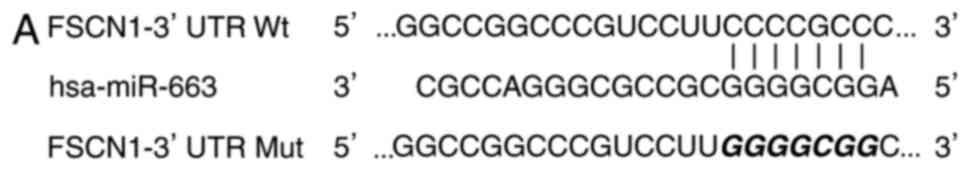Introduction
Colorectal cancer (CRC) originates from the
epithelial cells of the colon or rectum, and is the most frequently
diagnosed malignancy of the gastrointestinal tract (1,2).
Numerous risk factors involved in CRC initiation and progression
have been identified, including older age, hereditary components,
obesity, excess alcohol and red meat consumption, smoking and a
lack of physical exercise (3–6).
Despite rapid development in the variety of treatment methods and
approaches that have been used for patients with CRC, including
surgical resection, radiotherapy and chemotherapy, the overall
survival of patients with CRC has not notably changed (7). A total of ~30–50% of patients with
CRC develop local tumor recurrence or distant metastasis following
surgical resection (8,9). Therefore, it is important to
elucidate the mechanisms underlying the initiation and progression
of CRC, and to investigate novel therapeutic strategies for
patients with CRC.
MicroRNAs (miRNAs/miRs) are an abundant group of
endogenous, non-coding and evolutionarily-conserved RNAs consisting
of 17 to 23 nucleotides in length (10). miRNAs posttranscriptionally
regulate gene expression by directly binding to the complementary
sequences in the 3′untranslated regions (3′UTRs) of their target
genes and inducing gene degradation and/or mRNA translation
inhibition (11,12). Previous studies have demonstrated
that miRNAs are involved in a number of cancer-associated
biological processes, including cell proliferation, apoptosis, the
cell cycle, invasion, migration and metastasis (13–15).
Notably, miRNA dysregulation has been reported in the majority of
types of human cancer, including bladder cancer (16), prostate cancer (17), glioma (18), gastric cancer (19) and osteosarcoma (20). Previous studies have revealed that
abnormally expressed miRNAs may be correlated with tumorigenesis
and tumor development (21,22).
Therefore, miRNAs may be developed as therapeutic targets for novel
treatment strategies against CRC.
In the present study, the miR-663 expression level
and its association with clinicopathological factors in CRC was
investigated. In addition, the biological roles and underlying
mechanisms of miR-663 in the carcinogenesis and progression of CRC
were evaluated.
Materials and methods
Tissue specimens and cell lines
CRC tissues (n=48) and corresponding adjacent normal
tissues were collected from the Department of Surgical Oncology,
The Second Affiliated Hospital, Zhejiang University School of
Medicine (Hangzhou, China) between August 2012 and May 2014. No
patients were treated with systemic or local treatments prior to
surgical resection. All tissue samples were frozen in liquid
nitrogen immediately and stored at −80°C. The present study was
approved by the Ethical Committee of The Second Affiliated
Hospital, Zhejiang University School of Medicine. Informed consent
was obtained from all patients.
The 293T cell line, a human normal colon epithelial
cell line (FHC), and human CRC cell lines (SW620, SW480, LoVo,
HCT116, HT29) were purchased from the American Type Culture
Collection (Manassas, VA, USA). Cells were cultured in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) with 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.). All cells were maintained at 37°C in a
humidified environment with 5% CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
Total RNA from tissues and cells was extracted using
TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The
concentration of total RNA was determined using a NanoDrop ND-1000
Spectrophotometer (Thermo Fisher Scientific, Inc., Wilmington, DE,
USA). Total RNA was used to synthesize cDNA using a PrimeScript RT
kit (Takara Bio, Inc., Otsu, Japan). A SYBR PrimeScript miRNA
RT-qPCR kit (Takara Bio, Inc.) was used to analyze miR-663
expression, with U6 as an internal control. The thermocycling
conditions were as follows: 42°C for 5 min; 95°C for 10 sec,
followed by 40 cycles of 95°C for 5 sec, 55°C for 30 sec and 72°C
for 30 sec. Relative levels of fascin (FSCN1) mRNA were examined
using SYBR Green PCR Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.), with GADPH as an internal control. The
thermocycling conditions were as follows: 95°C for 10 min; followed
by 40 cycles of 95°C for 15 sec and 60°C for 1 min. The primers
were designed as follows: miR-663 forward,
5′-TGCGGAGGCGGGGCGCCGCGGG-3′ and reverse,
5′-CCAGTGCAGGGTCCGAGGT-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; FSCN1 forward,
5′-CTGGCTACACGCTGGAGTTC-3′ and reverse 5′-CTGAGTCCCCTGCTGTCTCCT−3′;
and GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse
5′-AGCCTTCTCCATGGTGGTGAAGAC−3′. The relative expression was
analyzed using the 2−ΔΔCq method (23).
miRNA mimics and small interfering
(si)RNA transfection
Oligonucleotides of human miR-663 mimics and miRNA
negative control (miR-NC) were synthesized by Shanghai GenePharma
Co., Ltd. (Shanghai, China). The miR-663 mimics sequence was
5′-AGGCGGGGCGCCGCGGGACCGC-3′ and the miR-NC sequence was
5′-UUCUCCGAACGUGUCACGUTT−3′. FSCN1 siRNA and negative control siRNA
(NC siRNA) were obtained from Guangzhou RiboBio Co., Ltd.
(Guangzhou, China). The FSCN1 siRNA sequence was
5′-AGCCCTGGGCGTGTAGTGTAA-3′ and the NC siRNA sequence was
5′-UUCUCCGAACGUGUCACGUTT-3′. Cells were transfected with miRNA
mimics (100 pmol) or siRNA (100 pmol) using Lipofectamine 2000
(Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. Following transfection for 48 h, RT-qPCR was used to
examine the transfection efficiency, according to the same protocol
described above.
Cell Counting Kit-8 (CCK8) assay
Cell proliferation was assessed using a CCK8 assay
(Dojindo Molecular Technologies, Inc., Kumamoto, Japan). The
transfected cells were collected and seeded in 96-well plates in
triplicate, at a density of 3,000 cells/well. Cells were incubated
at 37°C in a humidified environment with 5% CO2. The
CCK8 assay was performed at 24, 48, 72 and 96 h. A total of 10 µl
CCK8 solution was added to each well and, following 4 h of
incubation at 37°C, the absorbance at 450 nm was determined using a
microplate reader (Bio-Rad 550; Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Transwell invasion assay
Cell invasion was assessed using 24-well, 8-mm pore
size BD Matrigel invasion chambers (BD Biosciences, Franklin Lakes,
NJ, USA), according to the manufacturer's protocol. Transfected
cells were collected and re-suspended in FBS-free culture medium.
Subsequently, 1×105 cells were plated in the upper
chamber and the lower chamber was filled with culture medium
containing 20% FBS. Following incubation for 48 h, non-invaded
cells were removed using cotton swabs. Invaded cells were fixed
with 95% methanol at room temperature for 10 min and stained with
0.5% crystal violet at room temperature for 10 min. Cells in five
random fields were photographed and counted under an inverted
microscope (magnification, ×200; X71; Olympus Corporation, Tokyo,
Japan), and data are expressed as the average number of invaded
cells/field of view.
Target gene prediction
TargetScan (www.targetscan.org/vert_60) and PicTar (pictar.mdc-berlin.de) were used to predict potential
target genes of miR-663.
Luciferase reporter assay
The wild-type (Wt) and mutant (Mut) 3′UTR of FSCN1
was synthesized and subcloned into the pMIR-reporter (GenePharma,
Shanghai, China). 293T cells were seeded in 24-well plates in
triplicate at a density of 1.5×105 cells/well. Following
incubation overnight, cells were co-transfected with
pMIR-FSCN1-3′UTR Wt or pMIR-FSCN1-3′UTR Mut with miR-663 mimics or
miR-NC, using Lipofectamine 2000, according to the manufacturer's
protocol. Cells were harvested at 48 h post-transfection and
subjected to a Dual-Luciferase Reporter Assay System (Promega
Corporation, Madison, WI, USA). The results were normalized by
comparing with Renilla luciferase activity.
Western blot analysis
Total protein was extracted from cells using
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.) supplemented with phenylmethanesulfonyl fluoride and a
cocktail of protease inhibitors (Beyotime Institute of
Biotechnology, Haimen, China). The total protein concentration was
detected using the bicinchoninic acid method (Beyotime Institute of
Biotechnology). Equal quantities of protein (30 µg) were separated
using SDS-PAGE on a 10% gel, transferred to a polyvinylidene
fluoride membrane (EMD Millipore, Billerica, MA, USA) and blocked
with TBS containing 0.1% Tween-20 (TBST) and 5% non-fat dried milk
at room temperature for 2 h. Subsequently, the membranes were
incubated with mouse anti-human monoclonal FSCN1 antibody (1:1,000
dilution; cat. no. sc-21743; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) and mouse anti-human monoclonal GADPH antibody
(1:1,000 dilution; cat. no. sc-137179; Santa Cruz Biotechnology,
Inc.), at 4°C overnight.
Following washing three times with TBST every 10
min, membranes were further probed with goat anti-mouse horseradish
peroxidase-conjugated secondary antibody (1:2,000 dilution; cat.
no. sc-2005; Santa Cruz Biotechnology, Inc.) at room temperature
for 1 h and washed with TBST three times for 10 min. The protein
expression level was measured using Enhanced Chemiluminescence
Prime Western Blotting Detection Reagent (GE Healthcare Life
Sciences, Little Chalfont, UK). GAPDH was used as a reference.
Statistical analysis
All the data are expressed as mean ± standard
deviation. Data were analyzed using Student's t-tests or one-way
analysis of variance (ANOVA) using SPSS 17 software (SPSS Inc.,
Chicago, IL, USA). A Student-Newman-Keuls test was used as a post
hoc test following the ANOVA. P<0.05 was considered to indicate
a statistically significant difference.
Results
Down-regulation of miR-663 correlates
with clinicopathological features of human CRC
In order to elucidate miR-663 expression in CRC, its
expression was measured in CRC tissues and corresponding adjacent
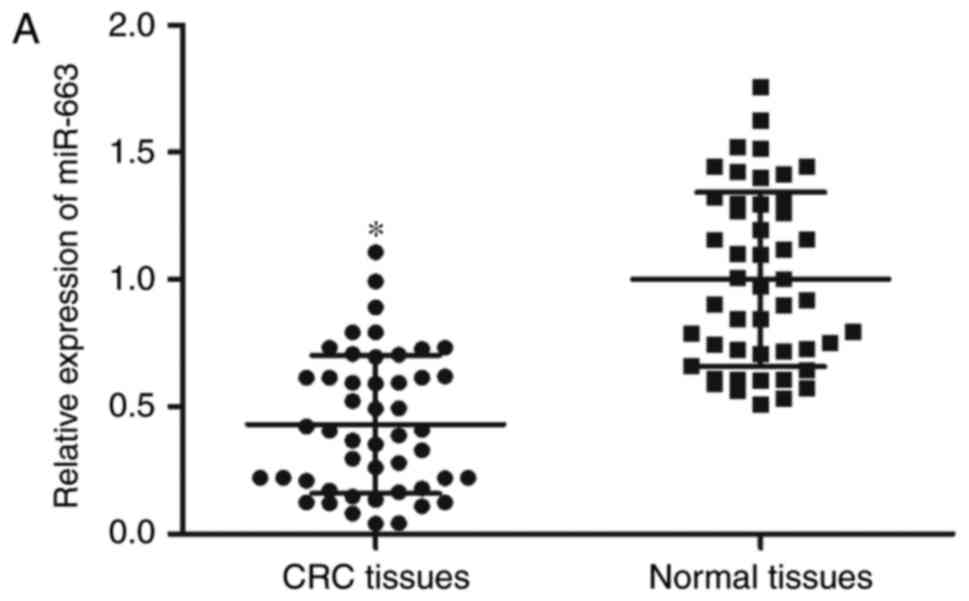
normal tissues using RT-qPCR analysis. As presented in Fig. 1A, miR-663 expression levels were
decreased in CRC tissues compared with adjacent normal tissues
(P<0.05). miR-663 expression was additionally determined in CRC
cell lines and a human normal colon epithelium cell line (FHC). As
presented in Fig. 1B, miR-663
expression in CRC cell lines was decreased compared with that in
the FHC cell line (P<0.05).
Subsequently, the association between miR-663
expression and clinicopathological features in patients with CRC
was analyzed. As presented in Table
I, decreased miR-663 expression in CRC tissues was
significantly correlated with tumor, node, metastasis (TNM) stage
(P=0.005) and lymph node metastasis (P=0.027), while there was no
correlation with gender, age and tumor size. The results of the
present study suggested that miR-663 may serve important roles in
CRC.
 | Table I.Correlation between miR-663
expression and clinicopathological features in colorectal
cancer. |
Table I.
Correlation between miR-663
expression and clinicopathological features in colorectal
cancer.
|
|
| miR-663
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | No. cases | Low | High | P-value |
|---|
| Sex |
|
|
| 0.074 |
|
Male | 31 | 21 | 10 |
|
|
Female | 17 | 7 | 10 |
|
| Age, years |
|
|
| 0.836 |
|
<60 | 16 | 9 | 7 |
|
|
≥60 | 32 | 19 | 13 |
|
| TNM stage |
|
|
| 0.005 |
|
I–II | 27 | 11 | 16 |
|
|
III–IV | 21 | 17 | 4 |
|
| Tumor size, cm |
|
|
| 0.762 |
|
<5 | 18 | 10 | 8 |
|
| ≥5 | 30 | 18 | 12 |
|
| Lymph node
metastasis |
|
|
| 0.027 |
| No | 27 | 12 | 15 |
|
|
Yes | 21 | 16 | 5 |
|
miR-663 suppresses cell proliferation
and invasion in CRC
In order to evaluate the functions of miR-663 in
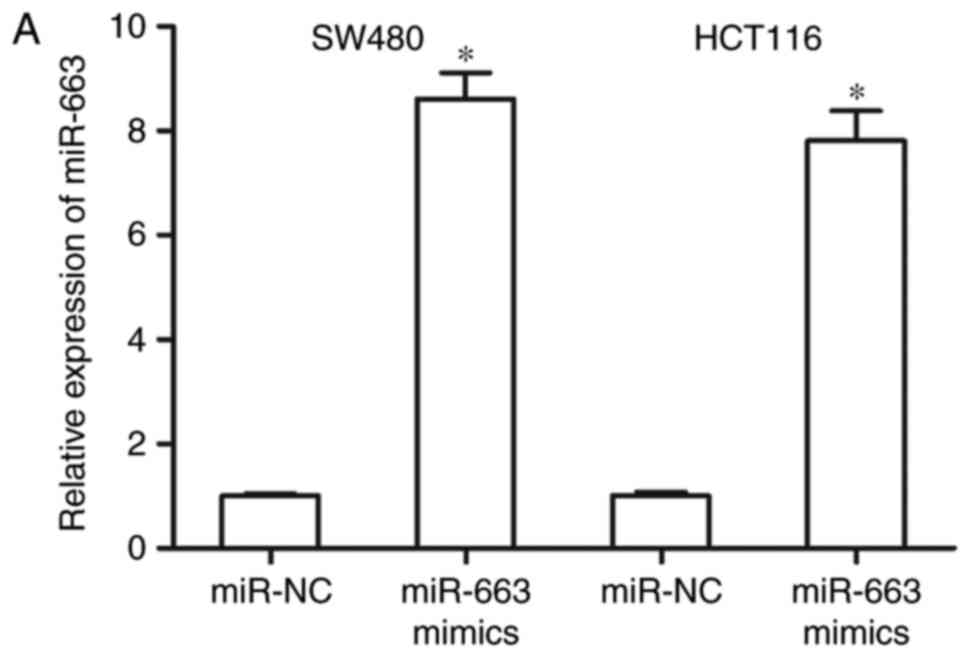
CRC, SW480 and HCT116 cells were transfected with miR-663 mimics or
miR-NC. Following transfection for 48 h, RT-qPCR analysis confirmed
that miR-663 expression was significantly increased in SW480 and
HCT116 cells transfected with miR-663 mimics (Fig. 2A; P<0.05).
CCK8 and Transwell invasion assay were used to
assess the effects of miR-663 overexpression on the cell
proliferation and invasiveness of CRC, respectively. As presented
in Fig. 2B, the proliferation of
SW480 and HCT116 cells was significantly decreased by miR-663
overexpression (P<0.05). The results of the Transwell invasion
assay demonstrated that invasive capacity was significantly limited
in SW480 and HCT116 cells transfected with miR-663 mimics compared
with cells transfected with miR-NC (Fig. 2C; P<0.05).
FSCN1 is a direct target of
miR-663
In order to examine the potential molecular
mechanisms underlying the role of miR-663 in the regulation of cell
proliferation and invasion in CRC, TargetScan and PicTar were used
to predicate its potential target genes. As presented in Fig. 3A, there are seven conserved binding
sites for miR-663 in the 3′UTR region of FSCN1.
A luciferase reporter assay was subsequently
performed in 293T cells co-transfected with miR-663 mimics or
miR-NC, and pMIR-FSCN1-3′UTR Wt or pMIR-FSCN1-3′UTR Mut. As
presented in Fig. 3B, upregulation
of miR-663 decreased the luciferase activity of pMIR-FSCN1-3′UTR Wt
(P<0.05), although not pMIR-FSCN1-3′UTR Mut, indicating that
miR-663 specifically targeted the 3′UTR of FSCN1.
FSCN1 is upregulated in CRC tissues
and inversely correlates with miR-663 levels in CRC tissues
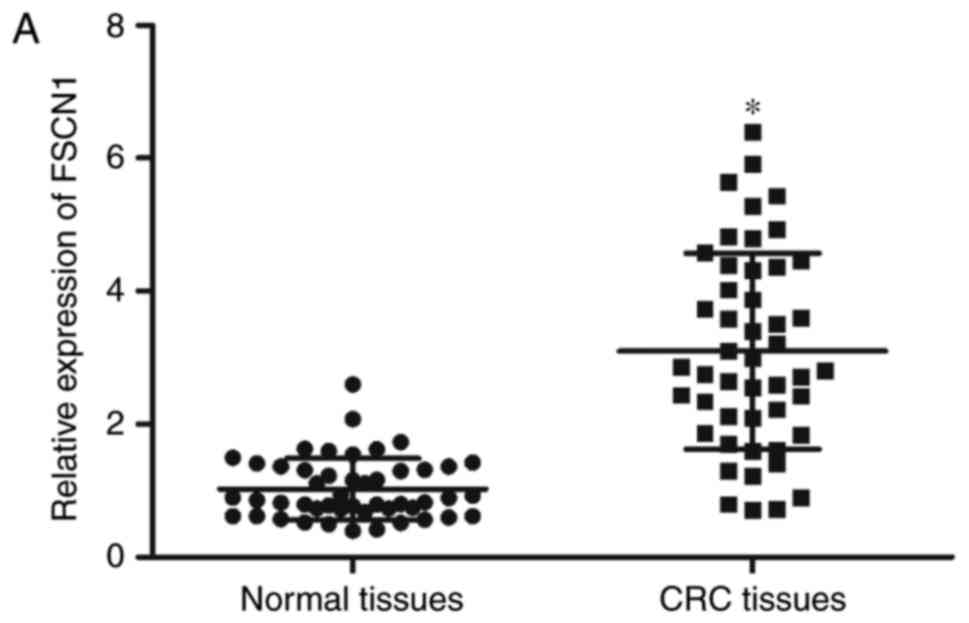
In order to further confirm that FSCN1 is a direct
target of miR-663, the FSCN1 expression levels were detected in CRC
tissues and corresponding adjacent normal tissues. The results
demonstrated that FSCN1 was expressed at high levels in CRC tissues
compared with adjacent normal tissues (Fig. 4A; P<0.05) and was negatively
correlated with miR-663 expression levels in CRC tissues (Fig. 4B; r=-0.5693; P<0.001).
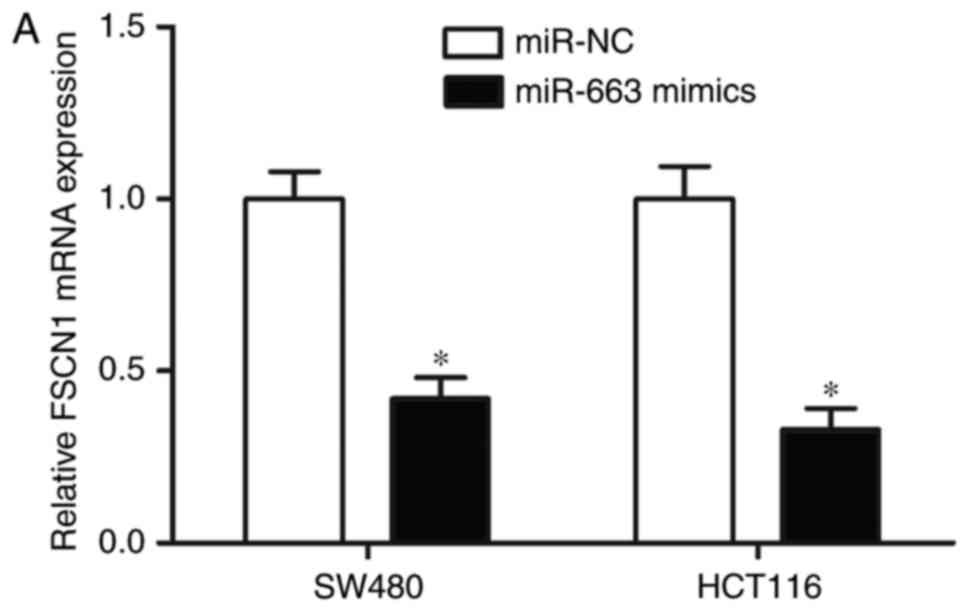
miR-663 negatively regulates FSCN1
expression in CRC cells
RT-qPCR and western blot analyses were used to
examine the alterations in endogenous FSCN1 mRNA and protein
expression in SW480 and HCT116 cells, following transfection with
miR-663 mimics or miR-NC. As presented in Fig. 5A and B, compared with miR-NC,
restoration of the expression of miR-663 reduced FSCN1 expression
in SW480 and HCT116 cells at the mRNA and protein levels
(P<0.05). The results of the present study suggested that
miR-663 negatively regulated FSCN1 expression in CRC cells by
directly targeting the 3′UTR of FSCN1.
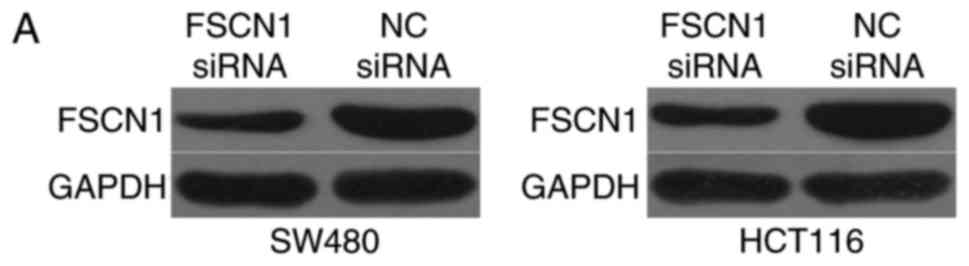
FSCN1 underexpression simulates the
tumor suppressor function of miR-663 mimics in CRC cells
As FSCN1 is a direct target of miR-663 in CRC, the
biological role of FSCN1 in the cell proliferation and invasiveness
of CRC was investigated. FSCN1 siRNA was transfected into SW480 and
HCT116 cells to decrease its expression (Fig. 6A). Subsequently, CCK8 and Transwell
invasion assays were performed. As presented in Fig. 6B and C, FSCN1 underexpression
suppressed the proliferation (P<0.05) and invasion (P<0.05)
of SW480 and HCT116 cells, which was consistent with the effects of
miR-663 mimics. The results of the present study suggested that the
tumor-suppressive role of miR-663 is mediated via downregulation of
FSCN1 in CRC.
Discussion
miR-663 has been observed to be aberrantly expressed
in numerous types of human cancer. For example, in glioblastoma,
miR-663 expression was inhibited in tumor tissues and cell lines
(24) and it was demonstrated to
be a poor prognostic marker in patients with glioblastoma (25). In pancreatic cancer, the expression
level of miR-663 was decreased and was significantly correlated
with TNM stage and the lymph node metastasis status of patients
(26). Pan et al (27) demonstrated that miR-663 expression
was decreased in gastric cancer cell lines compared with normal
cells. In papillary thyroid carcinoma, miR-663 was downregulated in
tumor tissues and cell lines; in addition, there was statistically
significant differences in the expression level of miR-663 with
regard to age and tumor size (28). However, in castration-resistant
prostate cancer, miR-663 was demonstrated to be overexpressed;
increased miR-663 expression was associated with Gleason score and
TNM stage, and was an independent prognostic predictor of clinical
recurrence (29). Previous studies
additionally demonstrated that miR-663 was increased in
nasopharyngeal carcinoma tissues (30), lung cancer (31), breast cancer (32) and hepatocellular carcinoma
(33).
miR-663 may act as either a tumor suppressor or
promoter in human malignancies. Li et al (24) reported that enforced miR-663
expression attenuated cell proliferation, migration and invasion of
glioblastoma through downregulation of transforming growth factor
(TGF)-β1. Shi et al (25,34)
demonstrated that ectopic miR-663 expression decreased the
proliferative and invasive capacities of glioblastoma cells by
targeting C-X-C chemokine receptor type 4 and phosphatidylinositol
4,5-bisphosphate 3-kinase catalytic subunit delta isoform. In
vitro and in vivo experiments demonstrated that
restoration of the expression of miR-663 inhibited cell
proliferation and invasion in pancreatic cancer via inhibition of
elongation factor 1-α2 (26). A
study by Wang et al (28)
demonstrated that miR-663 overexpression decreased papillary
thyroid carcinoma cell migration and invasion by directly targeting
TGF-β1. miR-663 was additionally identified to be an oncogene in a
number of types of human cancer. For example, in nasopharyngeal
carcinoma, downregulation of miR-663 suppressed cell proliferation
in vitro and tumor growth in vivo through negative
regulation of cyclin-dependent kinase inhibitor 1 (30). In hepatocellular carcinoma, miR-663
underexpression impaired cell proliferation and promoted apoptosis
under endoplasmic reticulum stress by targeting TGF-β1 (33). These conflicting findings indicated
that the expression and functions of miR-663 in tumors are diverse
and tissue-specific.
The present study used multi-dimensional approaches
to demonstrate that FSCN1 is a direct downstream target of miR-663.
Bioinformatics analysis indicated that FSCN1 was a potential target
gene of miR-663. Through the luciferase reporter assay, it was
observed that the 3′UTR of FSCN1 was directly targeted by miR-663.
It was additionally demonstrated that FSCN1 was significantly
upregulated in clinical CRC tissues and was inversely correlated
with the miR-663 expression level. Ectopic miR-663 expression
decreased endogenous FSCN1 expression at the mRNA and protein level
in CRC cells. Additionally, siRNA was used to specifically knock
down FSCN1 in CRC cells, demonstrated that it was able to simulate
the tumor suppressor functions induced by miR-663 overexpression in
the cell proliferation and invasion of CRC. The results of the
present study demonstrated that miR-663 may act as a tumor
suppressor in CRC by directly targeting FSCN1.
FSCN1, a 55-kDa globular protein, is an
actin-bundling protein and is well-established as an integral
component of invadopodia, which stabilize actin bundles in invasive
foot structures (35). A number of
previous studies have reported that FSCN1 is overexpressed in human
cancer, including prostate cancer (36), lung cancer (37), breast cancer (38), gastric carcinoma (39), esophageal cancer (40) and pancreatic cancer (41). Increased FSCN1 expression was
correlated with aggressive clinical course, poor prognosis and
shorter survival for patients with these types of cancer. FSCN1 has
been observed to be upregulated in human CRC. An increased
expression level of FSCN1 was significantly associated with reduced
overall survival and reduced disease-free survival; patients with
CRC exhibiting increased FSCN1 levels had worse overall survival
and disease-free survival compared with patients with low FSCN1
(42). FSCN1 expression in CRC may
be clinically useful for predicting metastasis and poor survival
(43). With the emerging
correlation of FSCN1 with aggressive CRC progression, FSCN1 may be
developed as a therapeutic target for patients with this disease.
The results of the present study demonstrated that miR-663/FSCN1
based reagents may be a novel therapeutic approach for CRC
patients.
In conclusion, the present study confirmed that
miR-663 acted as a tumor suppressor gene by inhibiting CRC cell
growth and invasion, via direct targeting of FSCN1. The results of
the present study provided novel evidence for the potential utility
of a miR-663/FSCN1-based targeted therapy in the treatment of
CRC.
References
|
1
|
East JE and Dekker E: Colorectal cancer
diagnosis in 2012: A new focus for CRC prevention-more serration,
less inflammation. Nat Rev Gastroenterol Hepatol. 10:69–70. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Andrews L: Dietary flavonoids for the
prevention of colorectal cancer. Clin J Oncol Nurs. 17:671–672.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Altobelli E, Lattanzi A, Paduano R,
Varassi G and di Orio F: Colorectal cancer prevention in Europe:
Burden of disease and status of screening programs. Prev Med.
62:132–141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sugarbaker PH: Colorectal cancer:
Prevention and management of metastatic disease. Biomed Res Int.
2014:7828902014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chan DS, Lau R, Aune D, Vieira R,
Greenwood DC, Kampman E and Norat T: Red and processed meat and
colorectal cancer incidence: Meta-analysis of prospective studies.
PLoS One. 6:e204562011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wei ZJ, Tao ML, Zhang W, Han GD, Zhu ZC,
Miao ZG, Li JY and Qiao ZB: Up-regulation of microRNA-302a
inhibited the proliferation and invasion of colorectal cancer cells
by regulation of the MAPK and PI3K/Akt signaling pathways. Int J
Clin Exp Pathol. 8:4481–4491. 2015.PubMed/NCBI
|
|
8
|
Amano R, Yamada N, Nakata B, Kimura K,
Yashiro M, Ohira M and Hirakawa K: Prognostic indicator for the
resection of liver metastasis of colorectal cancer. Surg Today.
44:1287–1292. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lieberman DA, Rex DK, Winawer SJ,
Giardiello FM, Johnson DA and Levin TR: Guidelines for colonoscopy
surveillance after screening and polypectomy: A consensus update by
the US multi-society task force on colorectal cancer.
Gastroenterology. 143:844–857. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lai EC: Micro RNAs are complementary to 3′
UTR sequence motifs that mediate negative post-transcriptional
regulation. Nat Genet. 30:363–364. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ostenfeld MS, Bramsen JB, Lamy P,
Villadsen SB, Fristrup N, Sørensen KD, Ulhøi B, Borre M, Kjems J,
Dyrskjøt L and Orntoft TF: miR-145 induces caspase-dependent and
-independent cell death in urothelial cancer cell lines with
targeting of an expression signature present in Ta bladder tumors.
Oncogene. 29:1073–1084. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lai VK, Ashraf M, Jiang S and Haider K:
MicroRNA-143 is a critical regulator of cell cycle activity in stem
cells with co-overexpression of Akt and angiopoietin-1 via
transcriptional regulation of Erk5/cyclin D1 signaling. Cell Cycle.
11:767–777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wu D, Zhou Y, Pan H, Zhou J, Fan Y and Qu
P: microRNA-99a inhibiting cell proliferation, migration and
invasion by targeting fibroblast growth factor receptor 3 in
bladder cancer. Oncol Lett. 7:1219–1224. 2014.PubMed/NCBI
|
|
16
|
Wu WB, Wang W, Du YH, Li H, Xia SJ and Liu
HT: MicroRNA-3713 regulates bladder cell invasion via MMP9. Sci
Rep. 6:323742016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Feng S, Zhu X, Fan B, Xie D, Li T and
Zhang X: miR-19a-3p targets PMEPA1 and induces prostate cancer cell
proliferation, migration and invasion. Mol Med Rep. 13:4030–4038.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu J, Cui H, Zhu Z and Wang L:
MicroRNA-200b-3p suppresses epithelial-mesenchymal transition and
inhibits tumor growth of glioma through down-regulation of ERK5.
Biochem Biophys Res Commun. 478:1158–1164. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang H, Xiong M, Hu Y, Sun Y and Ma Q:
MicroRNA-19b inhibits proliferation of gastric cancer cells by
targeting B-cell CLL/lymphoma 3. Oncol Rep. 36:2079–2086. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dong J, Liu Y, Liao W, Liu R, Shi P and
Wang L: miRNA-223 is a potential diagnostic and prognostic marker
for osteosarcoma. J Bone Oncol. 5:74–79. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:pp. 2257–2261.
2006; View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li Q, Cheng Q, Chen Z, Peng R, Chen R, Ma
Z, Wan X, Liu J, Meng M, Peng Z and Jiang B: MicroRNA-663 inhibits
the proliferation, migration and invasion of glioblastoma cells via
targeting TGF-β1. Oncol Rep. 35:1125–1134. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shi Y, Chen C, Zhang X, Liu Q, Xu JL,
Zhang HR, Yao XH, Jiang T, He ZC, Ren Y, et al: Primate-specific
miR-663 functions as a tumor suppressor by targeting PIK3CD and
predicts the prognosis of human glioblastoma. Clin Cancer Res.
20:1803–1813. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zang W, Wang Y, Wang T, Du Y, Chen X, Li M
and Zhao G: miR-663 attenuates tumor growth and invasiveness by
targeting eEF1A2 in pancreatic cancer. Mol Cancer. 14:372015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L,
Sun L, Liu J, Yang Z and Ran Y: Tumor-suppressive mir-663 gene
induces mitotic catastrophe growth arrest in human gastric cancer
cells. Oncol Rep. 24:105–112. 2010.PubMed/NCBI
|
|
28
|
Wang Z, Zhang H, Zhang P, Dong W and He L:
MicroRNA-663 suppresses cell invasion and migration by targeting
transforming growth factor beta 1 in papillary thyroid carcinoma.
Tumour Biol. 37:7633–7644. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Jiao L, Deng Z, Xu C, Yu Y, Li Y, Yang C,
Chen J, Liu Z, Huang G, Li LC and Sun Y: miR-663 induces
castration-resistant prostate cancer transformation and predicts
clinical recurrence. J Cell Physiol. 229:834–844. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yi C, Wang Q, Wang L, Huang Y, Li L, Liu
L, Zhou X, Xie G, Kang T, Wang H, et al: MiR-663, a microRNA
targeting p21(WAF1/CIP1), promotes the proliferation and
tumorigenesis of nasopharyngeal carcinoma. Oncogene. 31:4421–4433.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu ZY, Zhang GL, Wang MM, Xiong YN and
Cui HQ: MicroRNA-663 targets TGFB1 and regulates lung cancer
proliferation. Asian Pac J Cancer Prev. 12:2819–2823.
2011.PubMed/NCBI
|
|
32
|
Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, Yao
H, Song E, Chen Y, Wang M and Lin L: The overexpression of
hypomethylated miR-663 induces chemotherapy resistance in human
breast cancer cells by targeting heparin sulfate proteoglycan 2
(HSPG2). J Biol Chem. 288:10973–10985. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang Y, Liu J, Fan L, Wang F, Yu H, Wei W
and Sun G: miR-663 overexpression induced by endoplasmic reticulum
stress modulates hepatocellular carcinoma cell apoptosis via
transforming growth factor beta 1. Onco Targets Ther. 9:1623–1633.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Shi Y, Chen C, Yu SZ, Liu Q, Rao J, Zhang
HR, Xiao HL, Fu TW, Long H, He ZC, et al: miR-663 suppresses
oncogenic function of CXCR4 in glioblastoma. Clin Cancer Res.
21:4004–4013. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jayo A and Parsons M: Fascin: A key
regulator of cytoskeletal dynamics. Int J Biochem Cell Biol.
42:1614–1617. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Darnel AD, Behmoaram E, Vollmer RT, Corcos
J, Bijian K, Sircar K, Su J, Jiao J, Alaoui-Jamali MA and Bismar
TA: Fascin regulates prostate cancer cell invasion and is
associated with metastasis and biochemical failure in prostate
cancer. Clin Cancer Res. 15:1376–1383. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pelosi G, Pastorino U, Pasini F,
Maissoneuve P, Fraggetta F, Iannucci A, Sonzogni A, De Manzoni G,
Terzi A, Durante E, et al: Independent prognostic value of fascin
immunoreactivity in stage I nonsmall cell lung cancer. Br J Cancer.
88:537–547. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rodríguez-Pinilla SM, Sarrió D, Honrado E,
Hardisson D, Calero F, Benitez J and Palacios J: Prognostic
significance of basal-like phenotype and fascin expression in
node-negative invasive breast carcinomas. Clin Cancer Res.
12:1533–1539. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hashimoto Y, Shimada Y, Kawamura J,
Yamasaki S and Imamura M: The prognostic relevance of fascin
expression in human gastric carcinoma. Oncology. 67:262–270. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hashimoto Y, Ito T, Inoue H, Okumura T,
Tanaka E, Tsunoda S, Higashiyama M, Watanabe G, Imamura M and
Shimada Y: Prognostic significance of fascin overexpression in
human esophageal squamous cell carcinoma. Clin Cancer Res.
11:2597–2605. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Maitra A, Iacobuzio-Donahue C, Rahman A,
Sohn TA, Argani P, Meyer R, Yeo CJ, Cameron JL, Goggins M, Kern SE,
et al: Immunohistochemical validation of a novel epithelial and a
novel stromal marker of pancreatic ductal adenocarcinoma identified
by global expression microarrays: Sea urchin fascin homolog and
heat shock protein 47. Am J Clin Pathol. 118:52–59. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Alajez NM: Significance of BMI1 and FSCN1
expression in colorectal cancer. Saudi J Gastroenterol. 22:288–293.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Oh SY, Kim YB, Suh KW, Paek OJ and Moon
HY: Prognostic impact of fascin-1 expression is more significant in
advanced colorectal cancer. J Surg Res. 172:102–108. 2012.
View Article : Google Scholar : PubMed/NCBI
|