Introduction
Esophageal cancer (EC) is the most common
gastrointestinal tumor worldwide, with the highest incidence and
mortality rates observed in China, where most cases are esophageal
squamous cell carcinoma (ESCC) (1), and as the fourth cause of cancer
death, the 5-year survival rate remains at 15–40% after current
treatments (2–4). Indeed, the main causes of poor
prognosis for ESCC patients are local invasion and lymph node and
distant metastasis; thus, early detection is a key factor in
increasing survivability (3),
although the lack of effective and specificity markers still
plagues the diagnosis. Of note, since carcinogenesis is a
multi-step process, the diagnostic indices, such as epithelial
mesenchymal transition (EMT), invasion, metastases, and recurrence,
should dynamically change with the occurrence and development of
tumors (5).
For squamous neoplasms, such as ESCCs, EMT is a
crucial process, in which cancer cells lose polarity and reduce
adhesion, contributing to migration to and invasion of surrounding
issues (6). Therefore, discovering
the mechanism and key factors that control EMT in tumors would
provide strategies to solve the above dilemma. Recent data have
demonstrated that microRNAs (miRNAs) as non-coding small RNAs not
only exist widely in organisms, but also govern the expression of
different genes by binding to the 3′-untranslated region of target
genes and participating in a series of important processes,
including differentiation, proliferation, apoptosis and EMT
(7–9). However, study results are
conflicting, and microRNAs play roles as tumor-suppressors or
carcinogenic factors in various human malignancies (10). Studies have shown that miRNA-let-7
as a tumor suppressor, apparently has low expression in many tumor
types, such as head and neck, gastric, lung, ovarian and esophageal
cancers (11–16). Among the let-7 family, let-7a, have
been implicated in the inhibition of EMT in nasopharyngeal,
hepatocellular and rectal carcinoma, and breast cancer (17–20).
Specifically, as a highly conserved RNA-binding protein, Lin28,
which negatively regulates the maturation of let-7 by binding to
the terminal loop of its precursor, has also been positively
correlated with the cancer aggressiveness and poor prognosis of EC
patients (12,16,17).
Therefore, it is worth considering whether the changes of let-7a
have a potential indicative effect of EMT, even metastasis for
ESCC.
At this stage, the occurrence of EMT involves
multiple signal transduction pathways, such as Notch, TNF-α, TGF-β,
Wnt/β-catenin and others in tumors (19,21).
Given the many research theories in existence, there can be no
doubt that the Wnt signaling plays a critical role in deciding the
destiny of cancer cells. It has now been definitively established
that β-catenin migrates to the nucleus, interacts with
transcription factors TCF/LEF, and regulates the expression of
downstream target genes, such as cyclin D1, c-myc, etc., thereby
inducing a series of biological functions including self-renewal,
differentiation, proliferation and migration (22). However, controversial evidence
suggests that Wnt signaling consists of a series of oncogene and
tumor-suppressor genes. Nonetheless, the cascading effect of
downstream target miRNAs has attracted much attention in recent
years (23). Regarding ESCC, our
previous study identified that interleukin-23 (IL-23) promotes EMT
via the Wnt/β-catenin pathway (24). Additionally, recent research
revealed that the Wnt pathway could cooperate with Lin28, and the
abnormal activation of Wnt/β-catenin signaling plays a significant
role in the metastasis and recurrence of esophageal cancer
(16,19,25–27).
However, there is currently no evidence that let-7a acts as a
prognostic indicator via Wnt/Lin28 regulation in ESCC.
In this study, we aimed to explore the role of
Lin28/let-7a in regulation of migration, invasion and metastasis in
ESCC. We found that let-7a, out with other members, was negatively
correlated with higher TNM staging and recurrence of ESCC patients.
Moreover, we showed that Lin28 knock-down or let-7a mimic repressed
the invasion, EMT and metastasis. Furthermore, we identified an
important role of Wnt signaling in the maintaining Lin28 of ESCC
cells. We thus identify an important biochemical and functional
link between Lin28/let-7a with the Wnt pathway in ESCC
progression.
Materials and methods
Patients and tsissues
The Medical Ethics Committee of Jiangsu University
has approved this study. A total of 70 tumors and para-carcinoma
tissues were obtained from patients who had ESCC, had not undergone
chemotherapy or radiotherapy, had signed informed consent before
surgical resection, and had data that were collected from 2011 to
2013 at the Affiliated Hospital of Jiangsu University (Zhenjiang,
China). All tissue samples were confirmed by independent
pathologists, frozen in liquid nitrogen, and stored at −80°C.
Patients who developed any local recurrence or distant metastasis
diagnosed by computerized tomography scan within 3 years after
therapy are defined as recurrence (28).
Cell lines and RNA interference
The ESCC cell lines TE-1, ECA109 and KYSE-150, and
normal esophageal squamous epithelium cell line Het-1a were
purchased from the Cell Bank of the Chinese Academy of Sciences
(Shanghai, China). All cells were cultured in RPMI-1640 medium
containing 10% FBS, 100 IU/ml penicillin and streptomycin at 37°C,
5% CO2. All media were purchased from Gibco (Grand
Island, NY, USA). The following siRNA sequences were designed and
synthesized by GenePharma Company (GenePharma, Shanghai, China):
let-7a mimics 5′-UGAGGUAGUAGGUUGUAUAGUU-3′ (forward),
5′-CUAUACAACCUACUACCUCAUU-3′ (reverse); negative control (NC) for
mimics 5′-UUCUCCGAACGUGUCACGUTT-3′ (forward),
5′-ACGUGACACGUUCGGAGAATT-3′ (reverse); let-7a inhibitor
5′-AACUAUACAACCUACUACCUCA-3′; NC for inhibitor
5′-CAGUACUUUUGUGUAGUACAA-3′. Mimics (50 nM) or inhibitors (100 nM)
were transfected into ECA-109 cell lines with Lipofectamine 2000
reagents (Invitrogen, Shanghai, China) according to the
manufacturer's instructions. Untransfected groups were cultured
under normal conditions.
Molecular biology experiments
Total RNA was extracted from ESCC tissues and cells
with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to
the manufacturer's instructions. Relative concentrations of RNA
were quantified using the BioMate 3S Analyzer (Thermo Fisher
Scientific, Inc., San Jose, CA, USA), normalized to the expression
of U6 or GAPDH, and performed using a GeneAmp® PCR
systems 9700 (Bio-Rad, Hercules, CA, USA). The following sequences
of PCR primers (Invitrogen, Shanghai, China) were used: let-7a
(forward, 5′-GCCGCTGAGGTAGTAGGTTGTA-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′); let-7b (forward,
5′-TGAGGTAGTAGGTTGTGTGGTT-3′ and reverse,
5′-GCTGTCAACGATACGCTACCTA-3′); let-7c (forward,
5′-ACACTCCAGCTGGGTGAGGTAGTAGGTT-3′ and reverse,
5′-GGTGTCGTGGAGTCG-3′); U6 (primers: 5′-AACGCTTCACGAATTTGCGT-3′,
forward, 5′-CTCGCTTCGGCGCAGCACA-3′ and reverse,
5′-AACGCTTCACGAATTTGCGT-3′); Lin28 (forward,
5′-AAAGGAGACAGGTGCTAC-3′ and reverse, 5′-ATATGGCTGATGCTCTGG-3′);
Snail (forward, 5′-CTTCTCCTCTACTTCAGTCTCTTCC-3′ and reverse,
5′-TGAGGTATTCCTTGTTGCAGTATTT-3′); Slug (forward,
5′-AACAGAGCATTTGCAGACAGGTC-3′ and reverse,
5′-GCTACACAGCAGCCAGATTCC-3′); GAPDH (forward,
5′-TCAACGGATTTGGTCGTATTG-3′ and reverse,
5′-TGGGTGGAATCATATTGGAAC-3′); and VEGF-C (forward,
5′-TGTGTGTCCGTCTACAGATGTG-3′ and reverse,
5′-TCGGCAGGAAGTGTGATTGG-3′). Relative expression levels were
calculated using the 2−ΔΔCq method and the specificity
of PCR products was verified through 2% agarose gel
electrophoresis. The main steps of western blot analysis were
performed according to standard procedures as previously described
(24). The kits containing the
nuclear and cytoplasmic extraction reagents were purchased from
KeyGen Biotech, Co., Ltd. (Nanjing, China).
Cell proliferation assay
Cell proliferation was assessed by the Cell Counting
Kit-8 (CCK-8) reagent according to the manufacturer's protocol
(Biosharp, Hefei, China). The highly water-soluble tetrazolium salt
of CCK-8 kit, WST-8, is reduced by dehydrogenase activities in
cells to produce a yellow colored formazan dye, which is soluble in
the culture media and directly proportional to the number of living
cells. Briefly, the cells were seeded onto 96-well plates at a
density of 5×103 cells/ml and allowed to adhere for 24
h. After 24, 48 or 72 h of incubation, 10 µl of CCK-8 solution was
added to each well, and the cells were incubated at 37°C for 1 h.
The optical density (OD) was measured at an absorbance wavelength
of 450 nm in a microplate reader (Bio-Tek, Winooski, VT, USA). All
experiments were conducted at least in triplicate.
Cell migration and invasion assay
For migration analysis, 4×105 ECA109
cells/well were plated onto 24-well plates with complete medium.
After scratching with a 10-µl pipette tips, followed by
pretreatment with let-7a mimics or NC for 24 h, the cells were
washed twice with PBS, cultured in serum-free medium, and
photographed by a microscope (Olympus, Tokyo, Japan) after 24 and
48 h. Prior to the Transwell migration assay, ECA-109 cells were
cultured with or without let-7a mimic for 24 h. Subsequently, the
cells were digested, resuspended and seeded at a density of
~2×105 of cells/well into the upper chambers, harboring
a polycarbonate membrane (8 µm pore size; Corning Incorporated,
Corning, NY, USA) containing serum-free medium, while medium
containing 10% FBS was deposited in the lower chambers. Cells that
migrated through the membrane were fixed and stained with 0.1%
crystal violet (Solarbio, Beijing, China), and then counted in
three randomly selected fields at ×200 magnification. The results
are expressed as the percentage inhibition rate compared to
control.
ELISA and immunofluorescence
assays
As previously described, the cell culture
supernatants were collected, centrifuged, and immediately stored at
−80°C (24). ELISA (Lianke Bio,
Hangzhou, China) assay was utilized according to the manufacturer's
instructions. For immunofluorescence assays, the cells were fixed
and blocked, and successively incubated with primary antibodies
against E-cadherin (Cell Signaling Technology, Inc., Beverly, MA,
USA) and Vimentin (Boster, Wuhan, China) overnight at 4°C, and
subsequently incubated with Cy3 or FITC-conjugated secondary
antibodies (BD, Biosciences, CA, USA) at room temperature for 1 h.
The cells grown on coverslips were counterstained with DAPI
(Sigma-Aldrich, St. Louis, MO, USA) and randomly imaged using a
fluorescence microscope (Olympus).
Statistical analysis
The results obtained from multiple experiments were
reported as the means ± standard deviation. SPSS 16.0 software was
used for data handling, analysis, and presentation. Distinctions
between groups were analyzed by Student's t-test or one-way
analysis of variance (ANOVA) with post hoc Student-Newman-Keuls
test. Statistical significance was set as P<0.05.
Results
The expression of let-7a is repressed
in tissues of ESCC patients
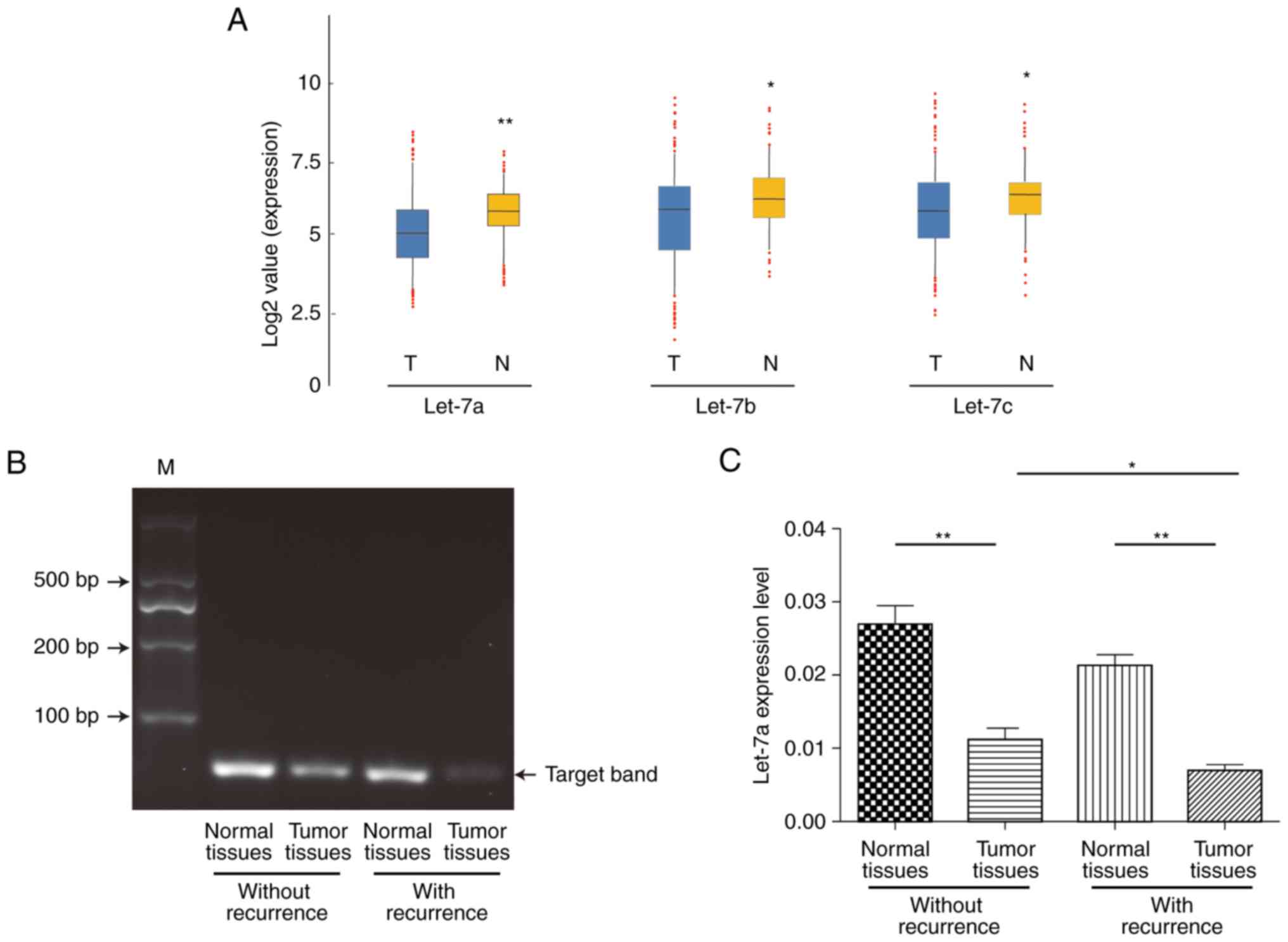
We first investigated the expression of let-7 family
members, including let-7a, let-7b and let-7c, in 70 pair of
surgically resected human ESCC and para-carcinoma normal tissues by
RT-qPCR analysis. The expression of let-7a, let-7b and let-7c were
apparently lower in tumors from ESCC patients, among which a
relative stable result showed that let-7a may be more
representative and focused (P=0.0089; P=0.0431; P=0.0397) (Fig. 1A). To verify the specificity of
RT-qPCR products, we then categorized the specimens into two groups
based on recurrence and detected these products by agarose gel
electrophoresis. As shown in Fig.
1B, the let-7a target specificity band emerged at the 100 bp
position and showed more expression of let-7a among para-carcinoma
normal tissues no matter whether recurrence from ESCC patients. We
next investigated the association between let-7a and recurrence.
The expression of let-7a was significantly decreased in the tumor
tissues with or without recurrence compared to that of matched
normal tissues (P<0.01; P=0.0033; P=0.0017). Furthermore, the
expression of let-7a was significantly lower in the tumor tissues
with recurrence than in those without recurrence (P<0.05;
P=0.0120; Fig. 1C). The
statistical results showed that the expression of let-7a in ESCC
tissues was decreased compared to that in the matched normal
tissues of 70 ESCC samples and was significantly correlated with
advanced stage and tumor recurrence (Table I), indicating that the low
expression of let-7a in most human ESCC cases might be involved in
the invasion, metastasis and poor prognosis of ESCC.
 | Table I.Correlation between let-7a and
clinic-pathological features of ESCC patients. |
Table I.
Correlation between let-7a and
clinic-pathological features of ESCC patients.
| Clinicopathological
features | Number | let-7a (fold
change) | P-value |
|---|
| Sex |
|
|
|
| Male | 40 | 0.39±0.02 | 0.5927 |
|
Female | 30 | 0.37±0.03 |
|
| Age, years |
|
|
|
| ≥50 | 56 | 0.38±0.02 | 1.0000 |
|
<50 | 14 | 0.38±0.05 |
|
| Diameter of tumor,
cm |
|
|
|
|
>4 | 42 | 0.38±0.02 | 0.8058 |
| ≤4 | 28 | 0.37±0.03 |
|
| Differentiation |
|
|
|
|
High | 31 | 0.39±0.03 | 0.9203 |
|
Middle | 17 | 0.38±0.03 |
|
|
Low | 22 | 0.37±0.03 |
|
| TNM stagea |
|
|
|
|
I/II | 47 | 0.41±0.02 | 0.0258 |
|
III/IV | 23 | 0.32±0.03 |
|
| Location |
|
|
|
|
Upper | 15 | 0.36±0.03 | 0.6796 |
|
Middle | 34 | 0.40±0.03 |
|
|
Lower | 21 | 0.37±0.04 |
|
| Recurrencea |
|
|
|
|
Yes | 41 | 0.33±0.02 | 0.0038 |
| No | 29 | 0.44±0.03 |
|
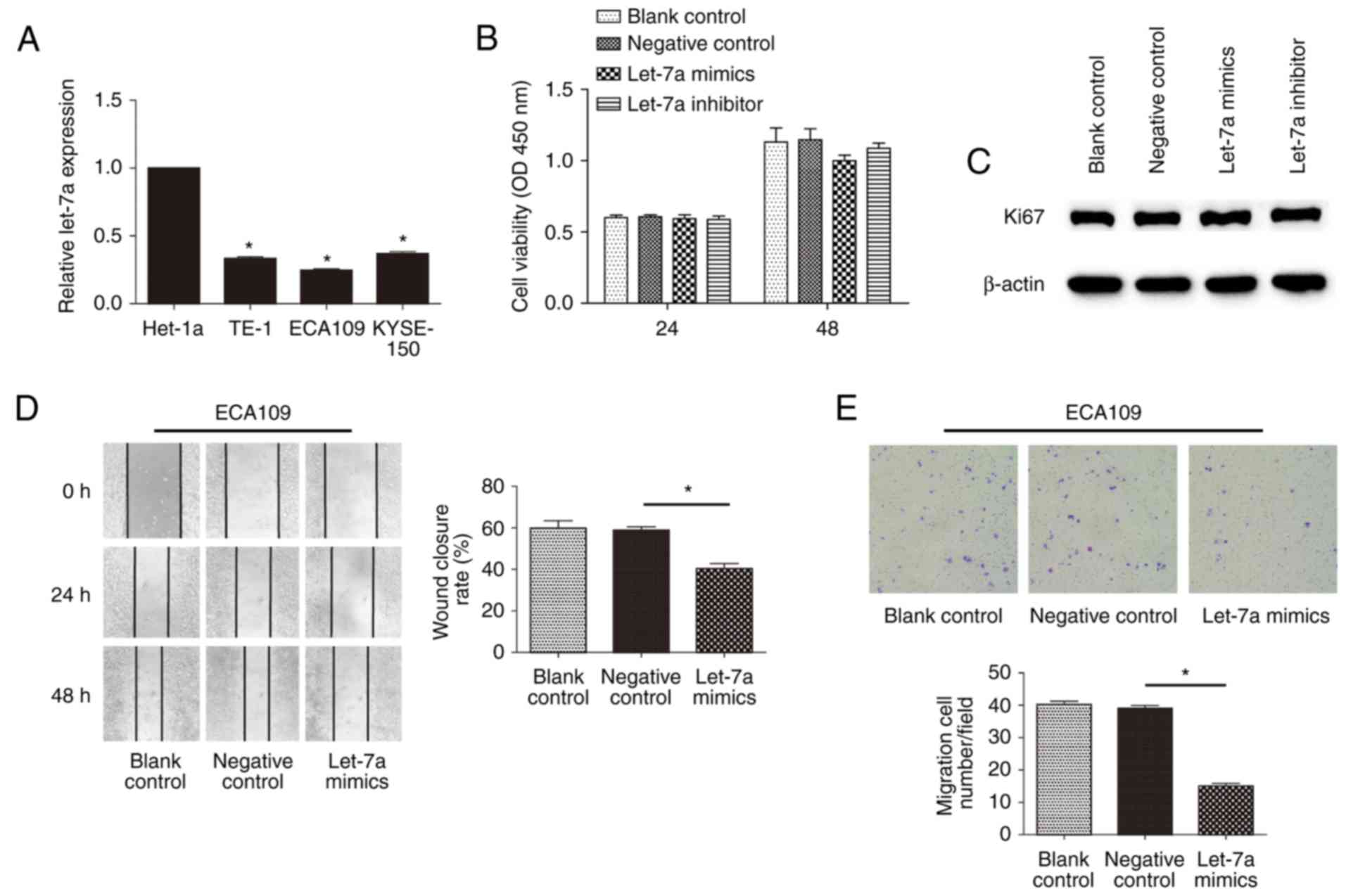
Repression of let-7a increases the
motility of ESCCs in vitro
To determine the relationship of let-7a expression
with the malignancy of ESCC, we next examined the expression of
let-7a among Het-1a cells and ESCC cell lines by RT-qPCR (Fig. 2A). Compared with Het-1a, the
expression of let-7a was significantly lower for ESCC cells
(P<0.01; P=0.0314; P=0.0130; P=0.0219). Nevertheless, there were
no significant differences among TE-1, ECA109 and KYSE-150 cells
(P>0.05; P=0.1192). Previous studies have indicated that let-7
represses the proliferation of ECA109 cells (29). To examine whether let-7a-mediated
inhibition affected cell motility, we determined proliferation by
CCK-8 assay in ESCC cells transfected with let-7a mimics or
inhibitors. Conversely, there was no significant difference among
treatment groups of ECA109 cells at 24 and 48 h (P>0.05;
P=0.0120) (Fig. 2B), including
TE-1 and KYSE-150 cells, even at 72 h (data not shown).
Furthermore, differences in let-7a expression in ECA109 cells
showed no significant change in Ki67 (a nuclear protein associated
with proliferation) production, compared to control (Fig. 2C). These data indicated that let-7a
has no effect on the proliferation of ESCCs. To address the effect
of let-7a on cell mobility in vitro, we performed the wound
healing and Transwell migration assays. The results showed that
let-7a mimics could markedly decrease the cell migration rate at
the edge of the scratch (Fig. 2D;
P=0.0232). Moreover, the migration assay also showed that the
invasiveness of cells was significantly restrained after treatment
with let-7a mimics (Fig. 2E;
P=0.0103). However, inhibitors of let-7a had no evident effects
(data not shown), potentially due to the extremely low inherent
expression of the molecules. These results indicated that let-7a
regulated the motility of ESCC cells in vitro, independent
of cell proliferation.
Lin28/let-7a regulates the progress of
EMT and spreading in ESCCs
EMT is a well-known inducer of tumor metastasis, and
we thus speculated that let-7a also acts as a repressor of this
progress. Although there were no significant changes in cell
morphologies after up or downregulated let-7a expression (Fig. 3A), the results of
immunofluorescence and immunoblotting assays showed that ECA109
cells treated with let-7a mimics had an inhibited Vimentin
expression and increased E-cadherin expression, indicating that
let-7a was involved in the stabilization of EMT (Fig. 3B). We also examined the effect of
Lin-28, as an inhibitor of let-7a, on EMT in the presence or not of
let-7a mimics. The overexpression of let-7a inhibited the
transcription of snail and slug, and the interference of Lin28
showed the same but relatively weaker effect; thus, the combined
treatment of both let-7a overexpression and Lin28 interference
caused cumulative effects (Fig.
3C; the downregulation of snail was 53.3±0.74; 72.2±0.81;
72.4±0.69% respectively; the downregulation of slug was 49.9±0.66;
54.2±0.88; 69.9±0.77%, respectively). However, the enhanced let-7a
expression did not affect the expression of Lin28. In addition, we
further investigated the levels of VEGF-C, VEGF-A, MMP2 and MMP9,
which are widely considered as key indicators of metastasis.
Similarly, the expression of VEGF-C and MMP9, but not VEGF-A and
MMP2, varied inversely with let-7a (Fig. 3D). ELISA results also showed
distinct decreases in the levels of VEGF-C (P=0.0303) and MMP9
(P=0.0402) expression in the culture supernatant of ECA109 cells,
except MMP2 (P=0.6734) (Fig. 3E).
These findings suggested that let-7a regulates not only the
transformation, but also the stromatolysis and vessels regeneration
with Lin28 in ESCCs.
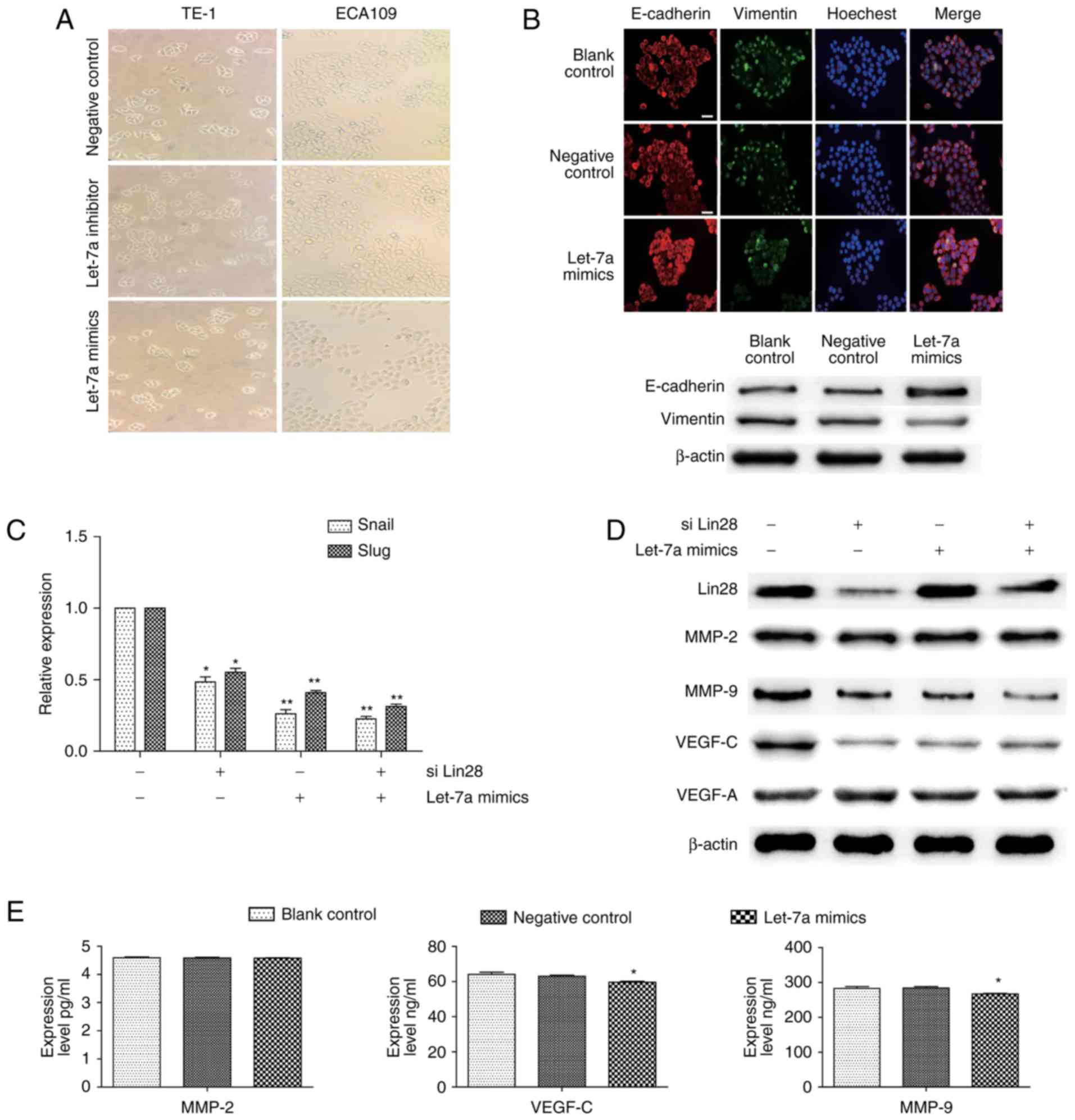 | Figure 3.Regulation of Lin28/let-7a on EMT and
spreading in ESCCs. (A) Representative images of cell morphology
performed in ESCC cells transfected with negative control, let-7a
inhibitors or let-7a mimic (magnification, ×100). (B) ECA109 cells
were transfected with let-7a mimics or NC for 24 h and then stained
for E-cadherin (red), Vimentin (green), and cell nuclei (blue)
(scale bar, 50 µm). The changes of E-cadherin and Vimentin were
measured by western blotting. (C) The relative expression of slug
and snail was detected by RT-qPCR after ECA109 cells were
transfected with siLin28 and (or) let-7a mimics. (D) The expression
changes of Lin28, VEGF-C, MMP2, MMP9, and VEGF-A after treatment
with siLin28 and (or) let-7a mimics for 48 h. (E) ELISA assay
measured the secreted VEGF-C, MMP2, and MMP9 after transfection or
not for 48 h in ECA109 cells. siLin28, siRNA-Lin28 treatment.
**P<0.01, *P<0.05. |
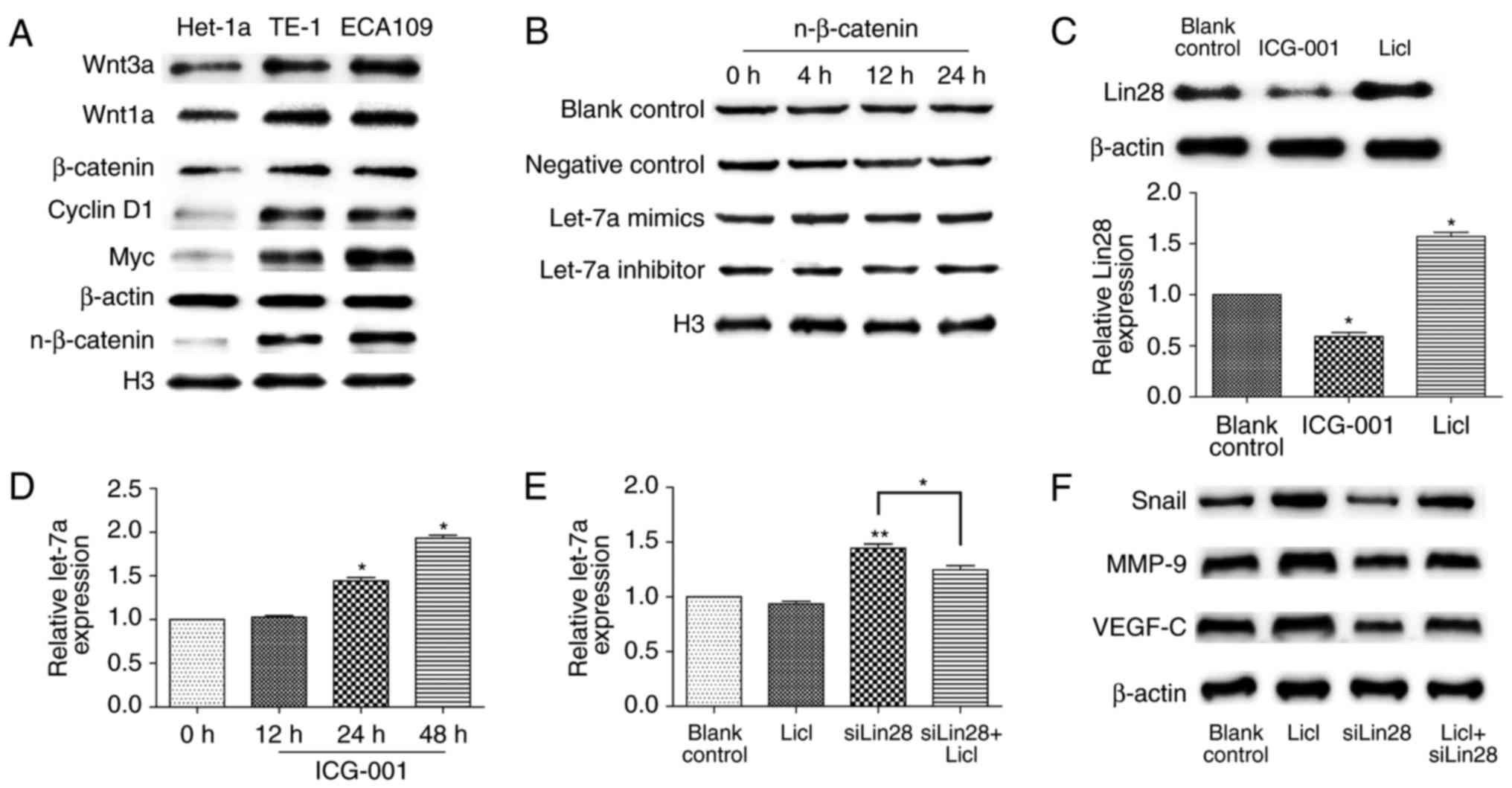
The hyper-activation of Wnt/β-catenin
suppresses let-7a by Lin28
The Wnt pathway is a key signaling pathway of
malignancy; therefore, we assessed the inherent level of β-catenin
among cells. The nuclear-localization expression of β-catenin was
higher in ECA109 and TE-1 cells compared to that in Het-1a cells
(Fig. 4A), although there was no
statistical significance between both ESCCs (data not shown), and
well-known downstream proteins, such as cyclin D1 and Myc, they
were synergistically overactive. A recent study demonstrated that
let-7a can inhibit EMT through the Wnt pathway in hepatocellular
carcinoma stem cells (19); thus,
we hypothesized that let-7a may exert the same role in ESCC.
Western blot analysis showed that nuclear β-catenin showed no
significant change in ECA109 cells after treatment with let-7a
mimics or inhibitors at different times (Fig. 4B). Since Lin28 is a direct
downstream gene of the Wnt pathway (25,26),
we next examined the effect of Wnt activation on Lin28 expression
in ESCCs. As shown in Fig. 4C,
LiCl (20 mM) upregulated Lin28 biosynthesis, while ICG-001 potently
prevented the accumulation of Lin28 at 10 µM. In contrast, the
expression of let-7a was considerably higher after the addition of
ICG-001 at 24 and 48 h (P<0.05; P=0.0301, with the upregulation
of 1.42±0.19 fold; P=0.0163, with the upregulation of 1.91±0.20
fold) (Fig. 4D). Next, we detected
the expression of let-7a after treatment with LiCl and/or
siRNA-Lin28, and compared with LiCl treatment, there was a
significant decrease of let-7a under the interference of Lin28
(P=0.0412) (Fig. 4E). Similarly,
the downregulation of Snail, VEGF-C, and MMP9 after Lin28 knockdown
was reverted by LiCl treatment (Fig.
4F). In conclusion, the activation of the Wnt pathway
suppresses let-7a by Lin28 in ESCCs.
Discussion
Let-7 has been recognized as one of the most
prominent miRNAs implicated in human malignancy (10–20).
The expression of let-7a, let-7b, and let-7c or other members was
previously identified as significantly reduced in tumors, and the
upregulation of these miRNAs inhibited the process of
proliferation, EMT or metastasis. (18–20).
In our study, we found low expression of let-7a, let-7b and let-7c
in ESCC tumors and that let-7a was significantly inversely
correlated with advanced stage, recurrence and poor prognosis.
Therefore, the identification of let-7a would definitely be helpful
for the clinical decision and management of ESCC. In vitro,
the upregulation of let-7a by mimics could markedly decrease the
migration and invasiveness rate, but it had no effect on
proliferation in ESCCs. According to the initial role of EMT for
metastasis, our experiment further revealed the accumulation of
E-cadherin and weaker accumulation of Vimentin, with distinct
decreases of snail and slug when let-7a was highly expressed.
Moreover, the regulation of VEGF-C and MMP9 were also dependent on
let-7a levels. This evidence indicated that the loss of let-7a has
potential to detect early signals of carcinogenic exposures for
ESCC.
The Wnt/β-catenin pathway is involved in the
development and homeostasis of many normal organs or tissues, thus
its dysfunction often leads to terrible consequences, such as
tumors (19,21–23,25–27).
Our previous study indicated that the hyper-activation of
Wnt/β-catenin pathway stimulated by IL-23 could promote EMT in
ESCCs (24). Although recent
evidence has shown that let-7a inhibited the Wnt pathway in HCC
stem cells (19), the same
character has not been confirmed in ESCCs. Furthermore, we found
that the inhibition of the Wnt pathway decreased the level of Lin28
and let-7a restoration. In addition, as the negative regulator of
let-7a, Lin28 was selected to observe the impact of Wnt signaling
on promoting stromatolysis and vessels regeneration. As expected,
after stimulation of the Wnt pathway by LiCl, malignancy was
disrupted by siRNA-Lin28 or let-7a-mimic pretreatment. In this
study, since the abnormal activation of Wnt/β-catenin signaling in
ESCC is inevitable, the status of let-7a mediated by Lin28 may
represent dynamic progression.
In conclusion, our results revealed that compared to
para-carcinoma tissues, the suppression of let-7a in tumors, is
closely associated with the invasion, metastasis and poor prognosis
of ESCC. In addition, Wnt/β-catenin/Lin28 signaling induced EMT,
and metastasis would occur through the elimination of let-7a. As a
screening test for miRNA expression, such as serum, could be easily
obtained, these findings provide a novel factor for the prognosis
of ESCC in the future.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (nos. 81572956 and 81370889),
and the Wu Jieping Medical Foundation (320.6755.15022).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Castillo A, Aguayo F, Koriyama C, Torres
M, Carrascal E, Corvalan A, Roblero JP, Naquira C, Palma M,
Backhouse C, et al: Human papillomavirus in esophageal squamous
cell carcinoma in Colombia and Chile. World J Gastroenterol.
12:6188–6192. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Chang D and Church J: Evaluating the
health-related quality of life of esophageal cancer patients. Pract
Radiat Oncol. 4:181–186. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ekman S, Dreilich M, Lennartsson J,
Wallner B, Brattström D, Sundbom M and Bergqvist M: Esophageal
cancer: Current and emerging therapy modalities. Expert Rev
Anticancer Ther. 8:1433–1448. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dreikhausen L, Blank S, Sisic L, Heger U,
Weichert W, Jäger D, Bruckner T, Giese N, Grenacher L, Falk C, et
al: Association of angiogenic factors with prognosis in esophageal
cancer. BMC Cancer. 15:1212015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jung HY, Fattet L and Yang J: Molecular
pathways: Linking tumor microenvironment to epithelial-mesenchymal
transition in metastasis. Clin Cancer Res. 21:962–968. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brabletz S and Brabletz T: The ZEB/miR-200
feedback loop-a motor of cellular plasticity in development and
cancer? EMBO Rep. 11:670–677. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chhabra R and Saini N: MicroRNAs in cancer
stem cells: Current status and future directions. Tumour Biol.
35:8395–8405. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rajasekaran S, Rajaguru P and Sudhakar
Gandhi PS: MicroRNAs as potential targets for progressive pulmonary
fibrosis. Front Pharmacol. 6:2542015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang G, Zhang W, Yu C, Ren J and An Z:
MicroRNA let-7: Regulation, single nucleotide polymorphism, and
therapy in lung cancer. J Cancer Res Ther. 11 Suppl 1:C1–C6. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Alajez NM, Shi W, Wong D, Lenarduzzi M,
Waldron J, Weinreb I and Liu FF: Lin28b promotes head and neck
cancer progression via modulation of the insulin-like growth factor
survival pathway. Oncotarget. 3:1641–1652. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ohshima K, Inoue K, Fujiwara A, Hatakeyama
K, Kanto K, Watanabe Y, Muramatsu K, Fukuda Y, Ogura S, Yamaguchi K
and Mochizuki T: Let-7 microRNA family is selectively secreted into
the extracellular environment via exosomes in a metastatic gastric
cancer cell line. PLoS One. 5:e132472010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boyerinas B, Park SM, Murmann AE, Gwin K,
Montag AG, Zillhardt M, Hua YJ, Lengyel E and Peter ME: Let-7
modulates acquired resistance of ovarian cancer to Taxanes via
IMP-1-mediated stabilization of multidrug resistance 1. Int J
Cancer. 130:1787–1797. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sugimura K, Miyata H, Tanaka K, Hamano R,
Takahashi T, Kurokawa Y, Yamasaki M, Nakajima K, Takiguchi S, Mori
M and Doki Y: Let-7 expression is a significant determinant of
response to chemotherapy through the regulation of IL-6/STAT3
pathway in esophageal squamous cell carcinoma. Clin Cancer Res.
18:5144–5153. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamano R, Miyata H, Yamasaki M, Sugimura
K, Tanaka K, Kurokawa Y, Nakajima K, Takiguchi S, Fujiwara Y, Mori
M and Doki Y: High expression of Lin28 is associated with tumour
aggressiveness and poor prognosis of patients in oesophagus cancer.
Br J Cancer. 106:1415–1423. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu Y, Li H, Feng J, Cui X, Huang W, Li Y,
Su F, Liu Q, Zhu J, Lv X, et al: Lin28 induces
epithelial-to-mesenchymal transition and stemness via
downregulation of let-7a in breast cancer cells. PLoS One.
8:e830832013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu A, Wu K, Li J, Mo Y, Lin Y, Wang Y,
Shen X, Li S, Li L and Yang Z: Let-7a inhibits migration, invasion
and epithelial-mesenchymal transition by targeting HMGA2 in
nasopharyngeal carcinoma. J Transl Med. 13:1052015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jin B, Wang W, Meng XX, Du G, Li J, Zhang
SZ, Zhou BH and Fu ZH: Let-7 inhibits self-renewal of
hepatocellular cancer stem-like cells through regulating the
epithelial-mesenchymal transition and the Wnt signaling pathway.
BMC Cancer. 16:8632016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li B, Chen P, Chang Y, Qi J, Fu H and Guo
H: Let-7a inhibits tumor cell growth and metastasis by directly
targeting RTKN in human colon cancer. Biochem Biophys Res Commun.
478:739–745. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang Y, Shi J, Chai K, Ying X and Zhou BP:
The role of snail in EMT and tumorigenesis. Curr Cancer Drug
Targets. 13:963–972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nusse R: Wnt signaling and stem cell
control. Cell Res. 18:523–527. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun X, He Y, Huang C, Ma TT and Li J:
Distinctive microRNA signature associated of neoplasms with the
Wnt/β-catenin signaling pathway. Cell Signal. 25:2805–2811. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen D, Li W, Liu S, Su Y, Han G, Xu C,
Liu H, Zheng T, Zhou Y and Mao C: Interleukin-23 promotes the
epithelial-mesenchymal transition of oesophageal carcinoma cells
via the Wnt/β-catenin pathway. Sci Rep. 5:86042015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tu HC, Schwitalla S, Qian Z, LaPier GS,
Yermalovich A, Ku YC, Chen SC, Viswanathan SR, Zhu H, Nishihara R,
et al: LIN28 cooperates with WNT signaling to drive invasive
intestinal and colorectal adenocarcinoma in mice and humans. Genes
Dev. 29:1074–1086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cai WY, Wei TZ, Luo QC, Wu QW, Liu QF,
Yang M, Ye GD, Wu JF, Chen YY, Sun GB, et al: The Wnt-β-catenin
pathway represses let-7 microRNA expression through transactivation
of Lin28 to augment breast cancer stem cell expansion. J Cell Sci.
126:2877–2889. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Ying J, Fan Y, Wu L, Ying Y, Chan
AT, Srivastava G and Tao Q: WNT5A antagonizes WNT/β-catenin
signaling and is frequently silenced by promoter CpG methylation in
esophageal squamous cell carcinoma. Cancer Biol Ther. 10:617–624.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gao J, Li N, Dong Y, Li S, Xu L, Li X, Li
Y, Li Z, Ng SS, Sung JJ, et al: miR-34a-5p suppresses colorectal
cancer metastasis and predicts recurrence in patients with stage
II/III colorectal cancer. Oncogene. 34:4142–4152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu Q, Lv GD, Qin X, Gen YH, Zheng ST, Liu
T and Lu XM: Role of microRNA let-7 and effect to HMGA2 in
esophageal squamous cell carcinoma. Mol Biol Rep. 39:1239–1246.
2012. View Article : Google Scholar : PubMed/NCBI
|