was incubated with HA-PADI4 in 1 ml buffer A (20 mM Tris-HCl, pH
8.0, 100 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.1% NP40, 5% glycerol, 10
µM MG-132, 1 µg/ml aprotinin, and 1 µg/ml leupeptin) for 6 h at
4°C. Following washing with buffer A, bound proteins were
fractionated by 10% SDS-PAGE and detected using western blot
analysis with anti-HA antibodies, as described in the western blot
analysis sub-section.
Western blot analysis
Proteins were separated by SDS-PAGE and transferred
onto polyvinylidene difluoride membranes. The membranes were
blocked with 5% milk in TBS buffer with 0.1% Tween-20 (TBS-T) at
room temperature for 1 h, and incubated with the primary antibodies
at room temperature for 1 h. The following primary antibodies were
used: Anti-FLAG (M2; cat. no. F3165; 1:5,000), anti-human influenza
hemagglutinin (HA)-tag (3F10; cat. no. 11867423001; 1:5,000),
obtained from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany);
anti-polyhistidine (His; cat. no. H-15; 1:1,000; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) and anti-SYVN1 rabbit
polyclonal antibodies (1:5,000), established in our laboratory as
previously described (37,45). Anti-PADI4 polyclonal antibodies
(1:1,000) were generated by injection of antigen peptides coding
PADI4 (aa 128–140) to a rabbit (Protein Purify, Gunma, Japan).
Following washing with TBS-T, the membranes were incubated with
horseradish peroxidase-conjugated anti-mouse (1:50,000; cat. no.
A5278; Sigma-Aldrich; Merck KGaA), anti-rabbit (1:75,000; cat. no.
A9169; Sigma-Aldrich; Merck KGaA) or anti-rat IgG secondary
antibody (1:75,000; cat. no. 112-035-062; Jackson ImmunoResearch
Laboratories, Inc., West Grove, PA, USA) at room temperature for 40
min. The protein bands were visualized using the enhanced
chemiluminescence Amersham ECL Select Western Blotting Detection
system (GE Healthcare Life Sciences, Chalfont, UK).
Results
Interactions between SYVN1 and
PADI4
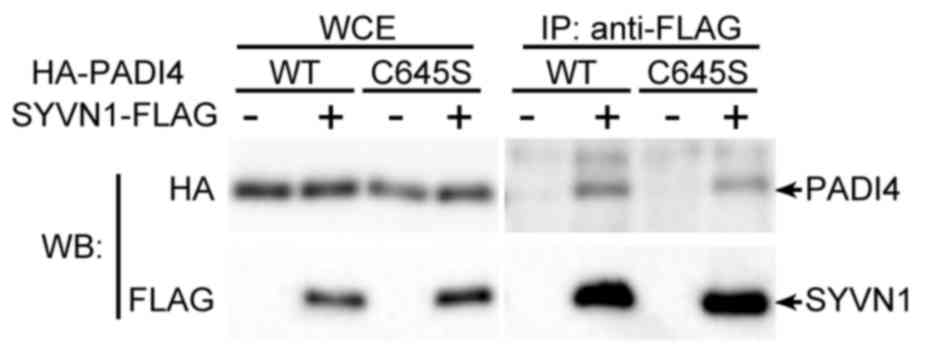
To assess the association between PADI4 and
ubiquitination, immunoprecipitation assays were performed to
determine whether PADI4 interacted with SYVN1, which represents the
E3 ligase in RA. HEK293 cells were transfected with FLAG-tagged
SYVN1 and HA-tagged PADI4 expression vectors. The complexes were
precipitated with anti-FLAG antibodies for SYVN1 and were detected
using anti-HA antibodies. FLAG-tagged SYVN1 was revealed to
interact with PADI4 (Fig. 1).
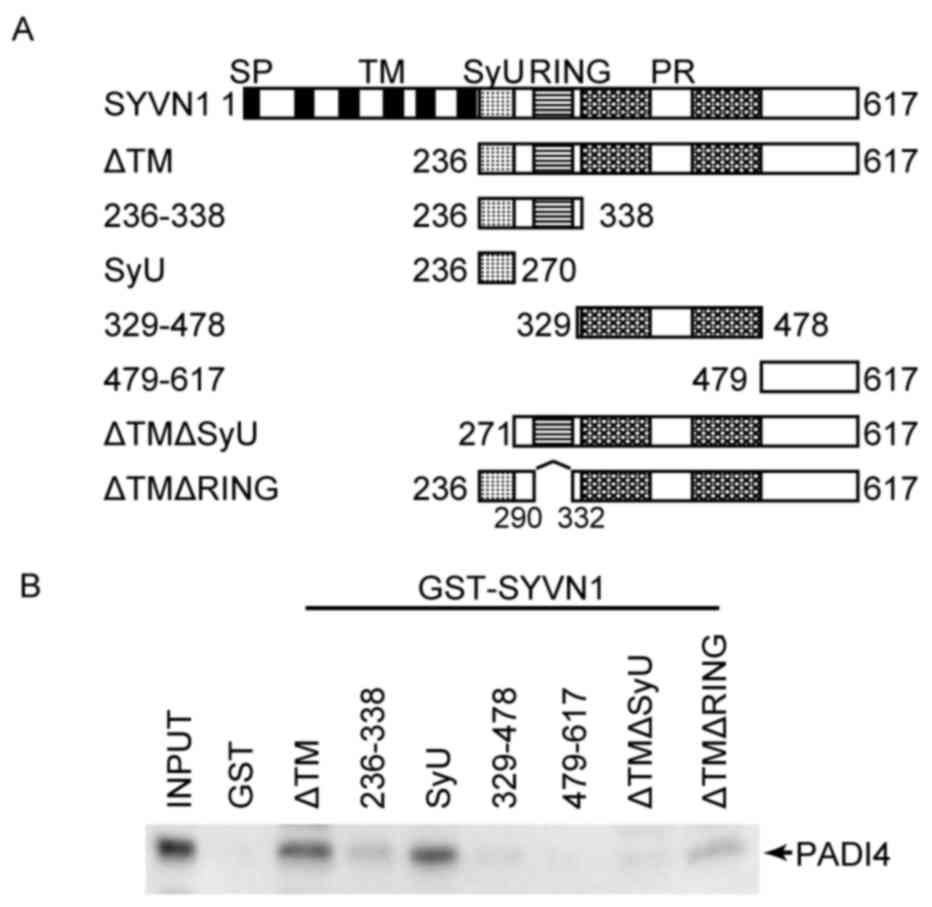
The PADI4 binding region in the SYVN1 protein was
then identified to further elucidate the association with PADI4.
SYVN1 contains 4 domains, the Really Interesting New Gene (RING)
finger domain, the SYVN1 unique domain (SyU) and two proline riche
domains. The SyU domain is responsible for interactions with p53
and PGC-1β (37,45). A series of deletion mutants of
SYVN1 (Fig. 2A) were expressed as
GST-fused proteins in E. coli. GST-fused SYVN1 deletion
mutants were incubated with HA-tagged PADI4, which was expressed
using the in vitro translation system. As a result,
GST-fused SYVN1 lacking the transmembrane domain (SYVN1 ΔTM) could
bind PADI4. The region from aa 236 to 338, which contains the RING
finger domain, and the SyU domain could bind PADI4. The region from
aa 339 to 478, which contains proline rich domains, may interact
with PADI4 weakly. In addition, the mutants lacking the RING finger
domain (SYVN1ΔTM ΔRING) and the SyU domain could bind with PADI4.
In contrast, the C-terminal region (aa 479 to 617) and SYVN1ΔTM
lacking the SyU domain (SYVN1ΔTM ΔSyU) did not interact with PADI4
(Fig. 2B). These results indicated
that SYVN1 may interact with PADI4 primarily via the SyU
domain.
Inhibition of ubiquitination by
PADI4
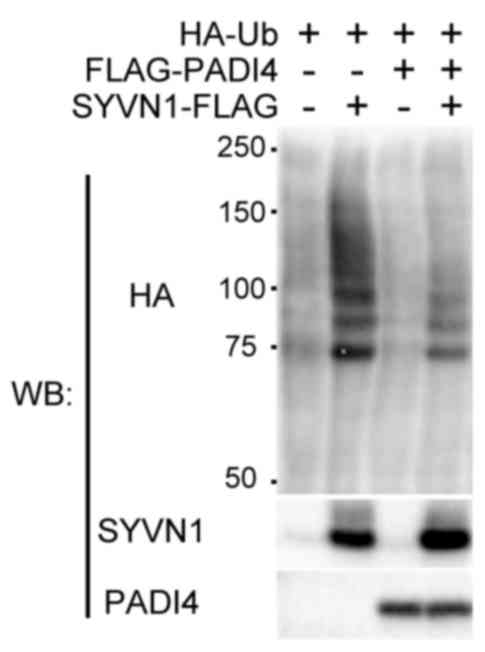
To assess the association of PADI4 with
ubiquitination, the effects of PADI4 overexpression on
ubiquitination levels in cells with or without SYVN1 was
investigated (36,39,41).
HEK293 cells were cotransfected with FLAG-PADI4 and SYVN1
expression vectors, and HA-tagged ubiquitin. In control cells,
ubiquitin ladder formations were detected, and SYVN1 expression
enhanced this ubiquitination. In contrast, co-expression with PADI4
reduced ubiquitination levels when compared with that observed in
control cells (Fig. 3).
Inhibition of the ubiquitination
activity of SYVN1 by PADI4
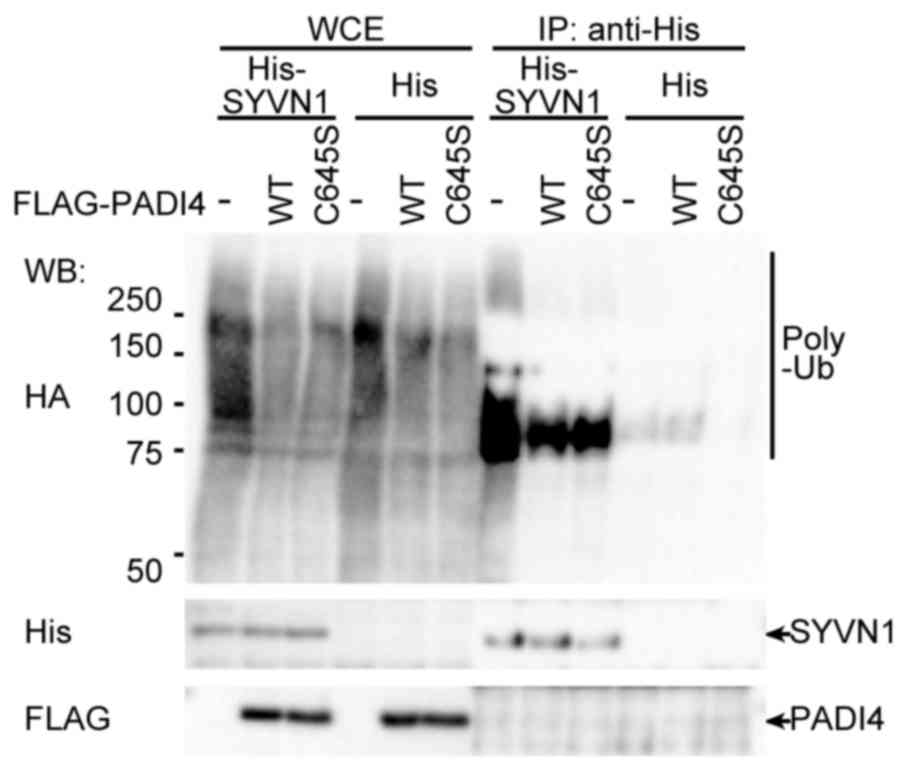
To confirm whether PADI4 inhibited ubiquitination by
SYVN1, in vivo ubiquitination assays were performed. HEK293
cells were transfected with His-tagged SYVN1 and FLAG-tagged PADI4
with HA-tagged ubiquitin. SYVN1 was purified with a Ni-sepharose,
and auto-ubiquitination was detected. The precipitated SYVN1 was
specifically ubiquitinated. PADI4 is a citrullination enzyme, and
the cysteine residue at amino acid position 645 in PADI4 is
essential for binding with calcium ions and its associated
enzymatic activity (26,47). Therefore, the effects of PADI4
enzymatic activity on ubiquitination were assessed using a PADI4
C645S mutant (26). The mutant
PADI4 was demonstrated to interact with SYVN1 in the same way as
the wild type (Fig. 1). The mutant
inhibited ubiquitination in HEK293 cells, decreasing ubiquitination
to the same level observed for wild type PADI4 (Fig. 4), indicating that the
stoichiometric mode rather than the enzymatic activity of PADI4 may
be important for inhibition of ubiquitination.
Discussion
Citrullination affects the structure and function of
protein substrates by altering the charge (25,48).
Such alterations are thought to induce autoimmune reactions,
generating autoantigens in RA. Citrullination is involved in cell
growth signals, therefore PADI4 is important for RA pathogenesis
(49). The present study revealed
that overexpression of PADI4 decreased ubiquitination levels in
HEK293 cells and that SYVN1 interacted with PADI4 via the SyU
domain and the C-terminal region. The results indicated that PADI4
may suppress the ubiquitination of proteins through associations
with E3 ligases. A PADI4 mutant, lacking enzymatic activity,
yielded similar results. Thus, PADI4 may prevent ubiquitination via
stoichiometric effects, not via enzymatic activity.
A number of post-translational modifications have
been reported to interact with each other (6). With regards to PADI and
citrullination, it has been demonstrated to interact with
acetylation and methylation. Arginine residues are substrates for
PADI and protein arginine methyltransferases, therefore
citrullination may antagonize methylation modifications and prevent
the decondensation of chromatin (25,50).
Similarly, ubiquitination and acetylation also compete for lysine
residues during the regulation of sodium channels (51). These regulatory mechanisms are
based on the competition for enzymatic target residues on the
common substrates. However, the association between citrullination
and ubiquitination may be mediated via an indirect pathway.
Notably, in type 1 diabetes, citrullination and ER stress signals
are implicated in the same inflammatory reaction. Binding
immunoglobulin protein (Bip)/glucose-related protein (GRP)-78, a
sensing protein that detects the accumulation of unfolded proteins
in the ER, translocates to the plasma membrane where it is modified
by PADI2. In this mechanism, the calcium influx induced by
inflammatory cytokines and ER stress may be able to activate PADI
(52). Citrullinated Bip secreted
from cells is recognized as an autoantigen, as previously reported
(52–54). Thus, in the present study,
inhibition of SYVN1-mediated ubiquitination by PADI4 may represent
a novel mechanism for the association between deiminase and
ubiquitin ligase.
Notably, PADI4 and SYVN1 regulate the same signaling
pathway. The present study demonstrated that SYVN1 binds to p53 in
the cytoplasm and negatively regulates p53 signaling via
ubiquitination. In addition, downregulation of SYVN1 expression
levels has been revealed to decrease the expression of p53 target
genes, such as p21 (37). In
contrast, upregulation of SYVN1 represses synoviocyte apoptosis in
patients with RA, leading to arthritis (37,38,55).
Additionally, PADI4 is recruited to target genes with p53 and
suppresses transactivation via citrullination of histone H3
(29,56–59).
In RA synoviocytes, activation of PADI4 function induces arthritis
by inhibiting apoptosis (23).
Thus, these reports suggest that activation of PADI4 and SYVN1
signals may enhance cell proliferation by inhibiting apoptosis.
However, the results of the present study demonstrated that PADI4
repressed ubiquitination by SYVN1. These results may be
inconsistent with the previous reports due to the presence of
multiple regulatory mechanisms involving PADI4 and SYVN1.
PADI4 may impair the repression of apoptosis via
SYVN1. PADI4 and SYVN1 induce cell proliferation via inhibition of
the p53 pathway and apoptosis. In addition, mice deficient in each
of these targets have exhibited resistance against the
collagen-induced arthritis (36)
and glucose-6-phosphate isomerase-induced arthritis models
(60), respectively. In addition,
synovium derived from patients with RA possess tumor-like
characteristics, including their growth ability and morphology
(61). However, the proliferation
of synoviocytes is not limitless and can be suppressed
spontaneously at some stages, unlike the growth of cancer cells
(62). One of the mechanisms
regulating these features may be Fas-mediated apoptosis in RA
synovium (62). Collectively, the
results indicated that PADI4 may mediate the balance between the
proliferation and death of cells through citrullination and
inhibition of ubiquitination by SYVN1. These findings may represent
a second mechanism for regulating the overgrowth of the synovium,
which is in contrast to tumor cells. Some patients with RA exhibit
vasculitis or severe extra-articular manifestations, classified as
malignant RA (MRA). The causes of MRA have not yet been clearly
defined (63). NETosis, the
release of neutrophil extracellular chromatin traps (NETs), has
been implicated in these severe rheumatoid disorders (64,65).
Autoantigens released outside of the cells during NETosis enhance
inflammatory responses as positive feedback, and citrullination
induced extracellular PADI4 is accelerated (66). However, the mechanisms mediating
the progression of these diseases have not been determined. It is
possible that apoptosis of synoviocytes may activate NETosis with
inflammatory cytokines derived from the synovium (64). Thus, apoptosis via the crosstalk
between PADI4 and SYVN1 may be one of the mechanisms limiting cell
proliferation in RA or regulating the exacerbation of RA
symptoms.
Another possibility is that inhibition of
ubiquitination may mimic the ER stress-repressed state. SYVN1 is
implicated in some stages of RA including, inflammation, fibrosis
and cartilage destruction through the ERAD system and quality
control of substrates (41,67–69).
The overexpression of SYVN1 in RA synovium may suppress apoptosis
via the activation of the ERAD system and cell proliferation
(41,55). Promoting the degradation of
unfolded proteins accumulated in the ER by overexpression of SYVN1
results in the reduction of ER stress, therefore ER stress may be
reduced or prevented in rheumatoid synovium highly expressing
SYVN1. This may lead to inhibition of apoptosis and proliferation
of synoviocytes. Namely, PADI4 may cause RA via suppression of the
ER stress pathway in addition to the production of autoantigens and
demethylation. A number of proteins, including fibrin, fibronectin,
vimentin and collagen are citrullinated by PADI in RA (20). The association between PADI family
proteins and substrates are unclear. PADI4 is mainly localized in
the nucleus and catalyzes histones, however, it has also been
detected in sera (24). In
addition, in the present study, PADI4 exhibited several functions
in addition to its role as a transcriptional repressor. SYVN1
catalyzes the ubiquitination of membrane-anchored proteins and
secreted proteins, including cytokines, extracellular matrix
proteins and receptors as substrates, and so SYVN1 and PADI4 may
catalyze the same substrates. In addition to PADI4, PADI2 has also
been implicated in RA. Further studies are required to understand
the crosstalk between ubiquitination and citrullination.
In some tumor tissues, PADI4-null mutations are
associated with resistance to apoptosis. This phenomenon may be
explained by the function of PADI4 as an inhibitor of cell
proliferation via citrullination of histone H4R3 under some
cellular stresses conditions, such as oxidative stress or radiation
(58). PADI4 has been predicted to
have the opposite effects in response to cellular stress. However,
the roles of PADI4 in ER stress have not been determined. In
contrast, SYVN1 has been implicated in oncogenesis and oxidative
stress (43). Collectively, these
results indicated that cell proliferation may be regulated by
PADI4-dependent SYVN1 suppression in response to several types of
stress.
The underlying crosstalk mechanisms between PADI4
and SYVN1 remain unclear, and so more extensive analysis is
required. PADI4 and SYVN1 serve important roles in a number of
diseases, including cancer and autoimmune diseases, such as RA.
Understanding of the crosstalk mechanisms associated with PADI4 and
SYVN1 may lead to the development of novel therapies for these
diseases. In addition, SYVN1 inhibitors have previously been
demonstrated to inhibit arthritis and fibrosis in vivo
(44–46,70).
Thus, these compounds may be useful in the treatment of diseases
involving PADI4 or citrullination.
Acknowledgements
The present study was supported by the Takeda
Science Foundation, the Naito Foundation, the Natural Science
Scholarship Daiichi-Sankyo Foundation of Life Science, Mitsubishi,
the Tanabe Pharma Corporation, Santen Pharmaceutical, the Bureau of
Social Welfare and Public Health, the Health Labour Sciences
Research Grant (grant no. H24-Nanchi-Wakate-010), the Japan Society
for the Promotion of Science KAKENHI (grant nos. 22790947,
24791006, 20249052, 23659176, 23659502, 26461476, 26670479,
26461478 and S1411011) and Supporting Positive Activities for
Female Researchers.
References
|
1
|
Harris ED Jr: Rheumatoid arthritis.
Pathophysiology and implications for therapy. N Engl J Med.
322:1277–1289. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Feldmann M, Brennan FM and Maini RN:
Rheumatoid arthritis. Cell. 85:307–310. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Aletaha D, Neogi T, Silman AJ, Funovits J,
Felson DT, Bingham CO III, Birnbaum NS, Burmester GR, Bykerk VP,
Cohen MD, et al: 2010 Rheumatoid arthritis classification criteria:
An American college of rheumatology/European league against
rheumatism collaborative initiative. Arthritis Rheum. 62:2569–2581.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gabriel SE: The epidemiology of rheumatoid
arthritis. Rheum Dis Clin North Am. 27:269–281. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Smolen JS and Aletaha D: Rheumatoid
arthritis therapy reappraisal: Strategies, opportunities and
challenges. Nat Rev Rheumatol. 11:276–289. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Yang XJ: Multisite protein modification
and intramolecular signaling. Oncogene. 24:1653–1662. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yamada R, Suzuki A, Chang X and Yamamoto
K: Citrullinated proteins in rheumatoid arthritis. Front Biosci.
10:54–64. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Martin-Mola E, Balsa A, Garcia-Vicuna R,
Gómez-Reino J, González-Gay MA, Sanmartí R and Loza E:
Anti-citrullinated peptide antibodies and their value for
predicting responses to biologic agents: A review. Rheumatol Int.
36:1043–1063. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Aggarwal R, Liao K, Nair R, Ringold S and
Costenbader KH: Anti-citrullinated peptide antibody assays and
their role in the diagnosis of rheumatoid arthritis. Arthritis
Rheum. 61:1472–1483. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
van Venrooij WJ and Pruijn GJ:
Citrullination: A small change for a protein with great
consequences for rheumatoid arthritis. Arthritis Res. 2:249–251.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tarcsa E, Marekov LN, Mei G, Melino G, Lee
SC and Steinert PM: Protein unfolding by peptidylarginine
deiminase. Substrate specificity and structural relationships of
the natural substrates trichohyalin and filaggrin. J Biol Chem.
271:30709–30716. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Vossenaar ER, Zendman AJ, van Venrooij WJ
and Pruijn GJ: PAD, a growing family of citrullinating enzymes:
Genes, features and involvement in disease. Bioessays.
25:1106–1118. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klareskog L, Rönnelid J, Lundberg K,
Padyukov L and Alfredsson L: Immunity to citrullinated proteins in
rheumatoid arthritis. Annu Rev Immunol. 26:651–675. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schellekens GA, de Jong BA, van den Hoogen
FH, van de Putte LB and van Venrooij WJ: Citrulline is an essential
constituent of antigenic determinants recognized by rheumatoid
arthritis-specific autoantibodies. J Clin Invest. 101:273–281.
1998. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schellekens GA, Visser H, de Jong BA, van
den Hoogen FH, Hazes JM, Breedveld FC and van Venrooij WJ: The
diagnostic properties of rheumatoid arthritis antibodies
recognizing a cyclic citrullinated peptide. Arthritis Rheum.
43:155–163. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mrabet D, Laadhar L, Haouet S, Sahli H,
Zouari B, Makni S and Sellami S: Anomalies of intra-synovial
citrullination: Is there any interest in the diagnosis of early
rheumatoid arthritis? Rheumatol Int. 33:787–791. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ikari K, Kuwahara M, Nakamura T, Momohara
S, Hara M, Yamanaka H, Tomatsu T and Kamatani N: Association
between PADI4 and rheumatoid arthritis: A replication study.
Arthritis Rheum. 52:3054–3057. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Foulquier C, Sebbag M, Clavel C,
Chapuy-Regaud S, Al Badine R, Méchin MC, Vincent C, Nachat R,
Yamada M, Takahara H, et al: Peptidyl arginine deiminase type
2(PAD-2) and PAD-4 but not PAD-1, PAD-3, and PAD-6 are expressed in
rheumatoid arthritis synovium in close association with tissue
inflammation. Arthritis Rheum. 56:3541–3553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang X, Xia Y, Pan J, Meng Q, Zhao Y and
Yan X: PADI2 is significantly associated with rheumatoid arthritis.
PLoS One. 8:e812592013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Anzilotti C, Pratesi F, Tommasi C and
Migliorini P: Peptidylarginine deiminase 4 and citrullination in
health and disease. Autoimmun Rev. 9:158–160. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jones JE, Causey CP, Knuckley B,
Slack-Noyes JL and Thompson PR: Protein arginine deiminase 4
(PAD4): Current understanding and future therapeutic potential.
Curr Opin Drug Discov Devel. 12:616–627. 2009.PubMed/NCBI
|
|
22
|
Suzuki A, Yamada R, Chang X, Tokuhiro S,
Sawada T, Suzuki M, Nagasaki M, Nakayama-Hamada M, Kawaida R, Ono
M, et al: Functional haplotypes of PADI4, encoding citrullinating
enzyme peptidylarginine deiminase 4, are associated with rheumatoid
arthritis. Nat Genet. 34:395–402. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chang X, Yamada R, Suzuki A, Sawada T,
Yoshino S, Tokuhiro S and Yamamoto K: Localization of
peptidylarginine deiminase 4 (PADI4) and citrullinated protein in
synovial tissue of rheumatoid arthritis. Rheumatology (Oxford).
44:40–50. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mastronardi FG, Wood DD, Mei J, Raijmakers
R, Tseveleki V, Dosch HM, Probert L, Casaccia-Bonnefil P and
Moscarello MA: Increased citrullination of histone H3 in multiple
sclerosis brain and animal models of demyelination: A role for
tumor necrosis factor-induced peptidylarginine deiminase 4
translocation. J Neurosci. 26:11387–11396. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Cuthbert GL, Daujat S, Snowden AW,
Erdjument-Bromage H, Hagiwara T, Yamada M, Schneider R, Gregory PD,
Tempst P, Bannister AJ and Kouzarides T: Histone deimination
antagonizes arginine methylation. Cell. 118:545–553. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang Y, Wysocka J, Sayegh J, Lee YH,
Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y,
et al: Human PAD4 regulates histone arginine methylation levels via
demethylimination. Science. 306:279–283. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wang Y, Li M, Stadler S, Correll S, Li P,
Wang D, Hayama R, Leonelli L, Han H, Grigoryev SA, et al: Histone
hypercitrullination mediates chromatin decondensation and
neutrophil extracellular trap formation. J Cell Biol. 184:205–213.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Li P, Li M, Lindberg MR, Kennett MJ, Xiong
N and Wang Y: PAD4 is essential for antibacterial innate immunity
mediated by neutrophil extracellular traps. J Exp Med.
207:1853–1862. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tanikawa C, Ueda K, Nakagawa H, Yoshida N,
Nakamura Y and Matsuda K: Regulation of protein Citrullination
through p53/PADI4 network in DNA damage response. Cancer Res.
69:8761–8769. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoo SA, You S, Yoon HJ, Kim DH, Kim HS,
Lee K, Ahn JH, Hwang D, Lee AS, Kim KJ, et al: A novel pathogenic
role of the ER chaperone GRP78/BiP in rheumatoid arthritis. J Exp
Med. 209:871–886. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Manié SN, Lebeau J and Chevet E: Cellular
mechanisms of endoplasmic reticulum stress signaling in health and
disease. 3. Orchestrating the unfolded protein response in
oncogenesis: An update. Am J Physiol Cell Physiol. 307:C901–C907.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hotamisligil GS and Erbay E: Nutrient
sensing and inflammation in metabolic diseases. Nat Rev Immunol.
8:923–934. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Park YJ, Yoo SA and Kim WU: Role of
endoplasmic reticulum stress in rheumatoid arthritis pathogenesis.
J Korean Med Sci. 29:2–11. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gao B, Lee SM, Chen A, Zhang J, Zhang DD,
Kannan K, Ortmann RA and Fang D: Synoviolin promotes IRE1
ubiquitination and degradation in synovial fibroblasts from mice
with collagen-induced arthritis. EMBO Rep. 9:480–485. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Connor AM, Mahomed N, Gandhi R, Keystone
EC and Berger SA: TNFα modulates protein degradation pathways in
rheumatoid arthritis synovial fibroblasts. Arthritis Res Ther.
14:R622012. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Amano T, Yamasaki S, Yagishita N,
Tsuchimochi K, Shin H, Kawahara K, Aratani S, Fujita H, Zhang L,
Ikeda R, et al: Synoviolin/Hrd1, an E3 ubiquitin ligase, as a novel
pathogenic factor for arthropathy. Genes Dev. 17:2436–2449. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yamasaki S, Yagishita N, Sasaki T,
Nakazawa M, Kato Y, Yamadera T, Bae E, Toriyama S, Ikeda R, Zhang
L, et al: Cytoplasmic destruction of p53 by the endoplasmic
reticulum-resident ubiquitin ligase ‘Synoviolin’. Embo J.
26:113–122. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yamasaki S, Yagishita N, Nishioka K and
Nakajima T: The roles of synoviolin in crosstalk between
endoplasmic reticulum stress-induced apoptosis and p53 pathway.
Cell Cycle. 6:1319–1323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Toh ML, Marotte H, Blond JL, Jhumka U,
Eljaafari A, Mougin B and Miossec P: Overexpression of synoviolin
in peripheral blood and synoviocytes from rheumatoid arthritis
patients and continued elevation in nonresponders to infliximab
treatment. Arthritis Rheum. 54:2109–2118. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Klaasen R, Wijbrandts CA, van Kuijk AW,
Pots D, Gerlag DM and Tak PP: Synovial synoviolin in relation to
response to TNF blockade in patients with rheumatoid arthritis and
psoriatic arthritis. Ann Rheum Dis. 71:1260–1261. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yagishita N, Yamasaki S, Nishioka K and
Nakajima T: Synoviolin, protein folding and the maintenance of
joint homeostasis. Nat Clin Pract Rheumatol. 4:91–97. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hasegawa D, Fujii R, Yagishita N,
Matsumoto N, Aratani S, Izumi T, Azakami K, Nakazawa M, Fujita H,
Sato T, et al: E3 ubiquitin ligase synoviolin is involved in liver
fibrogenesis. PLoS One. 5:e135902010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu T, Zhao F, Gao B, Tan C, Yagishita N,
Nakajima T, Wong PK, Chapman E, Fang D and Zhang DD: Hrd1
suppresses Nrf2-mediated cellular protection during liver
cirrhosis. Genes Dev. 28:708–722. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Nakajima F, Aratani S, Fujita H, Yagishita
N, Ichinose S, Makita K, Setoguchi Y and Nakajima T: Synoviolin
inhibitor LS-102 reduces endoplasmic reticulum stress-induced
collagen secretion in an in vitro model of stress-related
interstitial pneumonia. Int J Mol Med. 35:110–116. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fujita H, Yagishita N, Aratani S,
Saito-Fujita T, Morota S, Yamano Y, Hansson MJ, Inazu M, Kokuba H,
Sudo K, et al: The E3 ligase synoviolin controls body weight and
mitochondrial biogenesis through negative regulation of PGC-1β.
EMBO J. 34:1042–1055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yagishita N, Aratani S, Leach C, Amano T,
Yamano Y, Nakatani K, Nishioka K and Nakajima T: RING-finger type
E3 ubiquitin ligase inhibitors as novel candidates for the
treatment of rheumatoid arthritis. Int J Mol Med. 30:1281–1286.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Arita K, Hashimoto H, Shimizu T, Nakashima
K, Yamada M and Sato M: Structural basis for Ca(2+)-induced
activation of human PAD4. Nat Struct Mol Biol. 11:777–783. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Knuckley B, Bhatia M and Thompson PR:
Protein arginine deiminase 4: Evidence for a reverse protonation
mechanism. Biochemistry. 46:6578–6587. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vossenaar ER, Nijenhuis S, Helsen MM, Van
der Heijden A, Senshu T, van den Berg WB, van Venrooij WJ and
Joosten LA: Citrullination of synovial proteins in murine models of
rheumatoid arthritis. Arthritis Rheum. 48:2489–2500. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Raijmakers R, Zendman AJ, Egberts WV,
Vossenaar ER, Raats J, Soede-Huijbregts C, Rutjes FP, van Veelen
PA, Drijfhout JW and Pruijn GJ: Methylation of arginine residues
interferes with citrullination by peptidylarginine deiminases in
vitro. J Mol Biol. 367:1118–1129. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Butler PL, Staruschenko A and Snyder PM:
Acetylation stimulates the epithelial sodium channel by reducing
its ubiquitination and degradation. J Biol Chem. 290:12497–13503.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Rondas D, Crèvecoeur I, D'Hertog W,
Ferreira GB, Staes A, Garg AD, Eizirik DL, Agostinis P, Gevaert K,
Overbergh L and Mathieu C: Citrullinated glucose-regulated protein
78 is an autoantigen in type 1 diabetes. Diabetes. 64:573–586.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Skriner K, Adolph K, Jungblut PR and
Burmester GR: Association of citrullinated proteins with synovial
exosomes. Arthritis Rheum. 54:3809–3814. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Goëb V, Thomas-L'Otellier M, Daveau R,
Charlionet R, Fardellone P, Le Loët X, Tron F, Gilbert D and
Vittecoq O: Candidate autoantigens identified by mass spectrometry
in early rheumatoid arthritis are chaperones and citrullinated
glycolytic enzymes. Arthritis Res Ther. 11:R382009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Yamasaki S, Yagishita N, Tsuchimochi K,
Kato Y, Sasaki T, Amano T, Beppu M, Aoki H, Nakamura H, Nishioka K
and Nakajima T: Resistance to endoplasmic reticulum stress is an
acquired cellular characteristic of rheumatoid synovial cells. Int
J Mol Med. 18:113–117. 2006.PubMed/NCBI
|
|
56
|
Yao H, Li P, Venters BJ, Zheng S, Thompson
PR, Pugh BF and Wang Y: Histone Arg modifications and p53 regulate
the expression of OKL38, a mediator of apoptosis. J Biol Chem.
283:20060–20068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li P, Yao H, Zhang Z, Li M, Luo Y,
Thompson PR, Gilmour DS and Wang Y: Regulation of p53 target gene
expression by peptidylarginine deiminase 4. Mol Cell Biol.
28:4745–4758. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Tanikawa C, Espinosa M, Suzuki A, Masuda
K, Yamamoto K, Tsuchiya E, Ueda K, Daigo Y, Nakamura Y and Matsuda
K: Regulation of histone modification and chromatin structure by
the p53-PADI4 pathway. Nat Commun. 3:6762012. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu GY, Liao YF, Chang WH, Liu CC, Hsieh
MC, Hsu PC, Tsay GJ and Hung HC: Overexpression of peptidylarginine
deiminase IV features in apoptosis of haematopoietic cells.
Apoptosis. 11:183–196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Seri Y, Shoda H, Suzuki A, Matsumoto I,
Sumida T, Fujio K and Yamamoto K: Peptidylarginine deiminase type 4
deficiency reduced arthritis severity in a glucose-6-phosphate
isomerase-induced arthritis model. Sci Rep. 5:130412015. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Moran M, Fang C and Paul A: Rheumatoid
arthritis presenting as an invasive soft-tissue tumour. Arch Orthop
Trauma Surg. 122:538–540. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Nakajima T, Aono H, Hasunuma T, Yamamoto
K, Shirai T, Hirohata K and Nishioka K: Apoptosis and functional
Fas antigen in rheumatoid arthritis synoviocytes. Arthritis Rheum.
38:485–491. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Nobunaga M: Malignant rheumatoid
arthritis. Nihon Rinsho. 50:597–602. 1992.(In Japanese). PubMed/NCBI
|
|
64
|
Khandpur R, Carmona-Rivera C,
Vivekanandan-Giri A, Gizinski A, Yalavarthi S, Knight JS, Friday S,
Li S, Patel RM, Subramanian V, et al: NETs are a source of
citrullinated autoantigens and stimulate inflammatory responses in
rheumatoid arthritis. Sci Transl Med. 5:178ra402013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dwivedi N, Upadhyay J, Neeli I, Khan S,
Pattanaik D, Myers L, Kirou KA, Hellmich B, Knuckley B, Thompson
PR, et al: Felty's syndrome autoantibodies bind to deiminated
histones and neutrophil extracellular chromatin traps. Arthritis
Rheum. 64:982–992. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Spengler J, Lugonja B, Ytterberg A Jimmy,
Zubarev RA, Creese AJ, Pearson MJ, Grant MM, Milward M, Lundberg K,
Buckley CD, et al: Release of active peptidyl arginine deiminases
by neutrophils can explain production of extracellular
citrullinated autoantigens in rheumatoid arthritis synovial fluid.
Arthritis Rheumatol. 67:3135–3145. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Li L, Shen Y, Ding Y, Liu Y, Su D and
Liang X: Hrd1 participates in the regulation of collagen I
synthesis in renal fibrosis. Mol Cell Biochem. 386:35–44. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Toh ML, Gonzales G, Koenders MI, Tournadre
A, Boyle D, Lubberts E, Zhou Y, Firestein GS, van den Berg WB and
Miossec P: Role of interleukin 17 in arthritis chronicity through
survival of synoviocytes via regulation of synoviolin expression.
PLoS One. 5:e134162010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Burr ML, van den Boomen DJ, Bye H,
Antrobus R, Wiertz EJ and Lehner PJ: MHC class I molecules are
preferentially ubiquitinated on endoplasmic reticulum luminal
residues during HRD1 ubiquitin E3 ligase-mediated dislocation. Proc
Natl Acad Sci USA. 110:pp. 14290–14295. 2013; View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Bianchini E, Fanin M, Mamchaoui K, Betto R
and Sandonà D: Unveiling the degradative route of the V247M
α-sarcoglycan mutant responsible for LGMD-2D. Hum Mol Genet.
23:3746–3758. 2014. View Article : Google Scholar : PubMed/NCBI
|