Introduction
Renal cell carcinoma (RCC) is among the top 10 most
lethal types of cancer and is the cause of >10,000 cases of
mortality every year (1).
According to the most recent statistics published by the American
Cancer Society, ~65,340 individuals were predicted to be diagnosed
with RCC in 2018, whereas the estimated mortality rate was ~14,970
cases (1). Patients with RCC
frequently encounter recurrence and/or metastasis in late stages,
leading to poor clinical outcome despite the improved treatments
for RCC currently in use (2,3). The
5-year survival rate for patients with kidney cancer with distant
metastasis is only 12%, whereas it is 67% for patients with
localized cancer (1). At present,
~16% of patients are diagnosed with distant metastasis (1); however, there is no agreement
regarding the molecular mechanisms underlying RCC. Next-generation
sequencing technology has identified dozens of gene mutations
associated with various type of cancer; however, only mutations of
certain driver genes serve dominant roles in the development and
progression of cancer (4). A
number of genes associated with RCC have been identified, but the
heterogeneity of the condition increases the importance of
identifying novel genes and further understanding the molecular
mechanisms underlying RCC (5,6).
Ubiquitin-conjugating enzyme E2T (UBE2T) is a member
of the ubiquitin-proteasome family; this family is an efficient
protein-modification system that serves important roles in almost
all cell behaviors, including DNA replication, cell proliferation,
differentiation, apoptosis and angiogenesis (7,8).
UBE2T has been reported to bind to FA complementation group L
protein via its UBC domain, promoting the formation of the Fanconi
anemia (FA) core complex and activating the FA signaling pathway
(9–11). In addition, UBE2T has been reported
to cause FA, increasing the risk of acute myeloid leukemia or
head/neck squamous carcinomas (11–13).
Additionally, UBE2T has been identified to act as an oncogene in
numerous types of cancer. For example, UBE2T promotes cell
proliferation, invasion and metastasis in nasopharyngeal carcinoma
(14), and induces the
ubiquitination of p53 in hepatocellular carcinoma, promoting cell
growth (15). Elevated expression
of UBE2T also serves an oncogenic role in prostate cancer (16). Knockdown of UBE2T suppresses the
proliferation and invasion of osteosarcoma and gastric cancer cells
(17,18). Furthermore, cell cycle arrest and
apoptosis are induced by UBE2T silencing in bladder cancer
(19). In breast cancer, UBE2T
overexpression results in the degradation of BRCA1 and poor
clinical outcome (20).
Furthermore, UBE2T is overexpressed in lung cancer and is involved
in resistance to chemotherapeutic drugs (21–23).
At present, the role of the UBE2T gene in the development and
progression of RCC is yet to be investigated.
This study aimed to identify the role of UBE2T in
RCC; therefore, the expression of UBE2T was detected in RCC tissues
and cells, and its clinical relevance to RCC was analyzed.
Furthermore, the effects of UBE2T knockdown on the proliferation,
colony formation and invasion of RCC cells were studied in
vitro, and in vivo in a nude mouse model. Additionally,
the effects of UBE2T knockdown on the phosphorylation of PI3K, Akt
and mTOR were investigated via western blot analysis.
Materials and methods
Clinical samples and ethics
statement
A total of 52 fresh surgical tissues and matched
adjacent normal tissues from patients (15–62 years old, 36 males
and 16 females) diagnosed with RCC were collected from June 2014 to
July 2016 at the Department of Urology Surgery of First Affiliated
Hospital of Jiamusi University, flash frozen in liquid nitrogen and
stored at −80°C. Patients that did not receive chemotherapy or
radiotherapy prior to surgery were selected for this study.
Completed signed clinical information was collected. The
pathological stage of patients was established based on the TNM
classification system from the WHO (24). Total RNA and protein were extracted
and stored at −80°C, and used for reverse
transcription-quantitative PCR (RT-qPCR) and western blotting,
respectively. Patients were separated into high- and low-expression
groups for survival analysis based on their levels of UBE2T
expression; a fold change >2 in expression in tumor tissue
compared with in normal tissue was considered high, whereas a fold
change ≤2 was considered low.
Written informed consent was obtained from all
patients. All experiments were approved by the Institutional Review
Board of The First Affiliated Hospital of Jiamusi University.
Cell culture and transfection of small
interfering RNA (siRNA)
Human renal cancer cell lines (786-O, ACHN and
OSRC-2) and a non-cancer cell line (293) were purchased (Cell Bank
of the Chinese Academy of Sciences) and cultured in RPMI-1640
medium (Hyclone; GE Healthcare Life Sciences) with 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) in a humidified
atmosphere with 5% CO2. at 37°C. siRNA fragments
targeting human UBE2T (siUBE2T; sequence,
5′-GCAACTGTGTTGACCTCTATT-3′) and negative control (siNC; sequence,
5′-GCTTCGGATACGTTTCCTAAT-3′) were synthesized (Shanghai Telebio
Biomedical Co., Ltd.) and transfected into 786-O cells
(1×105) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). The dosage of siRNA
used was 1.0 µM, and the interval between transfection and
subsequent experiments was 6 h.
Construction of a UBE2T overexpression
plasmid (oeUBE2T) and transfection
The coding sequence for UBE2T (synthesized by Synbio
Technologies) was transferred into vector pcDNA3.1 (Invitrogen;
Thermo Fisher Scientific, Inc.) using EcoRI and NotI
restriction enzymes. Subsequently, ~1.5 µg of the recombined
plasmid pcDNA3.1-UBE2T or empty vector pcDNA3.1 were transfected
into 786-O cells (1×105) using Lipofectamine®
2000. After culturing for 48 h, total protein was extracted from
the treated cells, stored at −80°C and used for western
blotting.
Cell proliferation assay
siUBE2T- or siNC-transfected 786-O cells were seeded
into a 96-well plate at 1,000 cells/well. Following incubation for
48 h at 37°C, the culture medium was replaced and 20 µl MTT reagent
(5 mg/ml; Sigma-Aldrich; Merck KGaA) was added. After a further 4 h
at 37°C, supernatants were removed and 200 µl DMSO was added. The
absorbance was detected at a wavelength of 490 nm using a
microplate reader. Wortmannin (5 nM; Sigma-Aldrich; Merck KGaA) was
used to treat 786-O cells transfected with siUBE2T, siNC or oeUBE2T
for 24 h at 37°C.
Colony formation assay
siUBE2T-or siNC-transfected 786-O cells were seeded
in a 60-mm dish at 2,000 cells/dish. After 2 weeks at 37°C, 100%
methanol was used to fix the colonies for 2 min at room
temperature. Crystal violet staining solution (Sangon Biotech Co.,
Ltd.) was used to stain the fixed colonies for 15 min at room
temperature. The number of stained colonies was counted under a
Nikon microscope (magnification, ×100; Nikon Corporation); five
fields were selected randomly for evaluation.
Transwell assay
siUBE2T-or siNC-transfected 786-O cells were seeded
into the upper chambers of Transwell inserts (8.0-µm pore size; BD
Biosciences) with or without Matrigel matrix at 5×104
cells/well in DMEM containing 2% FBS. DMEM containing 20% FBS (500
µl) was placed in the lower chambers. Following incubation for 48 h
at 37°C, the cells in the bottom of each chamber were fixed with
100% methanol for 5 min at room temperature and stained with 0.1%
crystal violet staining solution for 15 min at room temperature.
Five fields were selected randomly and images were captured.
Subsequently, the cell number was counted under a Nikon microscope
(magnification, ×100; Nikon Corporation).
RT-qPCR
Total RNA was extracted from tissues and cell lines
using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.), and was treated with RNase-free DNase (Promega
Corporation). Total RNA (1 µg) was used for synthesis of first
strand of cDNA using a reverse transcription kit (Takara
Biotechnology Co., Ltd.). The protocol used was as follows: 42°C
for 50 min. qPCR was performed using a Hifair III One Step RT-qPCR
SYBR Green kit (Yeasen Biotech Co., Ltd.) using a Stepone Real-time
PCR system (Applied Biosystems; Thermo Fisher Scientific, Inc.).
qPCR was conducted as follows: 95°C for 5 min, then 40 cycles of
95°C for 10 sec and 60°C for 30 sec. GAPDH was used as the internal
control. The relative expression of each gene was calculated using
the 2−∆∆Cq method, in which ∆Cq=Cq(gene of
interest)-Cq(GAPDH) (25). The
primers used are presented in Table
I.
 | Table I.The primers for RT-qPCR for each
gene. |
Table I.
The primers for RT-qPCR for each
gene.
| Gene | Forward primer
(5′-3′) | Reverse primer
(5′-3′) |
|---|
| UBE2T |
CGAGCTCGTAGAAATATTAGGTGGA |
TCATCAGGGTTGGGTTCTGAC |
| GAPDH |
GAGAAGGCTGGGGCTCATTT |
AGTGATGGCATGGACTGTGG |
Western blot analysis
Total protein was extracted using a Nuclear and
Cytoplasmic Protein Extraction kit (Beyotime Institute of
Biotechnology) and quantified via the BCA method using an Enhanced
BCA Protein Assay kit (Beyotime Institute of Biotechnology).
Proteins (20 µg/lane) were separated via 10% SDS-PAGE and
transferred onto PVDF membranes. Membranes were blocked with 5%
nonfat milk at room temperature for 1 h. Subsequently, the PVDF
membranes were incubated with primary antibodies at 4°C for 12 h,
followed by washing with 0.05% TBS-Tween-20 three times. The
primary antibodies used during the study were specific for
phosphorylated (p)-AKT (1:500; cat. no. ab38449; Abcam), p-PI3K
(1:500; cat. no. ab138364; Abcam), p-mTOR (1:2,000; cat. no.
ab109268; Abcam), total AKT (1:2,000; cat. no. ab179463; Abcam),
total PI3K (1:2,000; cat. no. ab180967; Abcam), total mTOR
(1:1,000; cat. no. ab2732; Abcam), UBE2T (1:1,000 cat. no.
ab140611; Abcam), fibronectin (1:1,000; cat. no. ab45688; Abcam),
E-cadherin (1:600; cat. no. 14472; Cell Signaling Technology,
Inc.), N-cadherin (1:1,000; cat. no. 4061; Cell Signaling
Technology, Inc.), vimentin (1:500; cat. no. 3932; Cell Signaling
Technology, Inc.) and GAPDH (1:1,000; cat. no. 5174; Cell Signaling
Technology, Inc.). PVDF membranes were then incubated with
secondary antibodies for 1 h at 37°C, followed by washing with TBS
three times. The secondary antibodies included horseradish
peroxidase-conjugated sheep anti-mouse IgG (1:1,000; cat. no.
HAF007; R&D Systems, Inc.) and anti-rabbit IgG (1:1,000;
HAF008; R&D Systems, Inc.). Finally, the membranes were
analyzed using an ECL Chemiluminescence Detection kit (Beyotime
Institute of Biotechnology). PhotoShop CS6 software was used to
quantify protein expression following western blotting (Adobe
Systems, Inc.).
In vivo antitumor growth assay
A total of 16 BALB/c-nu mice (age, 7 weeks; weight,
20–25 g) were obtained from Shanghai SLAC Laboratory Animal Co.,
Ltd. and were allocated into two groups: siUBE2T and siNC, with 8
mice/group. Animals were maintained at a constant temperature of
25°C with ~50% humidity, under a regular 12:12-h light/dark
photoperiod with food and water available ad libitum.
Animals were inoculated into the right flank with 3×106
cancer cells following transfection for 24 h with siUBE2T or siNC
and monitored every day for 35 days. Tumor volume was calculated
twice a week using the following equation: V = (LxW2)/2,
where V is the tumor volume, L is the length and W is the width of
the tumor. The study was approved by the Animal Research Ethics
Committee of The First Affiliated Hospital of Jiamusi University.
On day 35, all mice were euthanized according to the protocols
approved by the Ethics Committee. All the experiments on mice were
performed within the guidelines of the Institutional Animal Care
and Use Committee of The First Affiliated Hospital of Jiamusi
University.
Statistical analysis
All experiments were repeated at least three times,
and the data are presented as the means ± standard deviation.
Differences between the groups were compared using one-way ANOVA
followed by Tukey's post hoc test. χ2 test was used to
analyze associations between UBE2T expression and the
clinicopathological factors of patients with RCC. The Kaplan-Meier
method and log-rank test were used to analyze the association
between UBE2T expression and the survival of patients with RCC.
P<0.05 was considered to indicate a statistically significant
difference.
Results
UBE2T exhibits clinical significance
in RCC
To investigate the role of UBE2T in RCC, a total of
52 RCC tissues and adjacent normal tissues were collected, and the
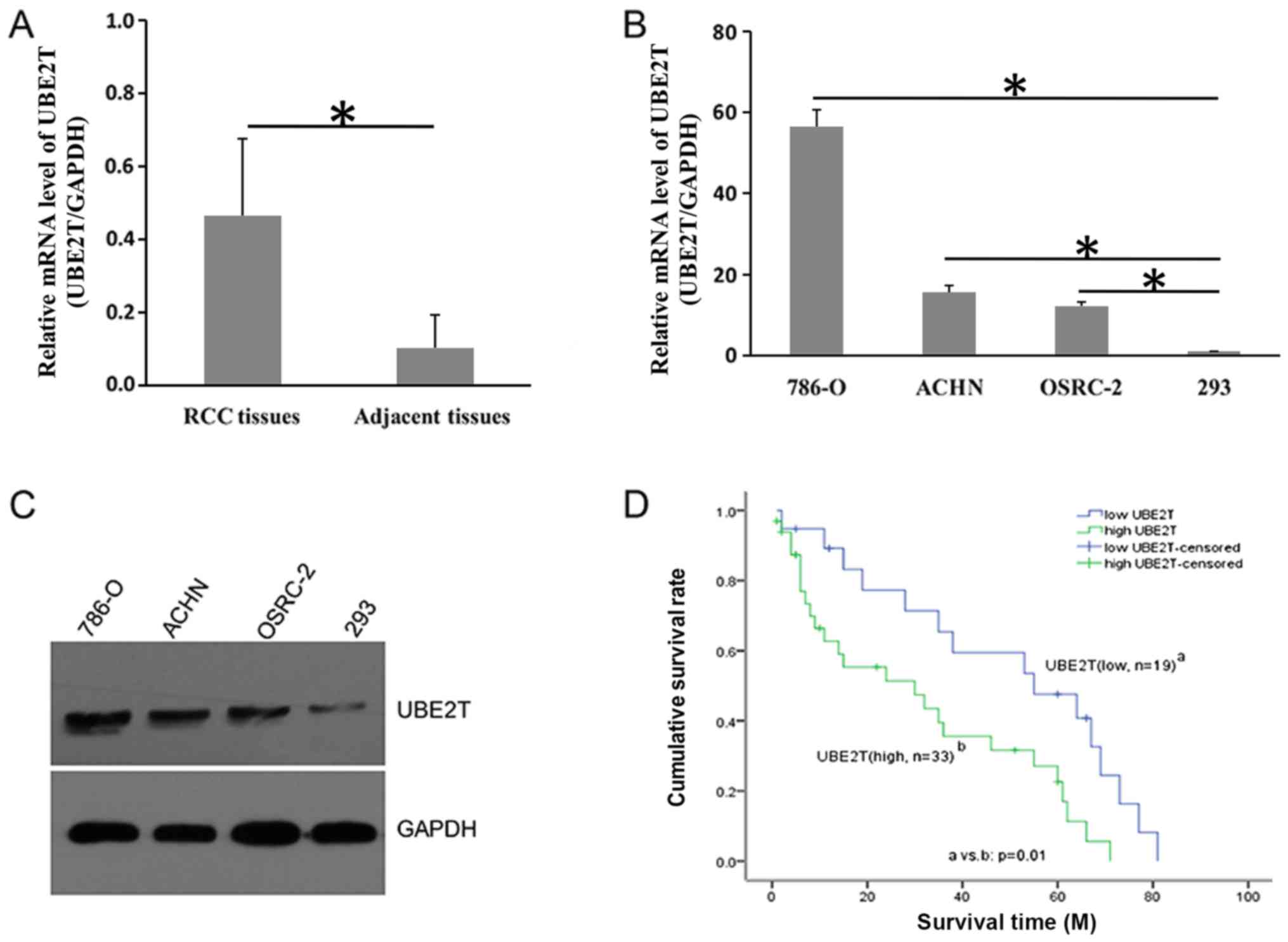
expression of UBE2T was determined via RT-qPCR. As presented in
Fig. 1A, UBE2T was overexpressed
in tumor tissues compared with adjacent normal controls. The mean
mRNA expression of UBE2T in tumor tissues was four-fold that in the
adjacent controls. Additionally, UBE2T expression was notably
increased at the mRNA and protein levels in RCC cell lines compared
within 293 cells (Fig. 1B and C).
Pathological association analysis revealed that UBE2T expression
was associated with TNM grade (P=0.001) and pathological stage
(P=0.001) in patients with RCC (Table
II). Furthermore, Kaplan-Meier analysis indicated that the
survival rate of patients with RCC with high UBE2T expression was
significantly decreased compared with patients with low UBE2T
expression (18.2 vs. 42.1%; P=0.01; Fig. 1D). These data suggested that UBE2T
exhibited clinical relevance for RCC and was associated with poor
prognosis.
 | Table II.Association of UBE2T expression with
clinicopathological factors in renal cell carcinoma. |
Table II.
Association of UBE2T expression with
clinicopathological factors in renal cell carcinoma.
|
| UBE2T
expression |
|
|---|
|
|
|
|
|---|
| Factor | Low (n=19) | High (n=33) | P-value |
|---|
| Sex |
|
| 0.477 |
|
Male | 9 | 19 |
|
|
Female | 10 | 14 |
|
| TNM stage |
|
| 0.001 |
|
I/II | 14 | 8 |
|
|
III/IV | 5 | 25 |
|
| Grade |
|
| 0.001 |
|
G1+G2 | 16 | 10 |
|
|
G3+G4 | 3 | 23 |
|
UBE2T regulates cell proliferation and
colony formation in RCC
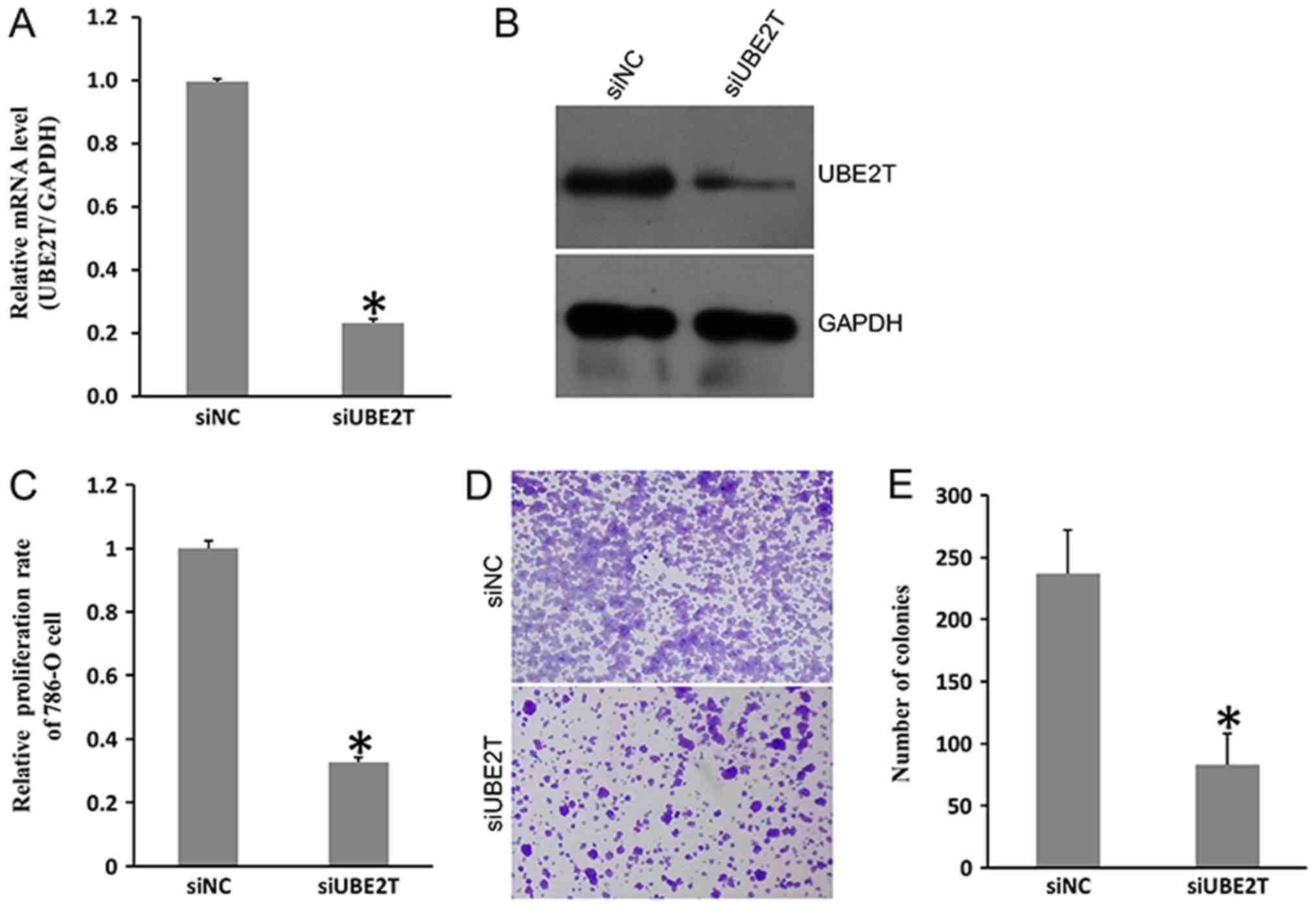
To investigate the function of UBE2T in RCC, 786-O
cells were transfected with siUBE2Tor siNC. As presented in
Fig. 2A and B, UBE2T was
successfully knocked down at the mRNA and protein level in 786-O
cells following siRNA transfection; the knockdown efficiency was
>70%. The proliferation rate of 786-O cells was significantly
decreased by ~65% following knockdown of UBE2T (Fig. 2C). The ability to form colonies was
also inhibited; as presented in Fig.
2D and E, the number of siUBE2T-treated 786-O cell colonies was
significantly decreased compared with the control. Conversely, the
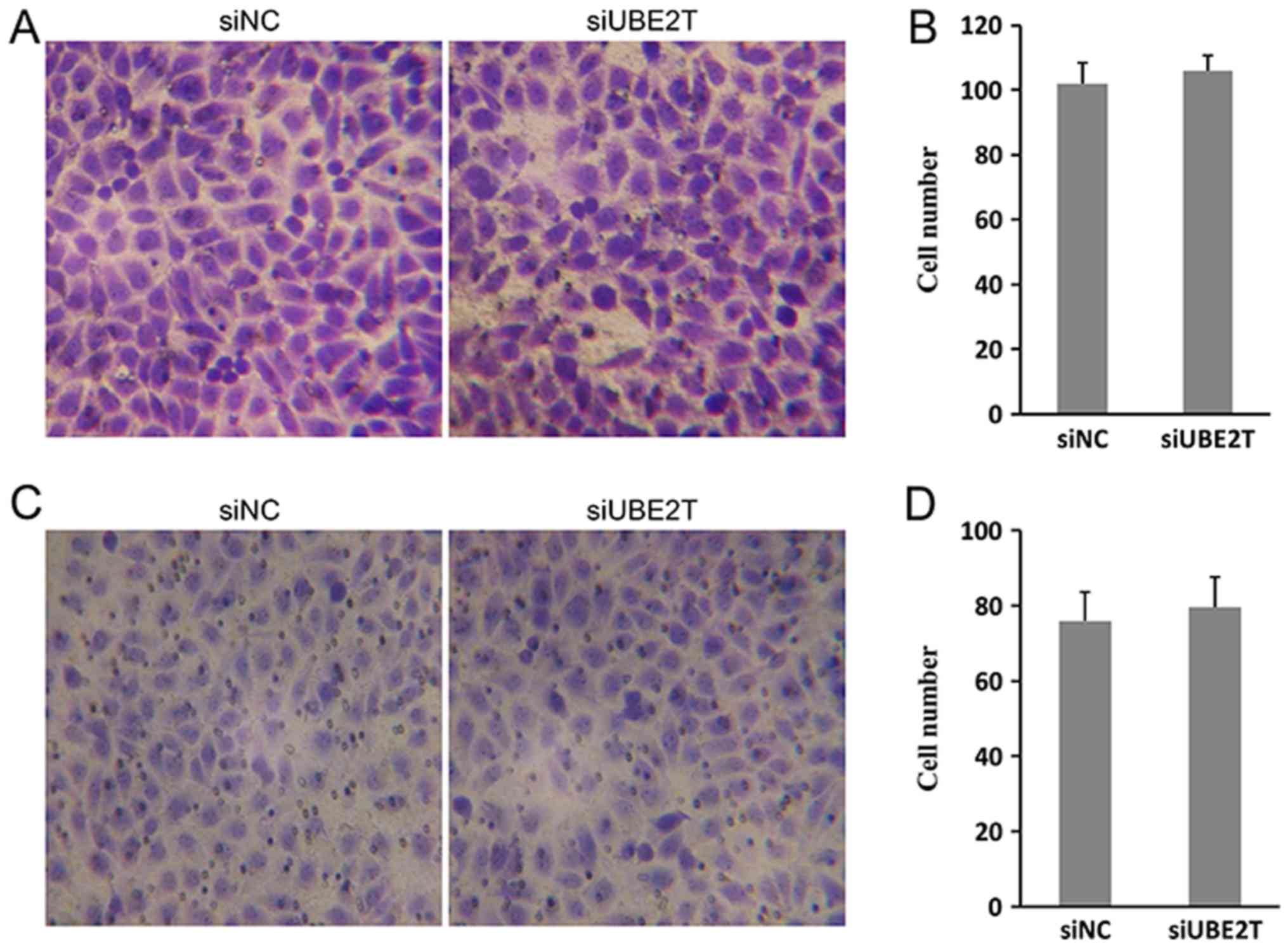
migration and invasion of 786-O cells was not significantly
affected by UBE2T (Fig. 3A-D). In
conclusion, the results indicated that UBE2T contributed to the
proliferation and growth, but not migration of 786-O cells.
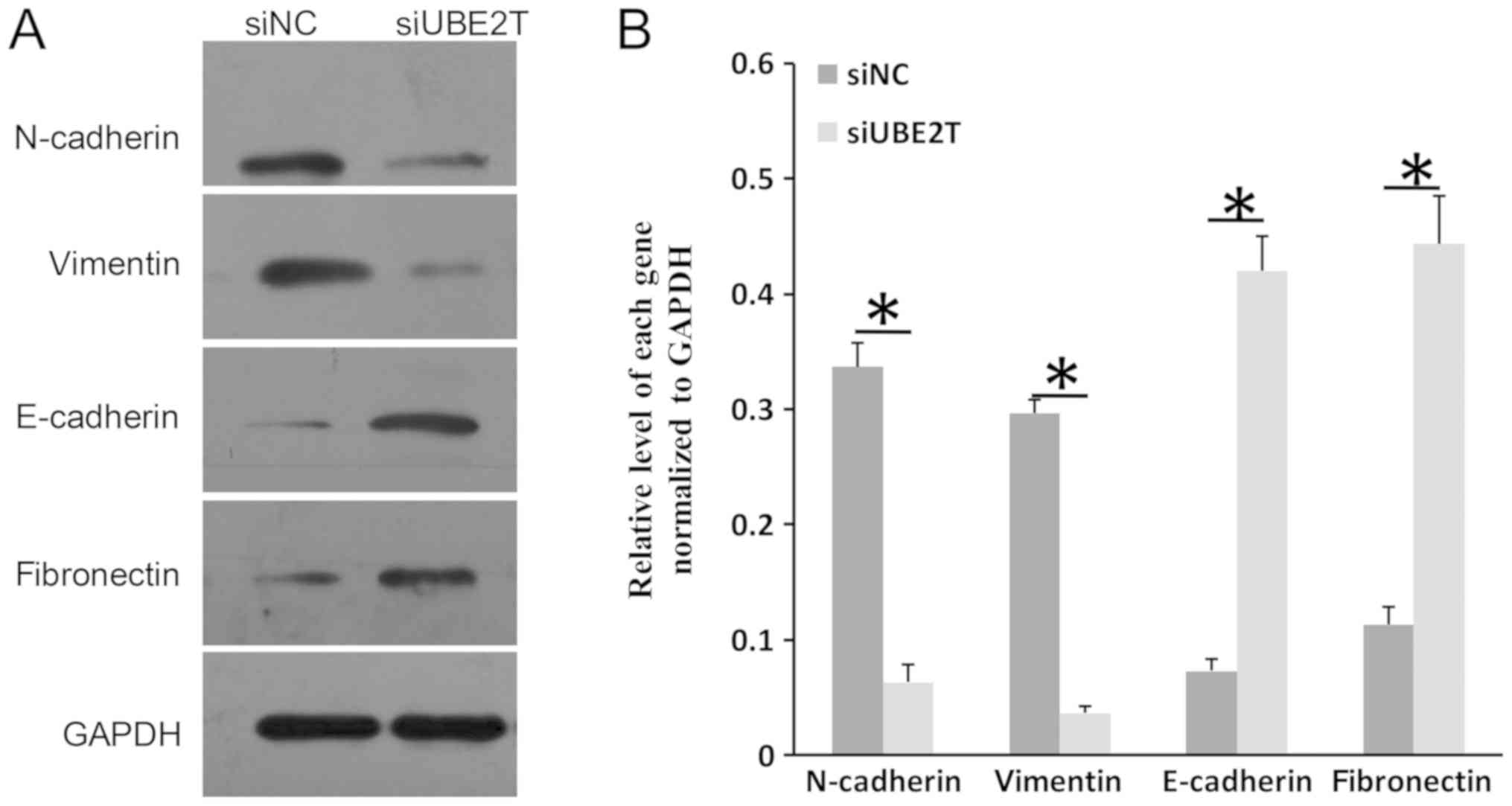
UBE2T regulates the expression of
epithelial-mesenchymal transition (EMT) markers in RCC
EMT is an important process during the
transformation of normal cells into tumor cells (26). Typical molecular markers of
epithelial cells are E-cadherin and fibronectin, whereas N-cadherin
and vimentin are frequently used markers of mesenchymal cells
(27,28). As presented in Fig. 4A and B, the expression of
E-cadherin and fibronectin was notably increased in 786-O cells
following UBE2T knockdown; conversely, the levels of N-cadherin and
vimentin were reduced. The results indicated that UBE2T may
participate in the EMT process in RCC.
UBE2T is critical for the activation
of PI3K/Akt/mTOR signaling in RCC
PI3K/Akt and the mTOR signaling pathway serve
important roles in embryonic development; however, these two
signaling pathways are frequently overactivated in tumors (29,30).
In the present study, it was demonstrated that the levels of PI3K,
Akt and mTOR phosphorylation were markedly decreased following
UBE2T knockdown in 786-O cells (Fig.
5A). Conversely, the levels of total PI3K and total AKT were
not altered; however, total mTOR expression was notably decreased.
Wortmannin, a specific Akt inhibitor, did not further reduce the
phosphorylation levels of PI3K, Akt and mTOR following UBE2T
knockdown. Additionally, the cell proliferation rate in the
siUBE2T-transfected group was not significantly different to that
in the siUBE2T/wortmannin or siNC/wortmannin groups (Fig. 5B). Conversely, oeUBE2T transfection
upregulated the expression of UBE2T compared with the control, and
increased the levels of p-PI3K, p-Akt and p-mTOR compared with siNC
(Fig. 5A); however, the effects on
phosphorylation were reversed by wortmannin, which also
significantly decreased cell proliferation even when UBE2T was
overexpressed in 786-O cells (Fig.
5B). Therefore, these data suggested that UBE2T was involved in
regulation of the activation of the PI3K/Akt and mTOR signaling
pathways in 786-O cells.
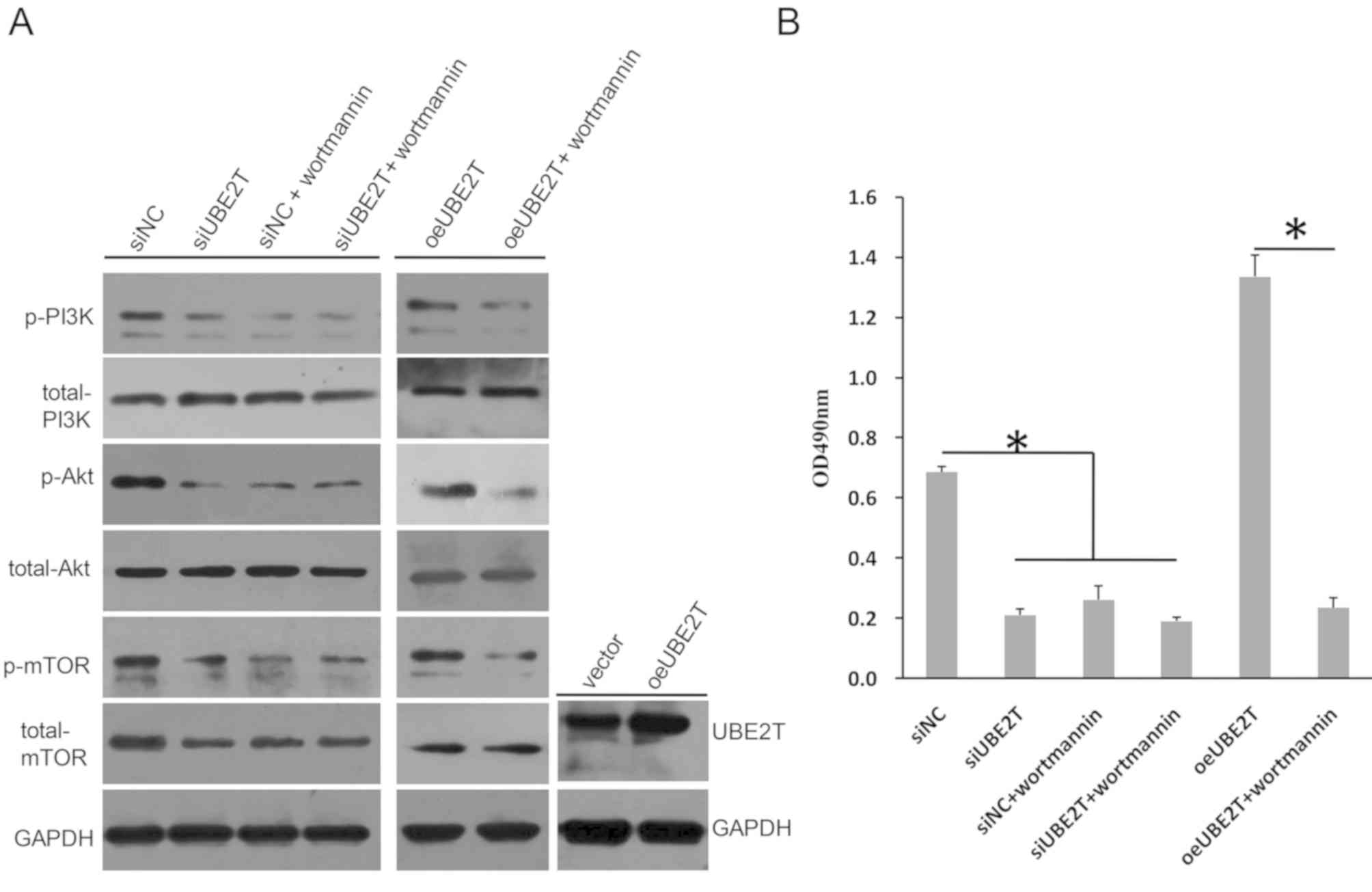 | Figure 5.UBE2T knockdown regulates
PI3K/AKT/mTOR signaling. (A) Phosphorylation levels of PI3K, Akt
and mTOR were decreased in 786-O cells following knockdown of
UBE2T, but were increased when UBE2T was overexpressed; the
expression of total PI3K or total Akt was not notably altered,
whereas total mTOR levels were markedly decreased following UBE2T
knockdown. Wortmannin reversed the effects of oeUBE2T. (B)
Proliferation of 786-O cells was inhibited by siUBE2T, but enhanced
by oeUBE2T in a Wortmannin-sensitive manner. *P<0.05. NC,
negative control; oe, overexpression plasmid; p-, phosphorylated;
si, small interfering RNA; UBE2T, ubiquitin-conjugating enzyme E2;
vector, empty vector. |
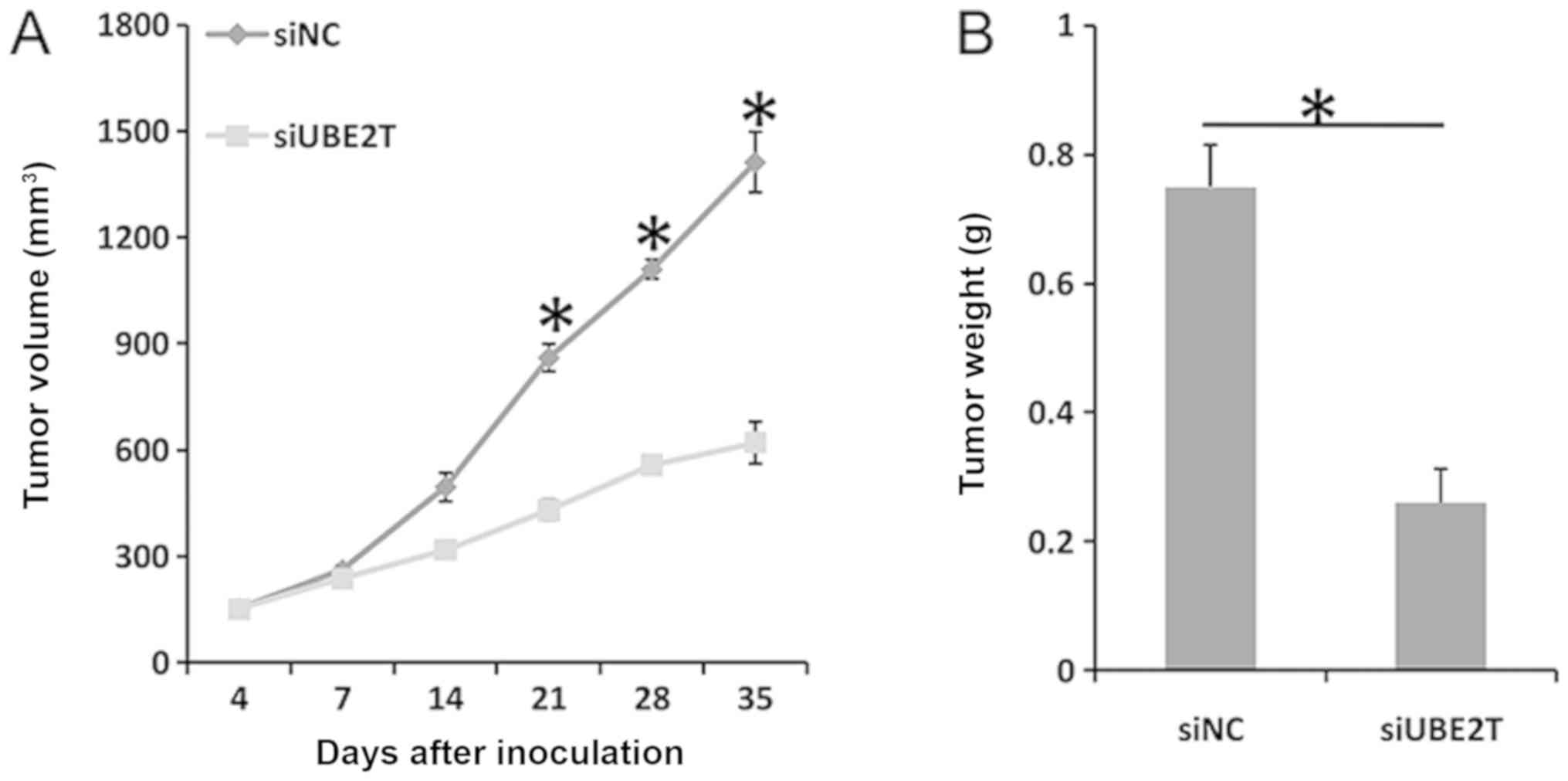
UBE2T knockdown suppresses xenograft
tumor growth in vivo
As presented in Fig.
6A, tumor growth was significantly suppressed following UBE2T
knockdown. The mean tumor volume in si-UBE2T mice was significantly
decreased compared with the control on day 35 (1,412.5
mm3 vs. 620 mm3; P<0.05). In addition, the
mean tumor weight was significantly decreased following UBE2T
knockdown (0.26 g vs. 0.75 g; P<0.05; Fig. 6B). Therefore, it was suggested that
UBE2T contributed to xenograft tumor growth in vivo.
Discussion
RCC is a common malignant tumor characterized by
poor prognosis, late diagnosis, frequent recurrence and
heterogeneity (1–3). The primary therapy for patients with
RCC is surgical removal followed by chemotherapy or radiotherapy;
however, traditional therapeutics have not improved the quality of
life of patients with RCC (31,32).
Therefore, there is a requirement for further investigation into
RCC. At present, a series of genes have been reported to serve
important roles during the tumorigenesis of RCC; however, the
critical genes vary between each report (33,34),
potentially due to the heterogeneity of RCC.
To the best of our knowledge, the present study is
the first to indicate that UBE2T may be a potential candidate
predictive factor in RCC. As aforementioned, UBE2T was
overexpressed in clinical tumor tissues and was negatively
associated with the survival of patients with RCC. The levels of
UBE2T were significantly associated with pathological
characteristics such as tumor stage and grade. These data suggested
the clinical relevance of UBE2T in RCC. This was consistent with a
previous study in which UBE2T was reported to be an independent
predictive factor in gastric cancer (35). A larger cohort of clinical RCC
samples is required to further validate this finding. UBE2T was
also reported to be upregulated in tumor cell lines compared with
that in control cells. UBE2T knockdown was reported to inhibit cell
proliferation and colony formation in 786-O cells in vitro.
In vivo, UBE2T contributed to tumor growth. Tumor cells
possess the ability to undergo potentially unlimited proliferation
(36). Therefore, it was
hypothesized that UBE2T served an important role in the progression
of RCC. This was consistent with previous findings; for example,
previous studies identified that UBE2T overexpression promotes cell
proliferation and metastasis in gastric cancer (18,35).
Metastasis is another characteristic of cancer (36); however, in the present study, UBE2T
expression did not appear to affect the migration and invasion of
786-O cells.
EMT is an important process during the
transformation of normal cells to cancer cells, which has been
reported to promote the progression of various types of cancer
(37). EMT is typically associated
with cell migration and invasion in cancer cells; however, in the
present study, altered expression of EMT markers was observed in
the absence of changes in migration or invasion following siUBE2T
transfection. Common molecular markers in EMT include E-cadherin,
N-cadherin, vimentin and fibronectin. In the study, the expression
of the epithelial markers E-cadherin and fibronectin was
upregulated following UBE2T knockdown; whereas the mesenchymal
markers N-cadherin and vimentin were downregulated, suggesting that
UBE2T regulated the expression of EMT markers. UBE2T may regulate
the ubiquitin level of EMT markers in RCC, leading to differential
degradation; however, further research is necessary to determine
the exact role of UBE2T in EMT in RCC. In conclusion, UBE2T
promoted the proliferation but not metastasis of RCC cells, and it
may be involved in EMT processes in RCC.
In the present study, it was demonstrated that UBE2T
was involved in RCC; however, the mechanisms underlying the effects
of UBE2T on cell proliferation in RCC remain unclear. PI3K/Akt is a
very important signaling pathway during embryonic development,
which frequently induces positive effects on cell growth or
proliferation. It has been reported to be overactivated in tumors,
with increased phosphorylation of PI3K and Akt acting as the
critical event (38). In the
present study, the phosphorylation levels of PI3K and Akt were
increased following UBE2T overexpression and decreased following
UBE2T knockdown. These findings suggested that UBE2T may regulate
PI3K/Akt signaling in RCC. Wortmannin, a specific inhibitor of Akt,
was demonstrated to downregulate the phosphorylation levels of PI3K
and Akt even when UBE2T was overexpressed in 786-O cells. The
proliferation-promoting role of UBE2T was also suppressed by
wortmannin. These data further supported the hypothesis that UBE2T
regulated PI3K/Akt signaling in RCC.
Hu et al reported that UBE2T activated the
Akt/glycogen synthase kinase 3β/β-catenin signaling pathway in
nasopharyngeal carcinoma (14).
UBE2T was also demonstrated to promote cell proliferation via the
regulation of PI3K/Akt signaling in osteosarcoma (17). Therefore, it was predicted that
UBE2T may activate PI3K/Akt signaling in RCC, as was observed.
Additionally, the phosphorylation levels of mTOR were regulated by
UBE2T in 786-O cells. mTOR is an important signaling molecule
during cell growth (39), which
has been demonstrated to crosstalk with PI3K/Akt signaling in
cancer cells; for example, PI3K/Akt/mTOR signaling was reported to
exhibit positive effects during tumorigenesis in medulloblastoma
and thyroid cancer (38,40). Ubiquitin conjugating enzyme E2C
(UBE2C) is another member of the ubiquitin-proteasome family that
possesses similar functions to UBE2T (41). UBE2C was reported to induce EMT via
the PI3K/Akt signaling pathway (42). Activation of PI3K/Akt/mTOR
signaling has been revealed to promote EMT in numerous types of
cancer (43–45). As aforementioned, UBE2T was
observed to be involved in the expression of EMT-associated markers
in RCC. Therefore, based on the aforementioned studies and reported
findings, it was hypothesized that UBE2T promoted proliferation and
EMT in RCC by activating the PI3K/Akt/mTOR signaling pathway;
however, further study is required to determine the molecular
interactions between UBE2T and the PI3K/Akt/mTOR signal
pathway.
In conclusion, to the best of our knowledge, this
study is the first to report that UBE2T promoted tumorigenesis in
RCC by regulating the PI3K/Akt/mTOR signaling pathway. Therefore,
UBE2T may be a novel target in the treatment of RCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
FHM designed and guaranteed the integrity of the
whole study, and reviewed the manuscript. PH participated in the
design of the study and performed the majority of the experiments.
BK was involved in the statistical analysis of all data. YPL
conducted the study of the literature, participated in the design
of major experiments and edited the manuscript. WQH was involved in
the collection and analysis of clinical data, and drafted and
revised the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Review Board of First Affiliated Hospital of Jiamusi University and
written informed consent was obtained from each patient. Animal
experiments were approved by the Animal Research Ethics Committee
of The First Affiliated Hospital of Jiamusi University.
Patient consent for publication
Each patient provided consent for the publication of
any data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bhatt JR and FinelliI A: Landmarks in the
diagnosis and treatment of renal cell carcinoma. Nat Rev Urol.
11:517–525. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kamps R, Brandao RD, Bosch BJ, Paulussen
AD, Xanthoulea S, Blok MJ and Romano A: Next-generation sequencing
in oncology: Genetic diagnosis, risk prediction and cancer
classification. Int J Mol Sci. 18:E3082017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mirza Z, Schulten HJ, Farsi HM,
Al-Maghrabi JA, Gari MA, Chaudhary AG, Abuzenadah AM, Al-Qahtani MH
and Karim S: Impact of S100A8 expression on kidney cancer
progression and molecular docking studies for kidney cancer
therapeutics. Anticancer Res. 34:1873–1884. 2014.PubMed/NCBI
|
|
6
|
Brugarolas J: Molecular genetics of
clear-cell renal cell carcinoma. J Clin Oncol. 32:1968–1976. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Alpi AF, Chaugule V and Walden H:
Mechanism and disease association of E2-conjugating enzymes:
Lessons from UBE2T and UBE2L3. Biochem J. 473:3401–3419. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Machida YJ, Machida Y, Chen Y, Gurtan AM,
Kupfer GM, D'Andrea AD and Dutta A: UBE2T is the E2 in the fanconi
anemia pathway and undergoes negative autoregulation. Mol Cell.
23:589–596. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Alpi A, Langevin F, Mosedale G, Machida
YJ, Dutta A and Patel KJ: UBE2T, the fanconi anemia core complex,
and FANCD2 are recruited independently to chromatin: A basis for
the regulation of FANCD2 monoubiquitination. Mol Cell Biol.
27:8421–8430. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Richman KA, Lach FP, Abhyankar A, Donovan
FX, Sanborn EM, Kennedy JA, Sougnez C, Gabriel SB, Elemento O,
Chandrasekharappa SC, et al: Deficiency of UBE2T, the E2 ubiquitin
ligase necessary for FANCD2 and FANCI ubiquitination, causes FA-T
subtype of fanconi anemia. Cell Rep. 12:35–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mamrak NE, Shimamura A and Howlett NG:
Recent discoveries in the molecular pathogenesis of the inherited
bone marrow failure syndrome fanconi anemia. Blood Rev. 31:93–99.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hira A, Yoshida K, Sato K, Okuno Y,
Shiraishi Y, Chiba K, Tanaka H, Miyano S, Shimamoto A, Tahara H, et
al: Mutations in the gene encoding the E2 conjugating enzyme UBE2T
cause fanconi anemia. Am J Hum Genet. 96:1001–1007. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Alter BP: Fanconi anemia and the
development of leukemia. Best Pract Res Clin Haematol. 27:214–221.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hu W, Xiao L, Cao C, Hua S and Wu D: UBE2T
promotes nasopharyngeal carcinoma cell proliferation, invasion, and
metastasis by activating the AKT/GSK3β/β-catenin pathway.
Oncotarget. 7:15161–15172. 2016.PubMed/NCBI
|
|
15
|
Liu LP, Yand M, Peng QZ, Li MY, Zhang YS,
Guo YH, Chen Y and Bao SY: UBE2T promotes hepatocellular carcinoma
cell growth via ubiquitination of p53. Biochem Biophys Res Commun.
493:20–27. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen M, Kwon Y, Wang Y, Mao JH and Wei G:
Elevated expression of UBE2T exhibits oncogenic properties in human
prostate cancer. Oncotarget. 6:25226–25239. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Leng H, Chen H, Wang L, Jiang N,
Hua X and Yu B: Knockdown of UBE2T inhibits osteosarcoma cell
proliferation, migration, and invasion by suppressing the PI3K/Akt
signaling pathway. Oncol Res. 24:361–369. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Luo C, Yao Y, Yu Z, Zhou H, Guo L, Zhang
J, Cao H, Zhang G, Li Y and Jiao Z: UBE2T knockdown inhibits
gastric cancer progression. Oncotarget. 8:32639–32654.
2017.PubMed/NCBI
|
|
19
|
Gong YQ, Peng D, Ning XH, Yang XY, Li XS,
Zhou LQ and Guo YL: UBE2T silencing suppresses proliferation and
induces cell cycle arrest and apoptosis in bladder cancer cells.
Oncol Lett. 12:4485–4492. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ueki T, Park JH, Nishidate T, Kijima K,
Hirata K, Nakamura Y and Kataqiri T: Ubiquitination and
downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T
overexpression in human breast cancer cells. Cancer Res.
69:8752–8760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ramaekers CH, van den Beucken T, Meng A,
Kassam S, Thoms J, Bristow RG and Wouters BG: Hypoxia disrupts the
fanconi anemia pathway and sensitizes cells to chemotherapy through
regulation of UBE2T. Radiother Oncol. 101:190–197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perez-pena J, Corrales-sanchez V, Amir E,
Pandiella A and Ocana A: Ubiquitin-conjugating enzyme E2T (UBE2T)
and denticleless protein homolog (DTL) are linked to poor outcome
in breast and lung cancers. Sci Rep. 7:175302017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hao J, Xu A, Xie X, Hao J, Tian T, Gao S,
Xiao X and He D: Elevated expression of UBE2T in lung cancer tumors
and cell lines. Tumour Biol. 29:195–203. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lopez-Beltran A, Scarpelli M, Montironi R
and Kirkali Z: 2004 WHO classification of the renal tumors of the
adults. Eur Urol. 49:798–805. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Chen T, You Y, Jiang H and Wang ZZ:
Epithelial-mesenchymal transition (EMT): A biological process in
the development, stem cell differentiation, and tumorigenesis. J
Cell Physiol. 232:3261–3272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Figiel S, Vasseur C, Bruyere F, Rozet F,
Maheo K and Fromont G: Clinical significance of
epithelial-mesenchymal transition markers in prostate cancer. Hum
Pathol. 61:26–32. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Scanlon CS, Van Tubergen EA, Inglehart RC
and D'Silva NJ: Biomarkers of epithelial-mesenchymal transition in
squamous cell carcinoma. J Dent Res. 92:114–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tian T, Li XY and Zhang JH: mTOR signaling
in cancer and mTOR inhibitors in solid tumor targeting therapy. Int
J Mol Sci. 20:7552019. View Article : Google Scholar :
|
|
30
|
Faes S and Dormond O: PI3K and AKT:
Unfaithful partners in cancer. Int J Mol Sci. 16:21138–21152. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berquist SW, Yim K, Ryan ST, Patel SH,
Eldefrawy A, Cotta BH, Bradshaw AW, Meagher MF, Bindayi A, McKay
RR, et al: Systemic therapy in the management of localized and
locally advanced renal cell carcinoma: Current state and future
perspectives. Int J Urol. Apr 3–2019.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wiechno P, Kucharz J, Sadowska M,
Michalski W, Sikora-Kupis B, Jonska-Gmyrek J, Poniatowska G,
Nietupski K, Ossolinski K and Demkow T: Contemporary treatment of
metastatic renal cell carcinoma. Med Oncol. 35:1562018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hsieh JJ, Purdue MP, Signoretti S, Swanton
C, Albiges L, Schmidinger M, Heng DY, Larkin J and Ficarra V: Renal
cell carcinoma. Nat Rev Dis Primers. 3:170092017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
D'Avella C, Abbosh P, Pal SK and Geynisman
DM: Mutations in renal cell carcinoma. Urol Oncol. Nov
23–2018.(Epub ahead of print). View Article : Google Scholar
|
|
35
|
Yu H, Xiang P, Pan Q, Huang Y, Xie N and
Zhu W: Ubiquitin-conjugating enzyme E2T is an independent
prognostic factor and promotes gastric cancer progression. Tumour
Biol. 37:11723–11732. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Brabletz T, Kalluri R, Nieto MA and
Weinberg RA: EMT in cancer. Nat Rev Cancer. 18:128–134. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Dimitrova V and Arcaro A: Targeting the
PI3K/Akt/mTOR signaling pathway in medulloblastoma. Curr Mol Med.
15:82–93. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cargnello M, Tcherkezian J and Roux PP:
The expanding role of mTOR in cancer cell growth and proliferation.
Mutagenesis. 30:169–176. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Manfredi GI, Dicitore A, Gaudenzi G,
Caraglia M, Persani L and Vitale G: PI3K/Akt/mTOR signaling in
medullary thyroid cancer: A promising molecular target for cancer
therapy. Endocrine. 48:363–370. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Xie C, Powell C, Yao M, Wu J and Dong Q:
Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int
J Biochem Cell Biol. 47:113–117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang R, Song Y, Liu X, Wang Q, Wang Y, Li
L, Kang C and Zhang Q: UBE2C induces EMT through Wnt/βcatenin and
PI3K/Akt signaling pathways by regulating phosphorylation levels of
aurora-a. Int J Oncol. 50:1116–1126. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin Y, Xu K, Chen Q, Wang B, Pan J, Huang
S, Wei Y and Ma H: Simvastatin inhibits the development of
radioresistant esophageal cancer cells by increasing the
radiosensitivity and reversing EMT process via the PTEN-PI3K/AKT
pathway. Exp Cell Res. 362:362–369. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Rumman M, Jung KH, Fang Z, Yan HH, Son MK,
Kim SJ, Kim J, Park JH, Lim JH, Hong S and Hong SS: HS-173, a novel
PI3K inhibitor suppresses EMT and metastasis in pancreatic cancer.
Oncotarget. 7:78029–78047. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang GW, Tian X, Li Y, Wang ZQ, Li XD and
Zhu CY: Down-regulation of ETS2 inhibits the invasion and
metastasis of renal cell carcinoma cells by inducing EMT via the
PI3K/Akt signaling pathway. Biomed Pharmacother. 104:119–126. 2018.
View Article : Google Scholar : PubMed/NCBI
|