Introduction
Calcitonin gene-related peptide (CGRP) is a 37-amino
acid peptide with a variety of biological effects, and is widely
distributed in peripheral and central nervous system (1). Initially, it was reported that CGRP
is produced by sensory nerve cells and has effect on sympathetic
nervous pathways (2). With the
development of research, it has been confirmed that CGRP has an
important role in the cardiovascular system, respiratory system and
skeletal system (3). Furthermore,
there is growing concern about the association between CGRP and the
immune system. As one of the most important neurotransmitters of
the neuroimmune axis, CGRP works as a bridge connecting the nervous
system and immune system (4–6).
Furthermore, the research on CGRP and macrophages has become a hot
topic in the past decades. CGRP can induce macrophages to produce
cytokines through interaction with CGRP receptors. The calcitonin
receptor-like receptor and receptor activity-modifying protein 1
are present on the surface of bone marrow-derived macrophages
(7). Additionally, α-CGRP
receptors are present on alveolar macrophages (8). CGRP has an important role on the
inflammatory response mediated by lipopolysaccharide (LPS) in
macrophages (9,10) and CGRP is also be secreted by
macrophages (11). It has been
proved that certain classical pathways are involved in biological
processes, including inflammatory response, the cyclic adenosine
monophosphate pathway and nuclear factor-κB pathway (9–12).
Circular RNA (circRNA) is a special type of
non-coding RNA that was first identified 20 years ago; however, it
was not until recent years that the study of circRNA function and
structure became common (13,14).
CircRNA is characterized by the presence of a covalent bond linking
the 3′ and 5′ends generated by back splicing (15). Although circRNA can be derived from
introns, untranslated regions and gene fragments, it is
predominantly produced from exonic DNA encodes protein (15,16).
The function of circRNA remains unclear; however, research has
provided some information. CircRNA is widely expressed in human
cells and the abundance of it exists in cytoplasm (17,18).
The sequences of the majority of circRNAs are highly conserved, and
have the ability to mediate gene expression at the transcriptional
level or post-transcriptional level by altering their downstream
miRNAs expression (15,18). Furthermore, circRNA is more stable
than linear RNA in cells, as its unique structure is resistant to
degradation by exonucleases (16).
Recently, it was reported that circRNA can act as a ‘sponge’ for
microRNAs (miRNAs) (19). The
previous study demonstrated that cerebellar degeneration-related
protein 1 transcript antisense RNA (CDR1as) acts as a circular
miRNA-7 (miR-7) inhibitor, which harbors more than 70 conventional
miR-7 binding sites. When CDR1 as is overexpressed, miR-7 will
associate with it and the expression of miR-7 target genes
increases (20); this is termed
the ‘sponge effect’ or competing endogenous RNA mechanism, and this
effect has been validated in multiple studies (21–23).
In the present study, it was demonstrated that
interleukin-6 (IL-6) mRNA expression in RAW264.7 macrophages is
enhanced by CGRP in a dose- and time-dependent manner. Furthermore,
the expression of certain circRNAs, including mmu_circRNA_007893,
was demonstrated to be changed when macrophages were stimulated by
CGRP. Subsequently, the function and mechanism of
mmu_circRNA_007893 was investigated. The results of the current
study demonstrated that mmu_circRNA_007893 is involved in IL-6 mRNA
induction by CGRP in RAW264.7 macrophages.
Materials and methods
Cell culture
The RAW264.7 macrophages (Infrastructure of Cell
Line Resources, Beijing, China) were plated on 6-well plate culture
dishes at density of 2×105 cells per well in 2 ml of
Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.). The 6-well plates
were placed at 37°C in a humidified atmosphere of 95% air and 5%
CO2. Media were replaced 1 h before the experiments.
Verification of optimum concentration
and time point of CGRP on RAW264.7 cell lines
In the concentration experiments, the cells were
divided into 6 groups: 0.01, 0.1, 1, 10, 100 nM CGRP and the
control group. Then cells were treated with CGRP (Sigma-Aldrich;
Merck KGaA, Darmstadt, Germany) at the corresponding concentration.
After stimulation for 2 h, the total RNA was collected and the
expression of IL-6 in each group was detected by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
Similarly, in the time point experiments, the cells were divided
into 5 groups: 4, 3, 2, 1 h and the control group (0 h). Then,
cells were treated with 1 nM CGRP for the corresponding time. The
total RNA was collected and the expression of IL-6 in each group
was detected by RT-qPCR.
Preparation and analysis of the
circRNA microarrays
The cells were divided into 2 groups: Control group
and subject group. In the subject group, the cells were treated
with 1 nM CGRP for 2 h. In the control, the cells were untreated.
The total RNA of the 2 groups was isolated using TRIpure (Aidalb
Biotechnologies Co., Ltd., Beijing, China) and RNA in each group
was quantified using NanoDrop 2000 (Thermo Fisher Scientific, Inc.,
Wilmington, DE, USA). The samples were prepared and hybridized
based on the Arraystar standard protocols (Arraystar, Inc.,
Rockville, MD, USA). In brief, according to the Arraystar Super RNA
Labeling protocol (Arraystar, Inc.), total RNA of samples was
amplified and transcribed into fluorescent cRNA utilizing random
primers. The labeled cRNAs were hybridized onto the Arraystar Human
circRNA Array (8×15K, Arraystar, Inc.). After washing the slides,
the arrays were scanned with an Agilent G2505C Scanner. Then,
Agilent Feature Extraction software (version 11.0.1.1) was used to
analyze the acquired array images. Quantile normalization and
subsequent data processing were performed with the R software
package. After the circRNA microarray was completed, all the
circRNAs that had changed above 2 folds were displayed in the
circRNA profiling data (The data was provided by the Arraystar,
Inc.). Then, the circRNAs were selected with reference to the
networks using circBase (http://www.circbase.org), mirBase (http://www.mirbase.org) and miRDB (http://www.mirdb.org); the selected circRNA were
confirmed by RT-PCR and their PCR products were sequenced (Nuclea
Bio-tech Inc., Guangzhou, China).
Fluorescence in situ hybridization
(FISH)
The probe used to detect mmu_circRNA_007893 was
synthesized by BersinBio (Guangzhou, China) and its sequence was
CTCCAGCTTGCCTTCATCCAGTCCAA. The FISH experiment was performed with
the BersinBio™ circRNA FISH kit. The cells were fixed by 4%
paraformaldehyde at room temperature for 20 min and washed with
diethyl pyrocarbonate twice. Prior to permeabilization, the cells
were washed by 1X PBS and fixed by 4% paraformaldehyde at room
temperature for 10 min. Then the samples were washed by 1X PBS
three times and permeabilized at 4°C for 5 min. The probe labeled
with fluorescein isothiocyanate (100 ng/µl; BersinBio) and
hybridization solution (BersinBio) were mixed at the ratio of 1:19
and denaturated at 73°C for 8 min. Then, the mixture was added to
the samples and the samples were incubated overnight at 42°C. In
the present study, DAPI (5 µg/ml; Beyotime Institute of
Biotechnology, Shanghai, China) was used to stain the nucleus at
room temperature for 10 min. The images were observed with a
confocal microscope (Leica Microsystems GmbH, Wetzlar,
Germany).
circRNA interference
The small interfering RNAs (siRNAs) targeting
mmu_circRNA_007893 were designed and synthesized by Guangzhou
RiboBio Co., Ltd. (Guangzhou, China). The sequences of siRNA1-3
were showed as followed: siRNA1, 5′-GCCUUCAUCCAGUCCAAUGdTdT-3′
(sense) and 3′-dTdTCGGAAGUAGGUCAGGUUAC-5′ (antisense); siRNA2,
5′-CAGCUUGCCUUCAUCCAGUdTdT-3′ (sense),
3′-dTdTGUCGAACGGAAGUAGGUCA-5′ (antisense); and siRNA3,
5′-CCCUCCAGCUUGCCUUCAUdTdT-3′ (sense) and
3′-dTdTGGGAGGUCGAACGGAAGUA-5′ (antisense), respectively. Negative
siRNA was used as a control. The RAW264.7 macrophages were
transfected with siRNA using GenMute™ siRNA Transfection Kit and
Transfection Buffer (SignaGen Laboratories, Rockville, MD, USA)
according to the manufacturer's instructions. Then the cells were
incubated for 48 h at 37°C prior to the sequential tests.
miRNA overexpression and
inhibition
The mmu-miR-485-5p mimic and inhibitor were
synthesized by Invitrogen (Thermo Fisher Scientific, Inc.). The
sequences were as follows: Mimic, 5′-AGAGGCUGGCCGUGAUGAAUUC-3′
(sense) and 3′-UCUCCGACCGGCACUACUUAAG-5′ (antisense); inhibitor,
5′-UCUCCGACCGGCACUACUUAAG-3′. The culture media were replaced 1 h
before transfection. Then the cells were transfected with mimics or
inhibitor using Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) following the manufacturer's protocol and
incubated for 24 h at 37°C prior to the subsequent experiments.
Detection of IL-6 mRNA,
mmu_circRNA_007893, mmu-miR-485-5p by RT-qPCR
Total RNA from RAW264.7 cell line was extracted
using TRIzol reagent (Thermo Fisher Scientific, Inc.). The mRNA
samples were treated with DNase to eliminate contaminating genomic
DNA and reverse transcribed using the First Strand cDNA Synthesis
Kit (Takara Bio, Inc., Otsu, Japan). For circRNA analysis, total
RNA was reverse transcribed using random primers (Invitrogen;
Thermo Fisher Scientific, Inc.). miRNAs were extracted using small
RNAiso (Takara Bio, Inc.) and reverse transcribed by MicroRNA
First-Strand Synthesis Kits (Takara Bio, Inc.). GAPDH was chosen to
be the internal standard for mRNA and circRNAs analysis and U6 was
selected to the internal standard for miRNA analysis. The primers
used in the experiment as follows: IL-6,
5′-CCACTTCACAAGTCGGAGGCTTA-3′ (forward) and
5′-CCAGTTTGGTAGCATCCATCATTTC-3′ (reverse); mmu_circRNA_007893,
5′-CAGTAAGGGAGGGCAAGAA-3′ (forward) and 5′-GCCATAAGGTGGAATCTGC-3′
(reverse); mmu-miR-485-5p, 5′-GCTGGCCGTGATGAATTCAAA-3′ (forward);
and GAPDH, 5′-AAGAAGGTGGTGAAGCAGG-3′ (forward) and
5′-GAAGGTGGAAGAGTGGGAGT-3′ (reverse). The RT-qPCR was undertaken
with the SYBR® Premix Ex Taq™ II kit (Takara Bio, Inc.,
Otsu, Japan). Briefly, the total reaction system was 25 µl and the
RT-qPCR was performed using 40 cycles with a 10 min denaturation at
95°C, 15 sec annealing at 95°C and a 1 min extension at 60°C. The
2−ΔΔCq method was used to analyze the results (24).
Western blotting
Protein was extracted using radioimmunoprecipitation
assay buffer (50 mM Tris HCl, pH 7.4, 150 mM NaCl, 1% Nonidet P40
and 0.1% sodium dodecyl sulfate) and phenylmethanesulfonyl fluoride
at 4°C. Bicinchoninic acid protein assay kit (BestBio, Shanghai,
China) was used to determine the concentration. The concentration
of each sample was 500 ng/µl and all the samples were run on 10%
sodium dodecyl sulfate-polyacrylamide gel and electro-transferred
onto polyvinylidene fluoride membranes for 1 h at 4°C. After being
electro-transferred, the polyvinylidene fluoride membranes were
blocked by skimmed milk for 1 h at room temperature. Antibodies
against GAPDH (97166; 1:4,000; Cell Signaling Technology, Inc.,
Danvers, MA, USA) and IL-6 (sc-57315; 1:200; Santa Cruz
Biotechnology, Inc., Dallas, TX, USA) were incubated with blots
overnight at 4°C. Then the blots were incubated with secondary
antibody (anti-mouse IgG; ab6789; 1:5,000; Abcam, Cambridge, UK) at
room temperature for 1 h. Proteins were detected by
electrochemiluminescence (Beijing Dingguo Changsheng Biotechnology
Co., Ltd., Beijing, China).
Statistical analysis
The SPSS version 20.0 program (IBM Corp., Armonk,
NY, USA) was used for the statistical analysis. The data of two
groups were analyzed by the Student's t test. The data of three or
more groups were evaluated using analysis of variance and least
significant difference post hoc tests when the variance was normal,
otherwise Dunnett's T3 test was used. The results are reported as
the mean ± standard error. P<0.05 was considered to indicate a
statistically significant difference.
Results
CGRP induces IL-6 mRNA expression in
RAW264.7 macrophages
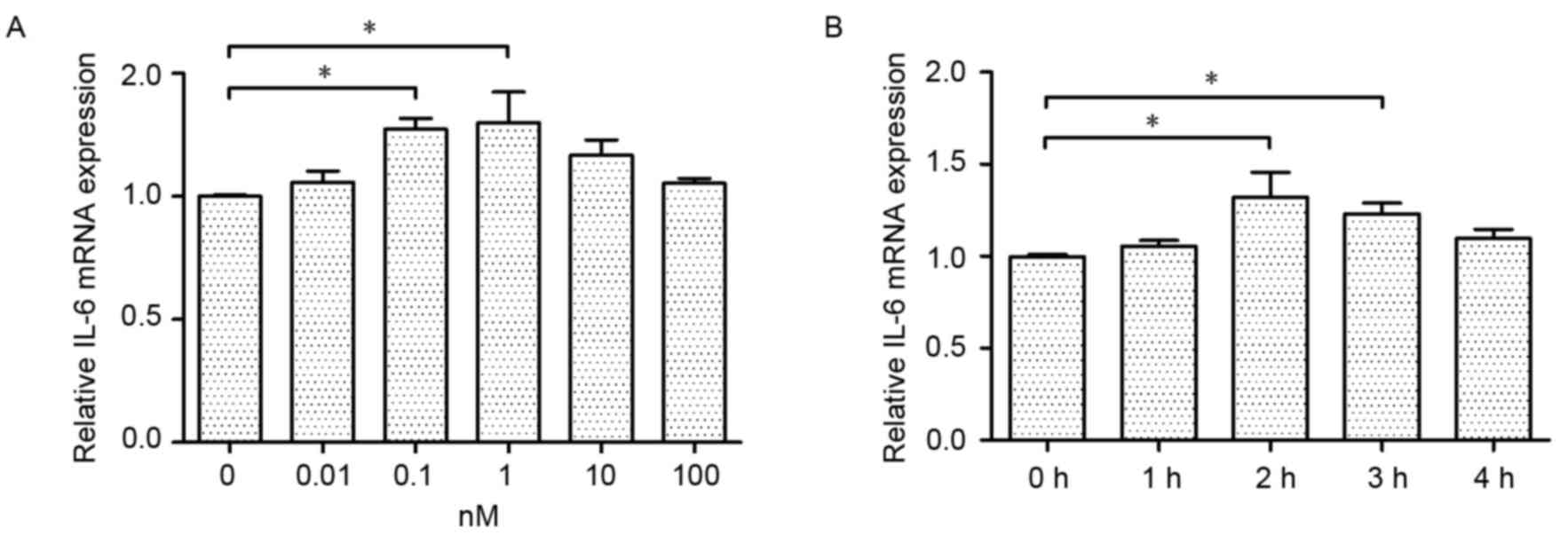
To identify the effect of CGRP on IL-6 mRNA
expression in RAW264.7 macrophages, cultured cells were treated for
different times, with different concentrations of CGRP. IL-6 mRNA
was measured by RT-qPCR. In the concentration experiments,
macrophages were stimulated with CGRP (0.01 to 100 nM) for up to 2
h. As presented in Fig. 1A, the
promotion effect of CGRP was observed at a concentration of 10 nM
and peaked at the concentration of 1 nM. The transient increase
effect was not different to the basal level at a concentration of
0.01 nM. Therefore, 1 nM CGRP was selected as the optimum
concentration for the subsequent experiments. Similarly, CGRP
caused a time-dependent change to IL-6 mRNA expression in RAW264.7
macrophages. The macrophages were treated with 1 nM CGRP for 1–4 h.
As presented in Fig. 1B, the
expression of IL-6 mRNA increased gradually from 1 to 2 h, and
remained stable for 1 h. Then, the IL-6 mRNA expression decreased
slightly. These results suggested that CGRP stimulates dose- and
time-dependent increases in IL-6 mRNA.
CGRP affects the expression of
circRNAs in RAW264.7 macrophages
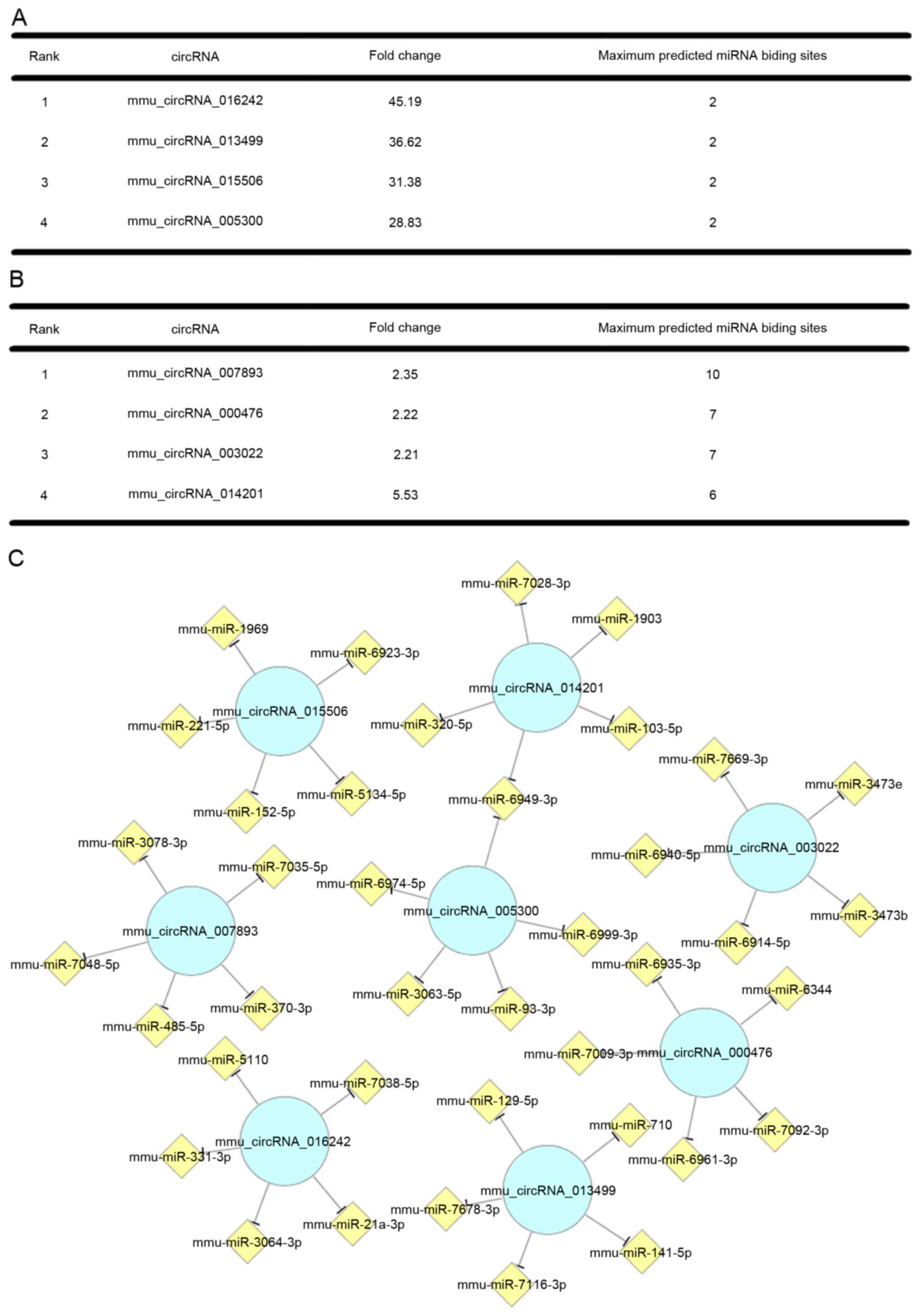
In the circRNA microarray, 349 circRNAs were changed
>2-fold between the control group and subject group (1 nm CGRP
for 2 h). Among them, 173 circRNAs were significantly upregulated.
The upregulated circRNAs were ranked according to the fold change
and the top four are listed in Fig.
2A. CircRNAs can interact with their target miRNAs by base
pairing and most have more than five miRNAs as potential targets.
Furthermore, each miRNA can bind to several circRNAs. Thus, it is
difficult to define which circRNA have the most important roles in
biological processes. Therefore, it was assumed that the circRNAs
that have more miRNA binding sites have more critical roles in the
circRNA-miRNA network (23).
According to this hypothesis, the upregulated circRNAs were ranked
by the number of the predicted miRNA binding sites and the top four
are listed in Fig. 2B.
Furthermore, the target miRNAs of the former eight circRNAs were
ranked according to mirSVR scores and the five highest ranking
miRNAs for each circRNA are presented in Fig. 2C. Cytoscape was used to construct
the circRNA-miRNA network.
CGRP promotes mmu_circRNA_007893
expression in RAW264.7 macrophages
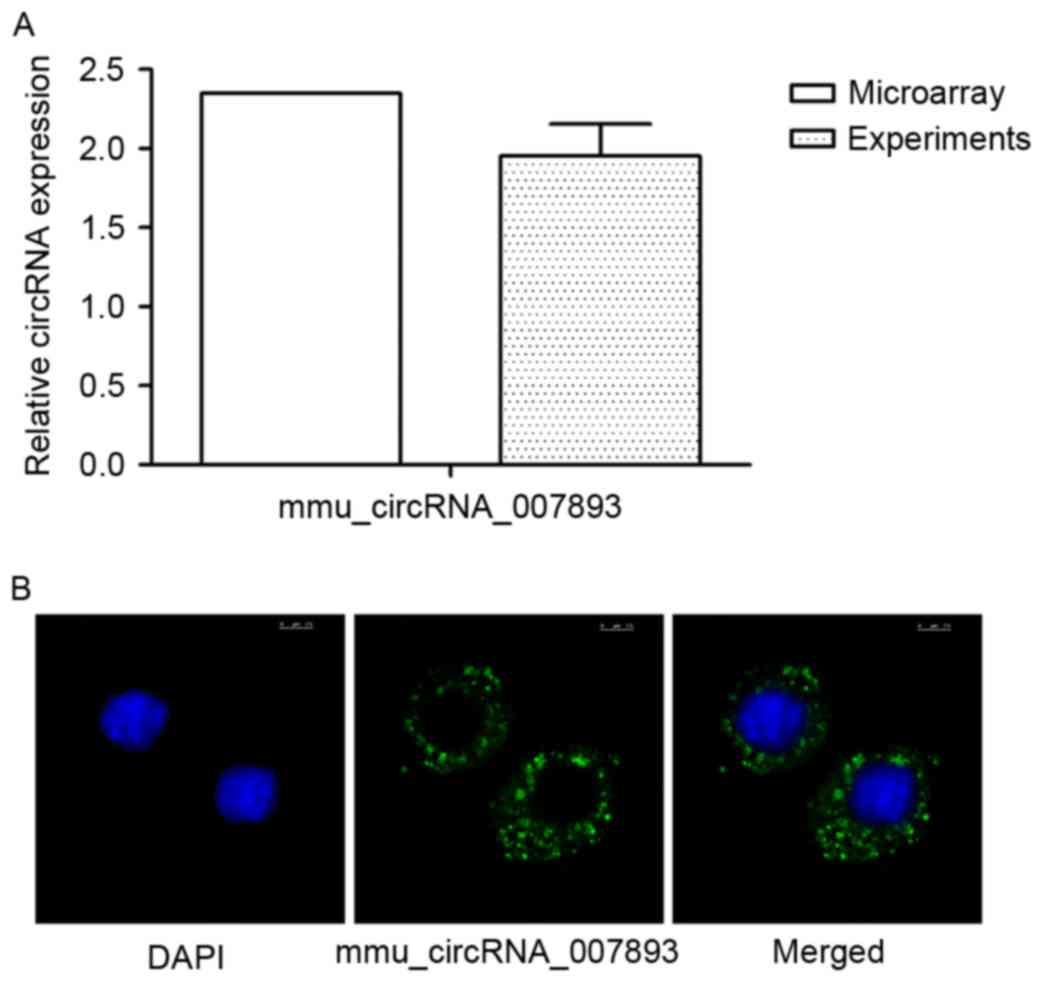
To confirm the validity of the data microarray, all
the circRNAs identified were also analyzed by RT-qPCR. However,
several circRNAs exhibited unchanged expression or the opposite
trend compared with the microarray results. As presented in
Fig. 3A, it demonstrated that
mmu_circRNA_007893 expression increased stably by ~2 fold relative
to the control group. The data was in accordance with the results
in the circRNAs microarray. The result indicated that CGRP promotes
mmu_circRNA_007893 expression in RAW264.7 macrophages. Furthermore,
according to the circRNA microarray, mmu_circRNA_007893 were
located mainly in the cytoplasm. To confirm the result, the
fluorescent probes complementary to the junction sequences was
attached to the circRNAs and fluorescence in situ hybridization
(FISH) was used. As shown in Fig.
3B, the location of the circRNA was mostly in the
cytoplasm.
CGRP promotes IL-6 expression by
mediated mmu_circRNA_007893 expression
Referring to the aforementioned results, CGRP
induced mmu_circRNA_007893 and IL-6 mRNA expression in RAW264.7
macrophages. It was then explored whether IL-6 is a downstream
mediator of mmu_circRNA_007893. Three different siRNAs were used to
reduce the circRNA expression. As demonstrated in Fig. 4A, all the siRNAs were effective and
siRNA3 was selected as the optimum siRNA for the subsequent
experiments. The IL-6 mRNA expression was detected following
circRNA inhibition using the siRNA. In the CGRP + siRNA group, the
cells were treated with siRNA3 for 48 h before CGRP stimulation. In
Fig. 4B, IL-6 mRNA expression
level declined correspondingly with the decrease of the circRNA.
IL-6 proteins were also detected using western blotting. The result
of western blot was consistent with that of RT-qPCR (Fig. 4C). Overall, the data indicates that
CGRP promotes IL-6 expression and mmu_circRNA_007893 is involved in
this effect.
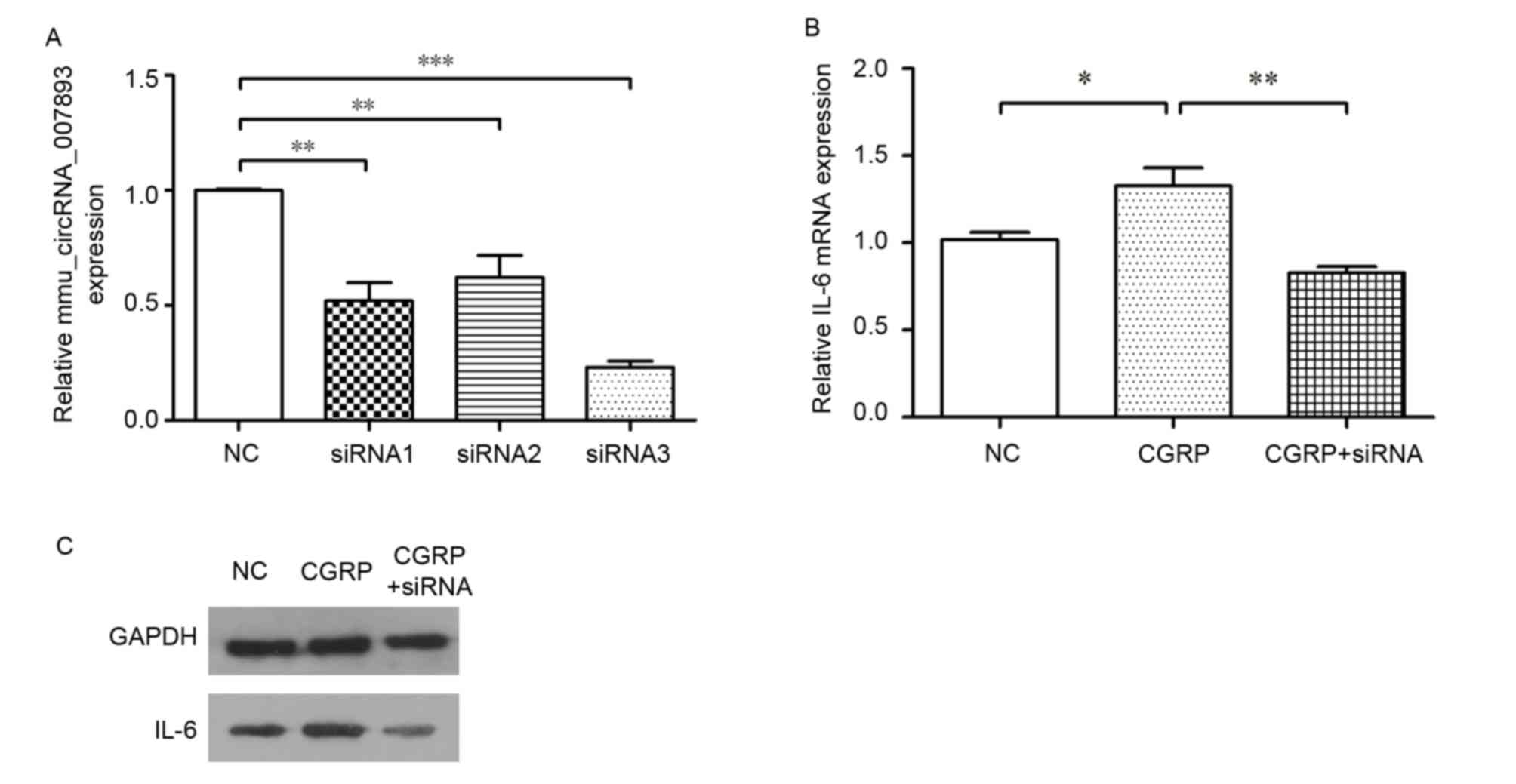 | Figure 4.Effects of mmu_circRNA_007893
inhibition on IL-6 mRNA expression in RAW264.7 macrophages. (A)
Cells were treated with three siRNAs targeting mmu_circRNA_007893.
circRNA expressions in siRNA1 groups (P=0.001), siRNA2 groups
(P=0.003) and siRNA3 groups (P<0.001) were significantly lower
than that in negative control groups. (B) IL-6 mRNA expression in
the CGRP-treated group was significantly higher than that in
negative control group (P=0.018). However, its expression level in
CGRP + siRNA groups decreased following knockdown of
mmu_circRNA_007893 using siRNA3, which was markedly lower than that
in the CGRP groups (P=0.002). The IL-6 mRNA and circRNA were
detected by reverse transcription-quantitative polymerase chain
reaction. The values are presented as the mean ± standard error.
*P<0.05, **P<0.01, ***P<0.001, n=3 for each group. (C)
IL-6 protein expression levels were analyzed by western blotting.
GAPDH was used as an internal control. mmu, Mus musculus;
circRNA, circular RNA; siRNA, small interfering RNA; NC, negative
control; CGRP, calcitonin gene-related peptide; IL-6,
interleukin-6. |
Role of mmu-miR-485-5p in
mmu_circRNA_007893-mediated IL-6 release
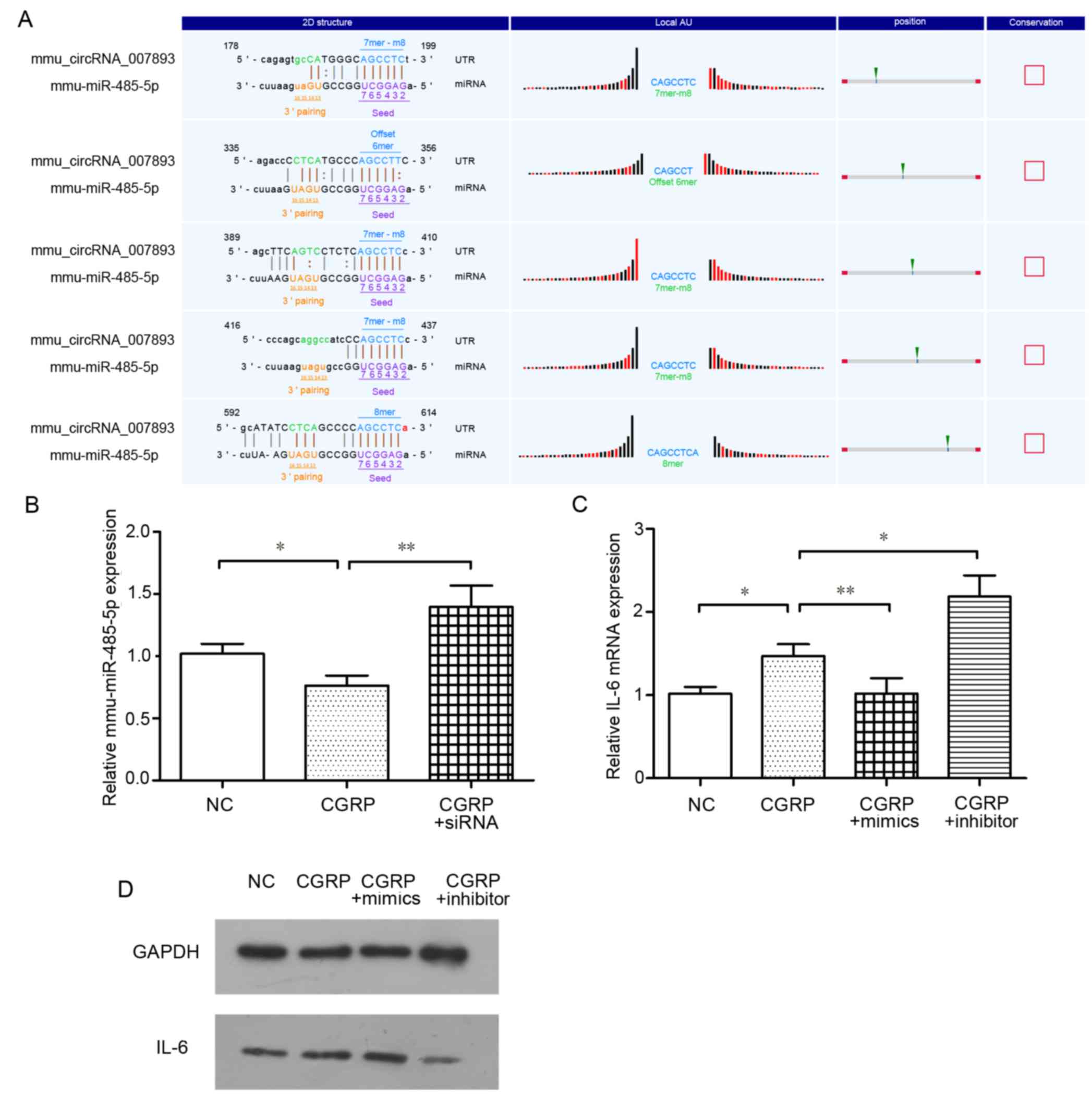
It is reported that circRNAs may act as a ‘sponge’
for miRNAs, and to thus influence mRNA expression. Therefore,
target miRNAs that may associate with circRNAs were identified
using bioinformatic networks and the circRNA microarray. According
to the circRNA microarrays, mmu-miR-485-5p was one of the
predicated target miRNAs of mmu_circRNA_007893. As demonstrated in
Fig. 5A, this mmu-miR-485-5p was
predicted to match five regions of the complete sequence of
mmu_circRNA_007893. To identify whether mmu-miR-485-5p took part in
the biological processes in thus study, the mmu-miR-485-5p
expression was determined in CGRP group and CGRP + siRNA group
cells. Referring to Fig. 5B, the
expression of miR-485-5p in the CGRP groups was significantly lower
than that in the negative control groups. However, with
mmu_circRNA_007893 expression knocked down by siRNA, the expression
of miR-485-5p was rescued. This result suggests that
mmu_circRNA_007893 reduces the expression level of mmu-miR-485-5p.
Subsequently, whether IL-6 mRNA is downstream of mmu-miR-485-5p was
investigated. miRNA mimics and inhibitor were transfected into
cells and termed the CGRP + mimics and CGRP + inhibitor groups,
respectively, for 24 h prior to CGRP stimulation. Transfection with
mimics resulted in the enforced expression of mmu-miR-485-5p
compared with the negative control, while the cells transfected
inhibitors had decreased miR-485-5p levels, as analyzed by RT-qPCR
(data not shown). As presented in Fig.
5C, IL-6 mRNA expression in the CGRP + mimics group was
significantly decreased compared with the CGRP treated group,
whereas, the IL-6 mRNA expression level was increased in the CGRP +
inhibitors group relative to that in the CGRP group indicating that
mmu-miR-485-5p attenuated the effect of CGRP on IL-6 induction.
IL-6 proteins were also evaluated (Fig. 5D). The results of western blotting
were in accordance with that of RT-qPCR. Taken together, these
results suggest that mmu_circRNA_007893 regulates IL-6 mRNA
expression by mediating the level of mmu-miR-485-5p.
Discussion
Previous studies on CGRP and macrophages have
focused on the potential pathways and kinases involved; however,
the importance of circRNAs and their functions remain largely
unclear. In the current study, mmu_circRNA_007893 was identified to
be was involved in IL-6 induction mediated by CGRP. Furthermore,
the data indicated that mmu-miR-485-5p, regulated by
mmu_circRNA_007893, is a negative regulator of IL-6 expression.
Overexpression of the miRNA attenuates the effect of CGRP on IL-6
expression. Therefore, the results of the present study expand the
previous understanding of the role of CGRP in macrophages and
provide novel insights into the endogenous mechanism of cytokine
induction.
It is reported that CGRP has an important role in
inflammatory cytokine secretion mediated by s in macrophage
(9,10). The results of the study suggest
that CGRP can stimulate RAW264.7 macrophages and induce IL-6 mRNA
expression. Furthermore, CGRP acts in dose- and time-dependent
manner. When the concentration of CGRP was 1 nM, the expression of
IL-6 mRNA peaked, and at higher or lower CGRP concentrations, IL-6
mRNA expression declined gradually. Similarly, IL-6 expression
reached the highest level after CGRP stimulation for 2 h, and
extending or reducing the stimulation duration resulted in reduced
expression of IL-6 compared with the level at 2 h. The results are
similar to the previous finding (7). The previous study reported that CGRP
can also increase the expression of IL-6 mRNA in bone
marrow-derived macrophages. However, the optimum concentration and
stimulation time as not entirely consistent with the results of the
present study, which may be attributes to the difference in the
cell type used (7).
Recent studies have demonstrated that circRNA
microarrays are a reliable and convenient method to research
circRNAs and their target miRNAs and genes (25–27).
Therefore, a circRNA microarray to identify the related circRNAs in
the current study. Initially, the circRNAs were sorted and
classified using the fold changes. Because certain studies have
reported that circRNA can act as miRNA sponges to suppress the
miRNA expression and thus, regulated the expression of target mRNAs
(16,20). Hence, it was assumed that the
circRNAs that harbored more predicted miRNA binding sites had more
important roles in altering the expression of their downstream
miRNA (23). Subsequently, the
circRNAs were sorted by the maximum number of predicted miRNA
binding sites. Furthermore, a circRNA-miRNA interaction network was
constructed. The miRNAs were selected according to their mirSVR
scores. The network provides a pivotal reference for further
investigation of the interaction of other differentially expressed
circRNAs and their potential targets.
In the current study, RT-PCR, nucleic acid
electrophoresis and PCR product sequencing were used to further
confirm the selected circRNAs. However, the data demonstrated that
not all the circRNA expression levels were in accordance with the
microarray analysis, and certain circRNAs were changed in the
opposite direction. Similarly, this phenomenon was also reported by
another study (28). This may be
due to sensitive detection method of circRNA microarrays, as even
minor variation or error can cause a marked difference. Thus, the
results obtained by the circRNA microarray can only be a reference
method and should be confirmed by further experiments.
The change of mmu_circRNA_007893 detected by RT-PCR
was consistent with that of the circRNA microarray. Furthermore,
interference with the expression of mmu_circRNA_007893 confirmed
the association between mmu_circRNA_007893 and IL-6 mRNA. Notably,
when the mmu_circRNA_007893 expression level was knocked down using
siRNA, the expression of IL-6 mRNA decreased. The result suggested
that CGRP regulates IL-6 mRNA expression by affecting the level of
mmu_circRNA_007893. However, other circRNAs identified in the
microarray may also be involved in the IL-6 mRNA increase and
should be examined further.
A previous study reported that circRNA ciRS-7 acts
as miR-7 sponge to suppress miR-7 activity, leading to increased
levels of miR-7 targets (20).
Several studies indicate that miR-485-5p act as a negative
regulator in gastric cancer (29,30).
It is also reported that miR-485-5p regulates T-cell acute
lymphocytic leukemia protein 1 expression in T-cell acute
lymphoblastic leukemia (31).
These findings demonstrated that other molecules may be mediated by
miR-485-5p and need to be identified. Although the present study
demonstrated that mmu-miR-485-5p can be regulated by
mmu_circRNA_007893, the results do not exclude the involvement of
other circRNAs or molecules that can regulate mmu-miR-485-5p
directly or indirectly.
Overall, the results demonstrated that CGRP induces
IL-6 mRNA expression, which is mediated by mmu_circRNA_007893, and
that mmu_circRNA_007893 functions as an endogenous miRNA sponge of
mmu-miR-485-5p. Various questions should be resolved in further
study. The direct association between mmu_circRNA_007893 and
mmu-miR-485-5p should be confirmed. Furthermore, the effect of CGRP
should be tested and detected in vivo.
Acknowledgements
This study was supported by Science & Technology
Bureau of Guangdong Province (grant no. 2013B021800278) and
Research Foundation of Science & Technology Bureau of Liwan
District of Guangzhou City (grant no. 2014126055).
References
|
1
|
Amara SG, Jonas V, Rosenfeld MG, Ong ES
and Evans RM: Alternative RNA processing in calcitonin gene
expression generates mRNAs encoding different polypeptide products.
Nature. 298:240–244. 1982. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Li Q and Peng J: Sensory nerves and
pancreatitis. Gland Surg. 3:284–292. 2014.PubMed/NCBI
|
|
3
|
Russell FA, King R, Smillie SJ, Kodji X
and Brain SD: Calcitonin gene-related peptide: Physiology and
pathophysiology. Physiol Rev. 94:1099–1142. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fernandes ES, Schmidhuber SM and Brain SD:
Sensory-nerve-derived neuropeptides: Possible therapeutic targets.
Handb Exp Pharmacol. 1–416. 2009.
|
|
5
|
Gasperisic R, Kovacic U, Cör A and
Skaleric U: Unilateral ligature-induced periodontitis influences
the expression of neuropeptides in the ipsilateral and
contralateral trigeminal ganglion in rats. Arch Oral Bio.
53:659–665. 2008. View Article : Google Scholar
|
|
6
|
Baliu-Piqué M, Jusek G and Holzmann B:
Neuroimmunological communication via CGRP promotes the development
of a regulatory phenotype in TLR4-stimulated macrophages. Eur J
Immunol. 44:3708–3716. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fernandez S, Knopf MA, Bjork SK and
McGillis JP: Bone marrow-derived macrophages express functional
CGRP receptors and respond to CGRP by increasing transcription of
c-fos and IL-6 mRNA. Cell Immunol. 209:140–148. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang W, Xv M, Yang WC, Wang N, Zhang XZ
and Li WZ: Exogenous α-calcitonin gene-related peptide attenuates
lipopolysaccharide-induced acute lung injury in rats. Mol Med Rep.
12:2181–2188. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu J, Chen M and Wang X: Calcitonin
gene-related peptide inhibits lipopolysaccharide-induced
interleukin-12 release from mouse peritoneal macrophages, mediated
by the cAMP pathway. Immunology. 101:61–67. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kurashige C, Hosono K, Matsuda H,
Tsujikawa K, Okamoto H and Majima M: Roles of receptor
activity-modifying protein 1 in angiogenesis and lymphangiogenesis
during skin wound healing in mice. FASEB J. 28:1237–1247. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma W, Dumont Y, Vercauteren F and Quirion
R: Lipopolysaccharide induces calcitonin gene-related peptide in
the RAW264.7 macrophage cell line. Immunology. 130:399–409. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Holzmann B: Antiinflammatory activities of
CGRP modulating innate immune responses in health and disease. Curr
Protein Pept Sci. 14:268–274. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cocquerelle C, Mascrez B, Hétuin D and
Bailleul B: Mis-splicing yields circular RNA molecules. FASEB J.
7:155–160. 1993.PubMed/NCBI
|
|
14
|
Surono A, Takeshima Y, Wibawa T, Ikezawa
M, Nonaka I and Matsuo M: Circular dystrophin RNAs consisting of
exons that were skipped by alternative splicing. Hum Mol Genet.
8:493–500. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Meczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Salzman J, Gawad C, Wang PL, Lacayo N and
Brown PO: Circular RNAs are the predominant transcript isoform from
hundreds of human genes in diverse cell types. PLoS One.
7:e307332012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ebbesen KK, Kjems J and Hansen TB:
Circular RNAs: Identification, biogenesis and function. Biochim
Biophys Acta. 1859:163–168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Thomas LF and Saetrom P: Circular RNAs are
depleted of polymorphisms at microRNA binding site. Bioinformatics.
30:2243–2246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Westholm JO, Miura P, Olson S, Shenker S,
Joseph B, Sanfilippo P, Celniker SE, Graveley BR and Lai EC:
Genome-wide analysis of drosophila circular RNAs reveals their
structural and sequence properties and age-dependent neural
accumulation. Cell Rep. 9:1966–1980. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J
and Ao Y: Circular RNA related to the chondrocyte ECM regulates
MMP13 expression by functioning as a MiR-136 ‘Sponge’ in human
cartilage degradation. Sci Rep. 6:225722016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen
MT and Ma QJ: Circular RNA expression alterations are involved in
OGD/R-induced neuron injury. Biochem Biophys Res Commun. 471:52–56.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dou C, Cao Z, Yang B, Ding N, Hou T, Luo
F, Kang F, Li J, Yang X, Jiang H, et al: Changing expression
profiles of lncRNAs, mRNAs, circRNAs and miRNAs during
osteoclastogenesis. Sci Rep. 6:214992016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Qu S, Song W, Yang X, Wang J, Zhang R,
Zhang Z, Zhang H and Li H: Microarray expression profile of
circular RNAs in human pancreatic ductal adenocarcinoma. Genom
Data. 5:385–387. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang YH, Yu XH, Luo SS and Han H:
Comprehensive circular RNA profiling reveals that circular
RNA100783 is involved in chronic CD28-associated CD8(+)T cell
aging. Immun Aging. 12:172015. View Article : Google Scholar
|
|
29
|
Jing LL and Mo XM: Reduced miR-485-5p
expression predicts poor prognosis in patients with gastric cancer.
Eur Rev Med Pharmacol Sci. 20:1516–1520. 2016.PubMed/NCBI
|
|
30
|
Kang M, Ren MP, Zhao L, Li CP and Deng MM:
miR-485-5p acts as a negative regulator in gastric cancer
progression by targeting flotillin-1. Am J Transl Res. 7:2212–2222.
2015.PubMed/NCBI
|
|
31
|
Correia NC, Melão A, Póvoa V, Sarmento L,
Gómez de Cedrón M, Malumbres M, Enguita FJ and Barata JT: microRNAs
regulate TAL1 expression in T-cell acute lymphoblastic leukemia.
Oncotarget. 7:8268–8281. 2016. View Article : Google Scholar : PubMed/NCBI
|