Introduction
Colorectal carcinoma (CRC) is a common malignancy of
the digestive tract (1), the
incidence of which is increasing annually in China (2). If patients with CRC metastasis in
advanced stages are not promptly treated, their average survival is
only 5–6 months (3,4), and the main causes of CRC-associated
mortality are invasion and metastasis (5,6).
Therefore, it is particularly important to identify novel tumor
markers to inhibit tumor metastasis.
In the occurrence and development of invasive
carcinoma from atypical hyperplasia and carcinoma in situ,
the destruction of epithelial integrity is an important event
(7). The first stage of tumor
invasion and metastasis includes destruction of the intact
epithelial structure and acquisition of stromal cell
characteristics; this process is known as epithelial-mesenchymal
transition (EMT). EMT is a phenomenon during which epithelial cells
convert to interstitial cells under specific physiological and
pathological conditions (8). It
has been reported that EMT is closely associated with tumor
invasion and metastasis, and that it serves a key role in in
situ infiltration and distant metastasis of various types of
cancer (9,10). Various cancer cells can undergo
partial or complete EMT (11–13).
MicroRNAs (miRNAs/miRs) are a series of endogenous
non-coding small RNA molecules, usually 18–25 nt in length
(14). miRNAs suppress protein
translation through binding to the 3′-untranslated region (3′UTR)
of target gene mRNA (15,16), miRNAs serve a key role in
translation inhibition following gene transcription. In recent
years, increasing experimental evidence has demonstrated that
abnormal miRNA expression in tumor cells is closely associated with
the occurrence of tumors. Different degrees of abnormal miRNA
expression (17–20) can be detected in all types of human
cancer, including CRC. In addition, miRNAs have been reported to
exert a strong regulatory effect on EMT (21–23).
Notably, miR-214 is considered a key hub that controls tumor
networks (24); however, the role
and mechanism of miR-214 in the development of CRC are currently
unclear.
In the present study, the expression of miR-214 was
detected in CRC tissues, and its target gene was identified.
Furthermore, the effects of miR-214 on viability and motility of
CRC cells were determined, and the underlying molecular mechanism
was analyzed.
Materials and methods
Tissue source
Between November 2016 and December 2017, 36 CRC and
adjacent normal tissues were collected from patients with CRC that
were admitted to The Affiliated Dongtai Hospital of Nantong
University (Dongtai, China). Written informed consent was obtained
from patients permitting their tissues to be used. The present
study was approved by the Ethics Committee of The Affiliated
Dongtai Hospital of Nantong University (approval no.
201501218).
Cell line
The LoVo human colon adenocarcinoma cell line
(American Type Culture Collection, Manassas, VA, USA) was
maintained in RPMI-1640 medium (M&C Gene Technology Co., Ltd.,
Beijing, China) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at 37°C in
an incubator containing 5% CO2.
Cell transfection
miR-214 mimics and miRNA mimics control were
purchased from Genomeditech (Shanghai, China). miR-214 mimics
sense, 5′-ACAGCAGGCACAGACAGGCAGU-3′, and antisense,
5′-UGCCUGUCUGUGCCUGCUGUUU-3′; mimics control sense,
5′-UUCUCCGAACGUGUCACGUTT-3′, and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. Using Lipofectamine® 3000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.), miR-214
mimics (50 nM; mimic group) or miRNA mimics control (50 nM; mock
group) were transfected into LoVo cells (60–80% confluence) at 37°C
for 24 h, and the control group were treated with PBS. In addition,
cells were treated with 50 ng/ml insulin-like growth factor-1
(IGF-1; AmyJet Scientific, Wuhan, China) for 24 h at 37°C, in order
to activate phosphoinositide 3-kinase (PI3K) (25).
Dual luciferase reporter assay
TargetScan version 7.2 (http://www.targetscan.org/vert_72/) was used to
predict the binding site between the 3′-UTR of transglutaminase 2
(TGM2) and miR-214. For the dual luciferase reporter experiments,
the 3′-UTR of TGM2 gene and a mutant (mut) 3′-UTR of TGM2, which
was constructed using a Site-Directed Mutagenesis kit (Stratagene;
Agilent Technologies, Inc., Santa Clara, CA, USA) according to
manufacturer's protocol, were amplified by PCR. The thermocycling
conditions were as follows: Initial denaturation at 95°C for 30
sec, followed by 18 cycles of denaturation at 95°C for 30 sec, 55°C
for 1 min and 68°C for 2 min, with a final elongation step at 68°C
for 5 min. Both PCR products were cloned into the psiCHECK-2 vector
(Promega Corporation, Madison, WI, USA) to generate TGM2-3′-UTR
plasmids, and the TGM2-3′-UTR mut plamids, respectively. Then, 293
cells (American Type Culture Collection, Manassas, VA, USA)
cultured in DMEM (HyClone; GE Healthcare Life Sciences; Logan, UT,
USA) with 10% FBS, 100 U/ml penicillin and 100 µg/ml streptomycin
at 37°C in an incubator with 5% CO2, were co-transfected
with miR-214 mimics/miRNA mimics (50 nM) and
TGM2-3′-UTR/TGM2-3′-UTR mut plasmids (50 ng/µl) using
Lipofectamine® 3000 reagent at 37°C for 24 h. After 24
h, the cells were lysed with 1X passive lysis buffer (50 µl) for 15
min at room temperature, and the suspension was then transferred
into a black enzyme plate. Then, the Luciferase assay reagent II
(100 µl) and 1X Stop&Glo® reagent (100 µl) (Promega
Corporation) were added to the cells, and luciferase activity was
detected using the GloMax® Discover Multimode Microplate
Reader (cat. no. GM3000; Promega Corporation) according to the
manufacturer's instructions. Luciferase activity was normalized to
Renilla luciferase.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from cells and tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). RT was conducted to synthesize cDNA from 2 µg RNA using H
BeyoRT II First Strand cDNA Synthesis kit (Beyotime Institute of
Biotechnology, Haimen, China), according to manufacturer's
protocol. The primer sequences are listed in Table I. cDNA was amplified using SYBR
Green qPCR Master Mix (MedChemExpress, Monmouth Junction, NJ, USA).
The conditions of amplification were as follows: Pre-denaturation
at 95°C for 10 sec, followed by 30 cycles of denaturation at 95°C
for 5 sec and 62°C for 25 sec, and a final elongation step at 70°C
for 30 min. The internal controls were U6 and GAPDH. Gene
expression was calculated and quantified using the
2−ΔΔCq method (26).
 | Table I.Primer sequences for reverse
transcription-quantitative polymerase chain reaction. |
Table I.
Primer sequences for reverse
transcription-quantitative polymerase chain reaction.
| Primer name | Sequence
(5′-3′) | Product size
(bp) |
|---|
|
miR-214-Forward |
ATAGAATTCTTTCTCCCTTTCCCCTTACTCTCC |
|
|
miR-214-Reverse |
CCAGGATCCTTTCATAGGCACCACTCACTTTAC | 235 |
| TGM2-Forward |
CCGAGGAGCTGGTCTTAGAG |
|
| TGM2-Reverse |
TCTTAGTGGAAAACGGGCCT | 236 |
|
E-cadherin-Forward |
TTTGAAGATTGCACCGGTCG |
|
| E-cadherin-
Reverse |
CAGCGTGACTTTGGTGGAAA | 180 |
| TIMP-2-Forward |
AGCACCACCCAGAAGAAGAG |
|
| TIMP-2-Reverse |
TGATGCAGGCGAAGAACTTG | 175 |
| MMP-2-Forward |
TGGCTACACACCTGATCTGG |
|
| MMP-2-Reverse |
GAGTCCGTCCTTACCGTCAA | 184 |
| MMP-9-Forward |
GAGACTCTACACCCAGGACG |
|
| MMP-9-Reverse |
GAAAGTGAAGGGGAAGACGC | 238 |
| U6-Forward |
CTCGCTTCGGCAGCACA |
|
| U6-Reverse |
AACGCTTCACGAATTTGCGT | 215 |
| GAPDH-Forward |
CCATCTTCCAGGAGCGAGAT |
|
| GAPDH-Reverse |
TGCTGATGATCTTGAGGCTG | 222 |
Western blot analysis
Total proteins were extracted from cells and tissues
using radioimmunoprecipitation assay buffer (Beijing Solarbio
Science & Technology Co., Ltd., Beijing, China). Protein
concentration was analyzed using Pierce Bicinchoninic Acid Protein
Assay kit (Pierce; Thermo Fisher Scientific, Inc.). The protein
lysate (25 µg) was then separated by 10% SDS-PAGE and transferred
to a polyvinylidene fluoride membrane (EMD Millipore, Billerica,
MA, USA). Subsequently, 5% non-fat milk was applied to block the
membrane at 37°C for 60 min, and the membrane was incubated with
anti-TGM2 (cat. no. ab216018, 1:800; Abcam, Cambridge, MA, USA),
anti-tissue inhibitor of metalloproteinases-2 (TIMP-2; cat. no.
ab180630, 1:1,000; Abcam), anti-matrix metalloproteinase (MMP)-2
(cat. no. ab37150, 1:1,200; Abcam), anti-MMP-9 (cat. no. ab73734,
1:600; Abcam), anti-E-cadherin (cat. no. ab15148, 1:800; Abcam),
anti-phosphorylated (p)-PI3K (cat. no. ab138364, 1:600; Abcam),
anti-PI3K (cat. no. MAB2686, 1:600; R&D Systems, Inc.),
anti-p-protein kinase B (Akt; cat. no. MAB887, 1:800; R&D
Systems, Inc.), anti-Akt (cat. no. MAB2055, 1:800; R&D Systems,
Inc.) and anti-GAPDH (cat. no. ab181602, 1:600; Abcam) at 4°C
overnight. The membrane was then incubated with corresponding
secondary antibodies [goat anti-mouse immunoglobulin (Ig) G
H&L, cat. no. ab6708, 1:6,000; goat anti-rabbit IgG H&L
(horseradish peroxidase), cat. no. ab6721, 1:7,000; Abcam) at 37°C
for 60 min. The proteins were visualized using an enhanced
chemiluminescence system (GE Healthcare, Chicago, IL, USA). The
protein levels were quantified using Bio-Rad ChemiDoc system with
Image Lab software (version 6.0 Bio-Rad Laboratories, Inc.,
Hercules, CA, USA).
Cell Counting kit-8 (CCK-8)
analysis
LoVo cells were inoculated in a 96-well plate
(3×103 cell/well) in an incubator at 37°C for 24 h.
After culturing, cells were treated with PBS (control group), or
were transfected with miRNA mimics (mock group) or miR-214 mimics
(mimics group) for 48 h. Subsequently, 10 µl CCK-8 reagent
(Beyotime Institute of Biotechnology) was added into each well, and
cells were incubated with CCK-8 reagent for 4 h at 37°C. Cell
viability was determined at 450 nm using a microplate absorbance
spectrophotometer (Bio-Rad Laboratories, Inc.).
Transwell analysis
Migration and invasion of cells were determined
using Transwell analysis. Cell invasion was measured using
Matrigel-coated Transwell chambers (BD Biosciences, Franklin Lakes,
NJ, USA), and cell migration was measured using uncoated Transwell
chambers (BD Biosciences). Invasion assay was performed using
Transwell inserts pre-coated with Matrigel (BD Biosciences).
RPMI-1640 medium supplemented with 12% FBS was added to the lower
chamber. The treated cells were digested with trypsin and a cell
(2×105 cells/ml) suspension was incubated in the upper
chamber at 37°C for 24 h. Subsequently, the cells were fixed with
4% paraformaldehyde at room temperature for 15 min and stained with
0.1% crystal violet for 20 min at room temperature. The stained
cells were observed under a light microscope (magnification,
×200).
Statistical analysis
SPSS 20.0 software was used to conduct statistical
analysis. Data are presented as the means ± standard deviation.
Differences among the groups were analyzed by one-way analysis of
variance followed by Tukey test. Differences between two groups
were analyzed by Student's t-test. The association between TGM2
expression and the clinicopathological features of patients with
CRC was investigated using χ2 test. The correlation
between miR-214 and TGM2 expression in CRC was determined using the
two-tailed Spearman nonparametric correlation test. P<0.05 was
considered to indicate a statistically significant difference. All
experiments were repeated independently at least three times.
Results
TGM2 was the target gene for miR-214
in CRC
The expression levels of miR-214 and TGM2 were
detected in CRC and adjacent normal tissues using RT-qPCR and
western blotting. Compared with in adjacent normal tissues, the
expression levels of miR-214 were decreased in CRC tissues;
however, TGM2 expression was increased (Fig. 1A-C). Two-tailed Spearman
nonparametric correlation test identified a negative correlation
between miR-214 and TGM2 expression in CRC tissues (Fig. 1D). When LoVo cells were transfected
with miR-214 mimics, miR-214 expression was enhanced, whereas the
mRNA and protein expression levels of TGM2 were suppressed
(Fig. 1E-G). According to
TargetScan (http://www.targetscan.org/vert_72/), miR-214 possessed
TGM2 binding sites (Fig. 1H);
therefore, the binding abilities of miR-214 and TGM2 were tested
using a dual luciferase reporter assay. Although relative
luciferase activity was reduced in cells exposed to miR-214 mimics
and TGM2-3′UTR, it remained relatively stable in cells
co-transfected with miR-214 + TGM2-3′UTR mut (Fig. 1I). Furthermore, TGM2 expression was
divided into a low expression group and a high expression group
according to the median value of relative expression, and the
results of a χ2 test revealed that TGM2 expression was
associated with lymph node metastasis; however, it was not
associated with age, sex, tumor location or tumor type (Table II).
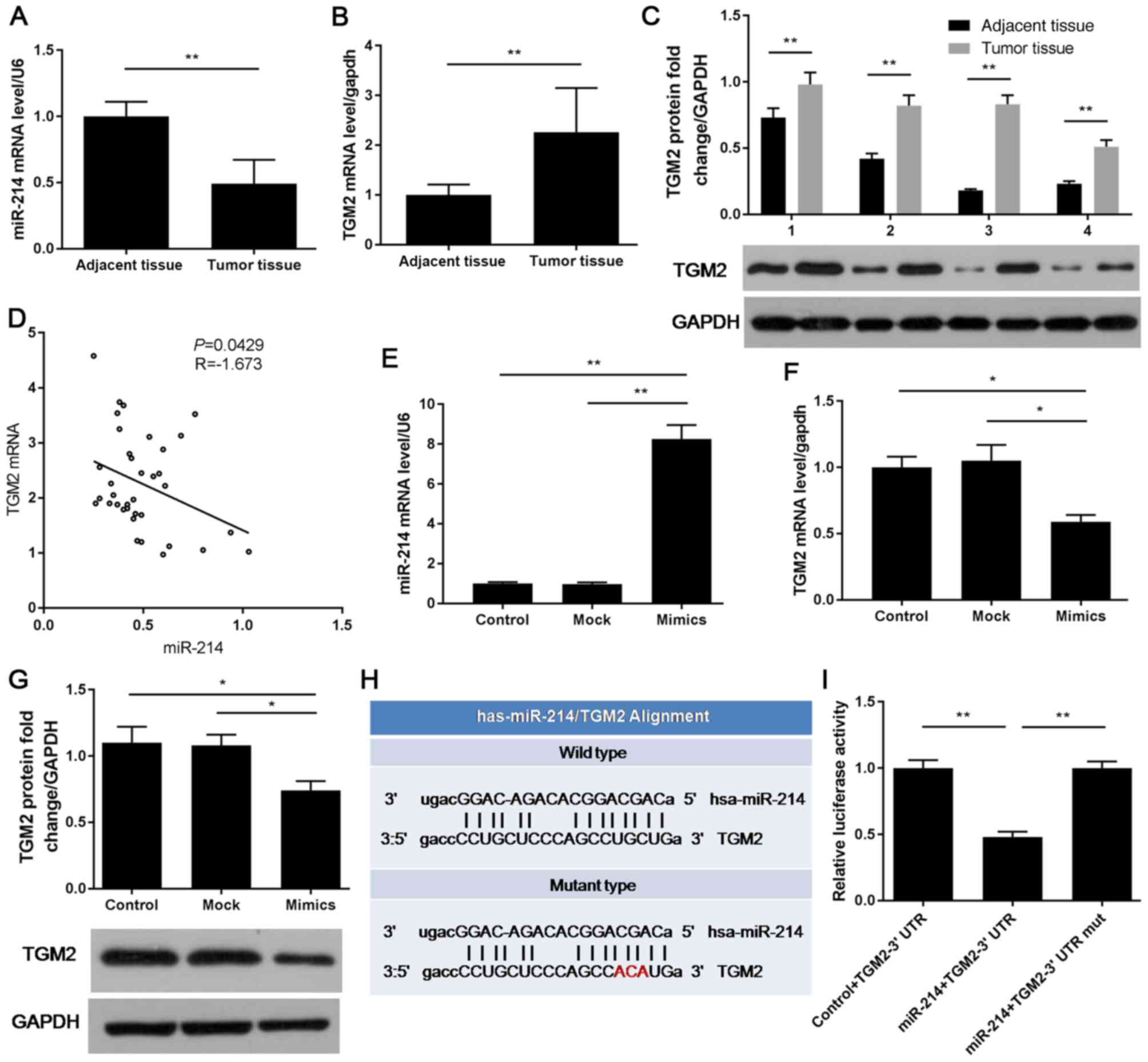 | Figure 1.TGM2 is a target gene of miR-214 in
CRC. (A-C) mRNA and protein expression levels of miR-214 and TGM2
in CRC and adjacent normal tissues, as determined by (A and B)
RT-qPCR and (C) western blotting. (D) Correlation between miR-214
and TGM2 expression in CRC, as determined by two-tailed Spearman
nonparametric correlation test. (E-G) LoVo cells were treated with
PBS (control group), or were transfected with miRNA mimics (mock
group) or miR-214 mimics (mimics group). Expression levels of
miR-214 and TGM2 were examined by (E and F) RT-qPCR and (G) western
blotting. (H) TGM2 binding site in miR-214 was predicted by
TargetScan. (I) Luciferase activity was measured by dual luciferase
reporter assay. *P<0.05, **P<0.01. 3′UTR, 3′-untranslated
region; CRC, colorectal carcinoma; miR-214, microRNA-214; mut,
mutant; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction; TGM2, transglutaminase 2. |
 | Table II.Association between TGM2 expression
and the clinicopathological features of patients with colorectal
cancer. |
Table II.
Association between TGM2 expression
and the clinicopathological features of patients with colorectal
cancer.
| Variables | Low TGM2 expression
(n=17) | High TGM2
expression (n=19) | P-value |
|---|
| Age (years) |
|
| 0.738 |
|
<60 | 8 | 10 |
|
|
≥60 | 9 | 9 |
|
| Sex |
|
| 0.549 |
|
Male | 10 | 13 |
|
|
Female | 7 | 6 |
|
| Tumor location |
|
| 0.955 |
|
Colon | 10 | 11 |
|
|
Rectum | 7 | 8 |
|
| Tumor size |
|
| 0.271 |
| <30
mm | 15 | 14 |
|
| ≥30
mm | 2 | 5 |
|
| Lymph node
metastasis |
|
| 0.019a |
| No | 12 | 6 |
|
|
Yes | 5 | 13 |
|
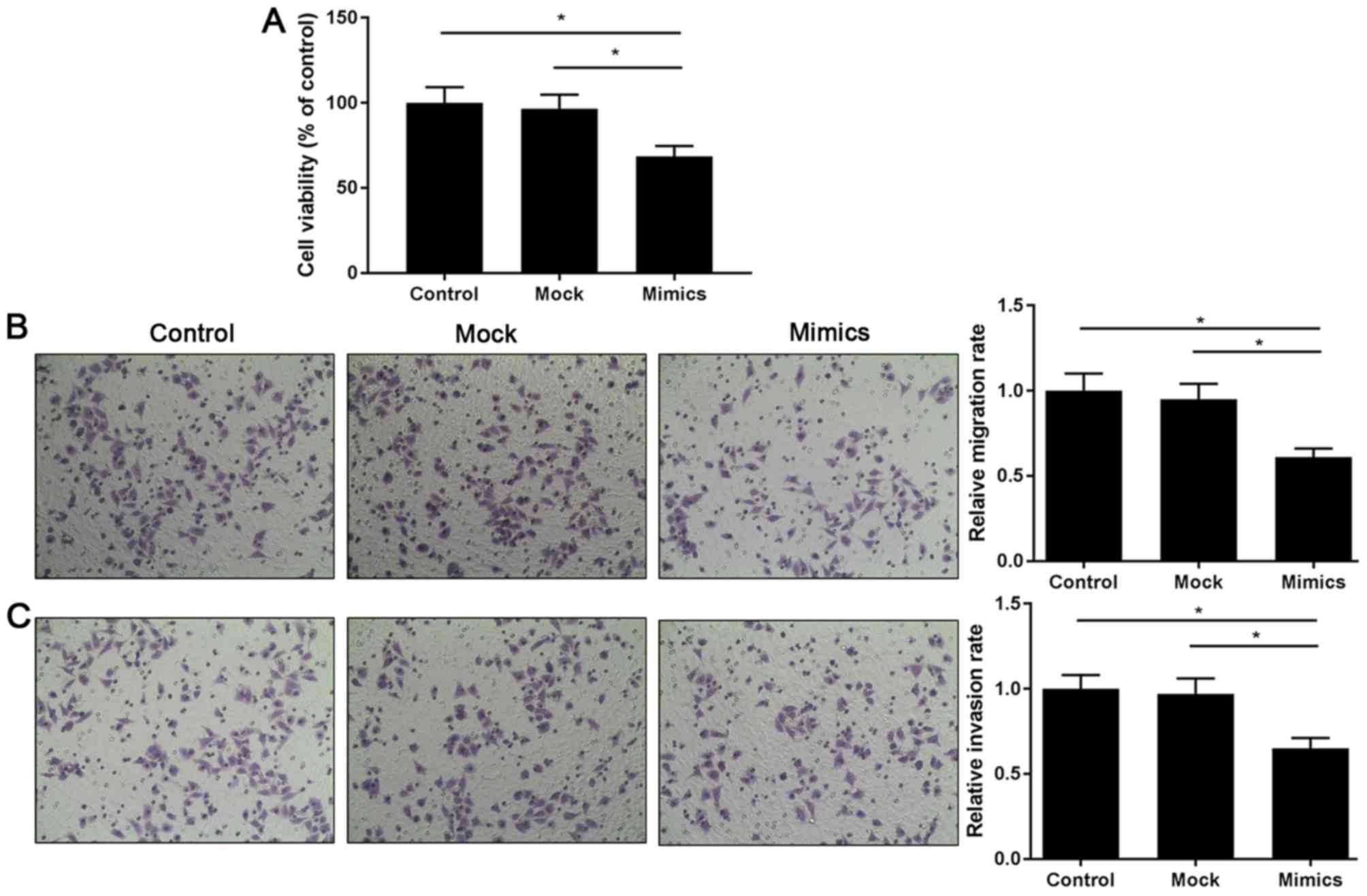
miR-214 suppresses the viability,
migration and invasiveness of LoVo cells
CCK-8 and Transwell assays were performed to explore
the effect of miR-214 on the viability, migration and invasion of
LoVo cells. CCK-8 results revealed that miR-214 overexpression
inhibited cell viability compared with in the mock group (Fig. 2A). Transwell results demonstrated
that when cells were exposed to miR-214 mimics, the relative rates
of migration (Fig. 2B) and
invasion (Fig. 2C) were markedly
reduced compared with in the mock group.
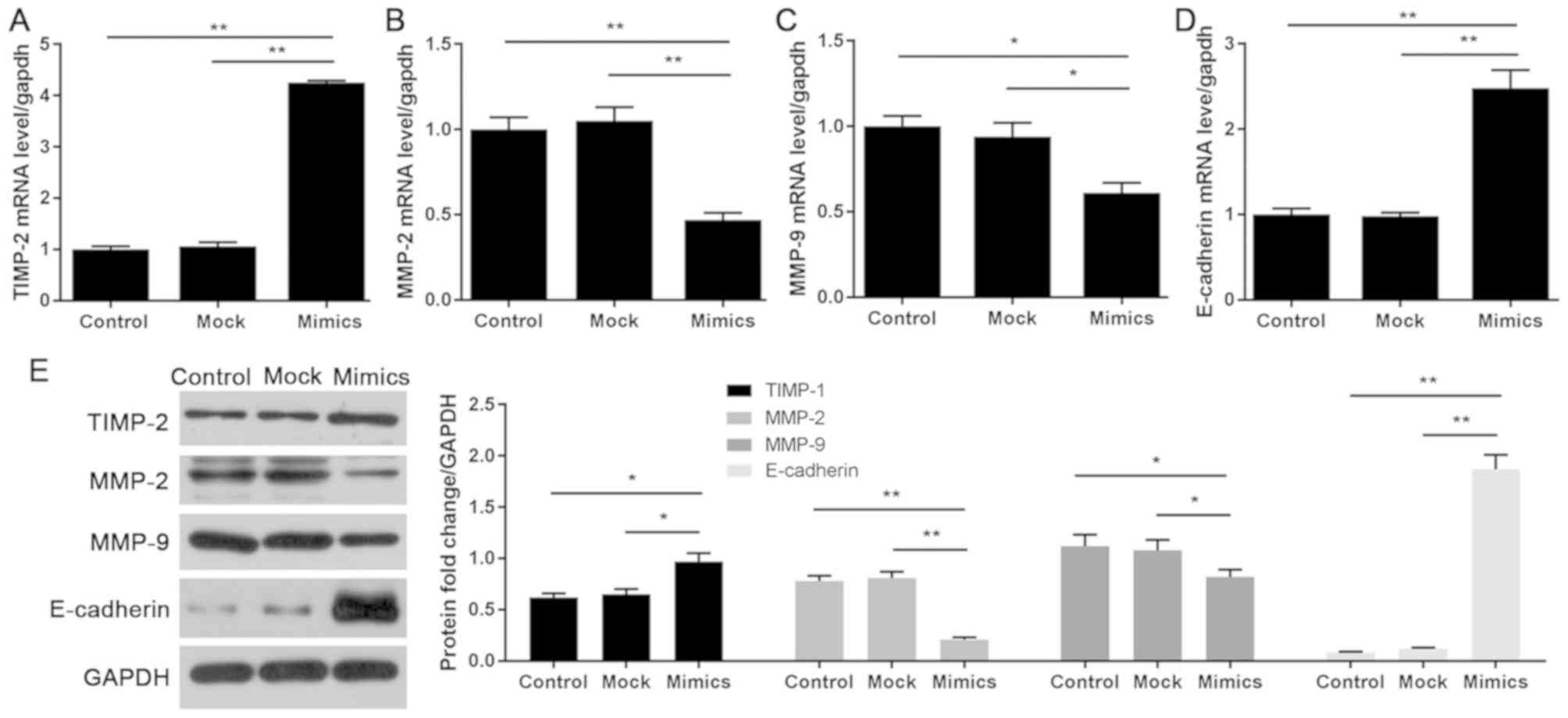
miR-214 regulates EMT-associated
factors in LoVo cells
To examine the molecular mechanism underlying the
effects of miR-214 on cell migration and invasion, the expression
levels of EMT-associated factors (TIMP-2, MMP-2, MMP-9 and
E-cadherin) were detected by RT-qPCR and western blotting. As
determined by RT-qPCR, the expression levels of TIMP-2 and
E-cadherin were enhanced in the mimics group; however, MMP-2 and
MMP-9 expression was inhibited compared with in the mock group
(Fig. 3A-D). In addition, as
determined by western blotting, miR-214 overexpression
significantly suppressed the protein expression levels of MMP-2 and
MMP-9, but promoted TIMP-2 and E-cadherin expression (Fig. 3E).
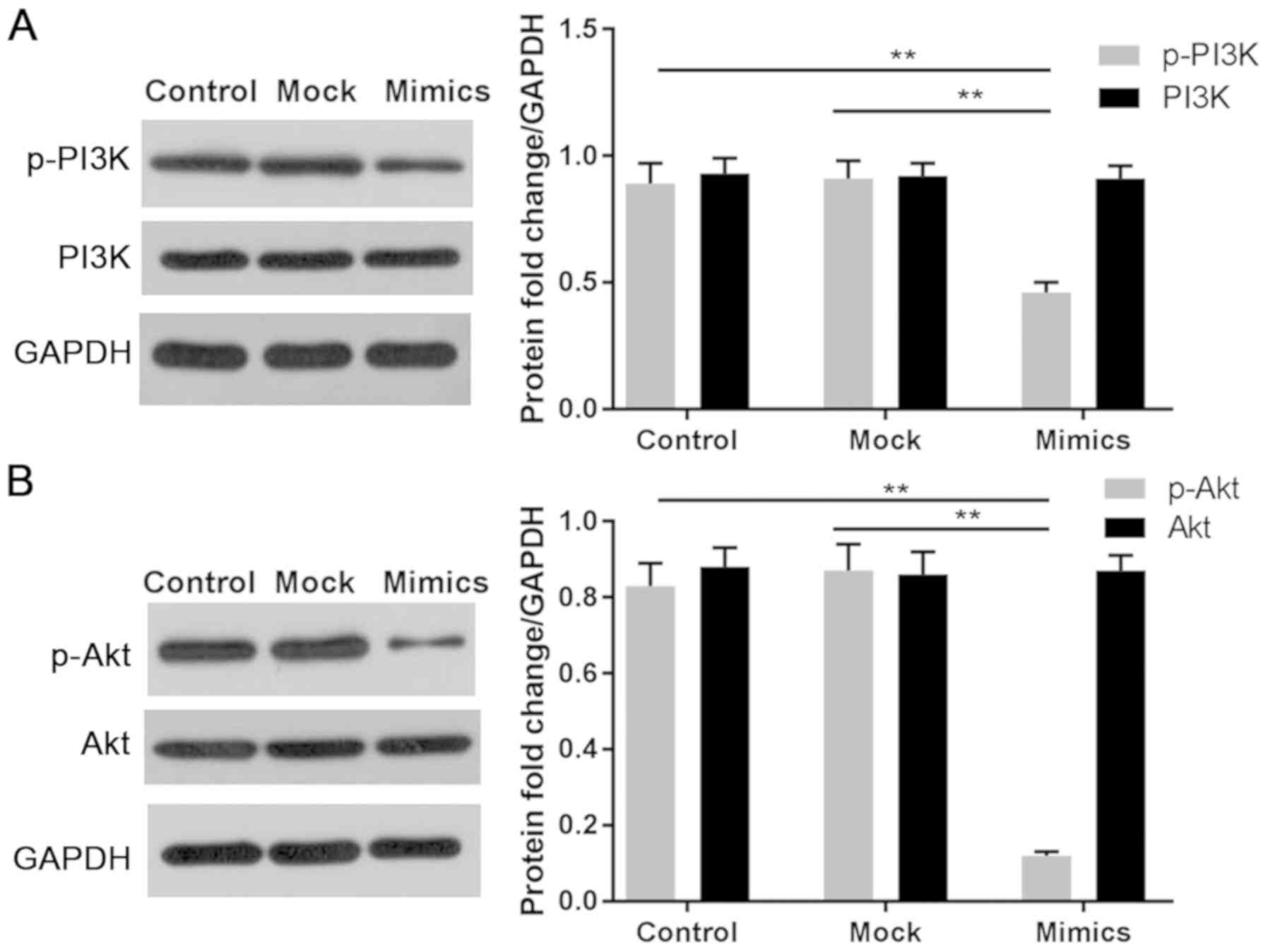
miR-214 blocks the PI3K/Akt signaling
pathway in LoVo cells
In order to assess the effects of miR-214 on
signaling in LoVo cells, the PI3K/Akt signaling pathway was
analyzed by western blotting. The results revealed that
phosphorylation of PI3K and Akt were significantly reduced when
cells were exposed to miR-214 mimics, compared with in the control
and mock groups (P<0.01). Nevertheless, the protein expression
levels of total PI3K and Akt remained stable in all groups
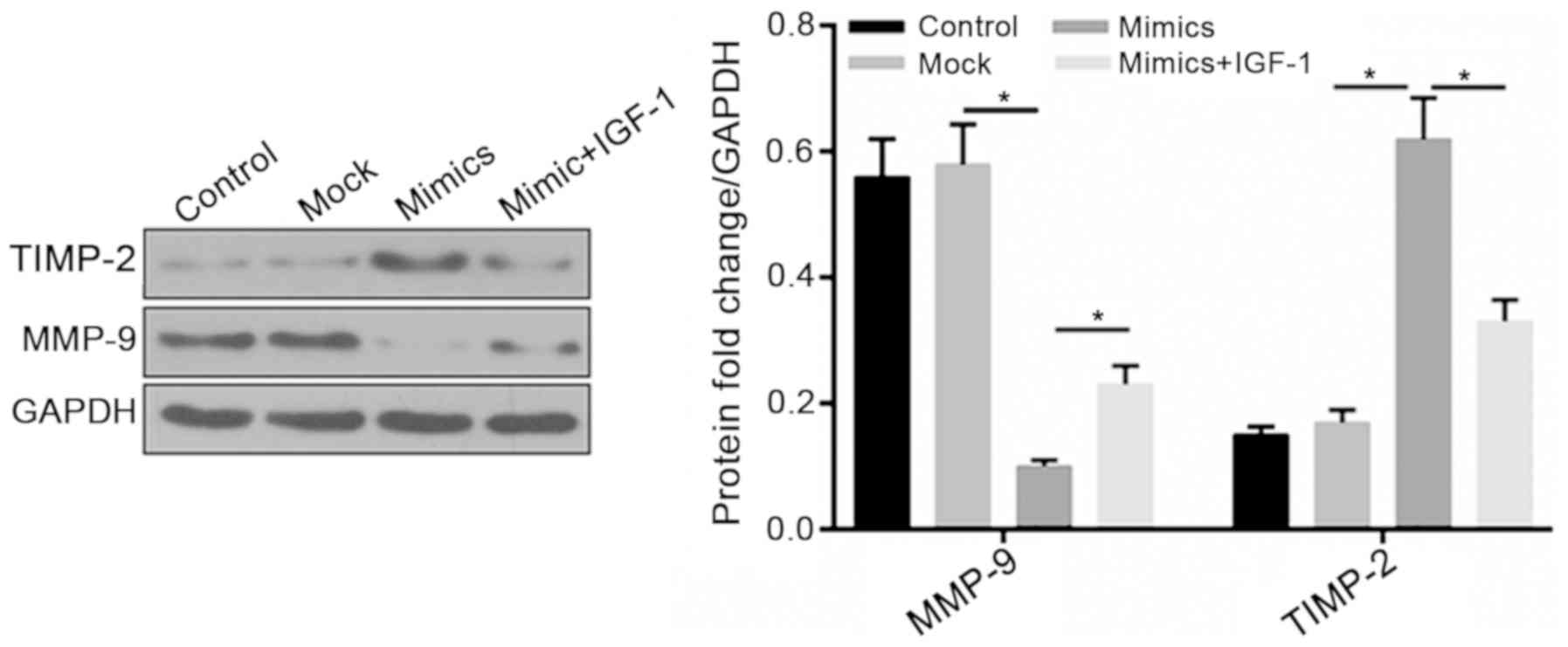
(Fig. 4A and B). In addition, the
PI3K activator IGF-1 was used to further detect the role of
PI3K/Akt signaling; the results revealed that MMP-9 expression was
upregulated, whereas TIMP-2 expression was decreased in the IGF-1 +
mimic group compared with in the mimic group. These findings
indicated that the effects of miR-214 on cell invasion and
migration may be partly associated with inhibition of PI3K/Akt
signaling (Fig. 5).
Discussion
miR-214 has been reported to be particularly active
in cancer, being abnormally expressed in several types of malignant
tumors (27–29). However, the expression of miR-214
varies in different tumors, and has certain tumor specificity.
Specifically, miR-214 has been demonstrated to have a high
expression in pancreatic cancer, oophoroma and melanoma (30–32),
and a low expression in esophageal squamous cell carcinoma, liver
cancer, breast cancer, cervical carcinoma and CRC (33–37).
In the present study, low miR-214 expression was detected in CRC
tissues, which was in accordance with the results of previous
studies (33,38).
As a member of the transglutaminase family, TGM2 is
composed of 687 amino acid residues, and is a calcium-dependent
multifunctional protein with a molecular weight of 78 kD (39). Numerous studies have demonstrated
that TGM2 expression is closely associated with the proliferation
and metastasis of various malignant tumor cells (40–43).
TGM2 expression is increased in breast cancer, ovarian cancer, lung
cancer and CRC (44–47). Similar to previous findings
(45), the present data also
demonstrated that TGM2 expression was increased in CRC tissues. It
is well known that different miRNAs can regulate the same mRNA
molecule, and that the same miRNA can regulate numerous mRNA
molecules. In this study, the results of a two-tailed Spearman
nonparametric correlation test identified a negative correlation
between miR-214 and TGM2 expression in CRC. Furthermore,
overexpression of miR-214 significantly inhibited the expression
levels of TGM2. Therefore, it may be speculated that TGM2 acts as a
target gene for miR-214 in CRC. According to TargetScan, miR-214
possesses a binding site to TGM2. Additionally, luciferase activity
was reduced in cells co-transfected with miR-214 + TGM2-3′UTR;
however, it remained stable in cells co-transfected with miR-214 +
TGM2-3′UTR mut compared with in the control + TGM2-3′UTR group.
These results confirmed that TGM2 was a target of miR-214.
Previous studies have reported that abnormal miRNA
expression exists in several human diseases, and that it serves a
pivotal role in the occurrence, development, invasion, metastasis
and angiogenesis of cancer (15,18–20).
Furthermore, miR-214 participates in the growth, invasion and
metastasis of cancer. Lu et al (48) demonstrated that miR-214 suppresses
cell invasion and migration in esophageal squamous cell cancer.
Zhao et al (49) also
revealed that miR-214 inhibits the growth and metastasis of lung
cancer cells, and Schwarzenbach et al (35) demonstrated that miR-214 inhibits
the proliferation and metastasis of cervical cancer and CRC cells.
Similarly, the present data demonstrated that miR-214 markedly
suppressed the viability, invasiveness and migration of LoVo cells.
Notably, decreased cell invasion and migration may be partially
caused by inhibited cell viability.
Li et al (50) reported that decreased expression of
miR-214 promotes intrahepatic cholangiocarcinoma metastasis via
regulating EMT-related factors (Twist and E-cadherin). Cai et
al (51) also demonstrated
that miR-194 increases cell metastasis and modulates the EMT
process by reducing E-cadherin levels and increasing MMP-2 levels
in CRC. Another study observed that miR-29b inhibits EMT via
affecting the expression of E-cadherin, MMPs and TIMPs (52). Therefore, it was hypothesized that
miR-214 may suppress the invasion and migration of LoVo cells via
regulating EMT-associated factors (E-cadherin, MMPs and TIMPs). As
expected, this study revealed that elevated expression of miR-214
markedly increased the expression levels of TIMP-2 and E-cadherin,
and reduced MMP-2 and MMP-9 expression. The results suggested that
miR-214 inhibited cell invasion and migration through
downregulating MMP-2 and MMP-9 expression and by upregulating
TIMP-2 and E-cadherin expression.
The PI3K/Akt signaling pathway has been increasingly
studied in CRC. The pathway serves a critical role in the
development of CRC and may be used for the development of novel
drugs: PI3K/Akt pathway may potentially be used as a drug target
for CRC, as pathway inhibitors may promote apoptosis and inhibit
proliferation of CRC cells. Activation of this pathway can inhibit
the apoptosis of CRC cells, and promote cell proliferation,
invasion and metastasis (53–55).
A previous have reported that GDC-0941, a novel class I PI3K
inhibitor, can enhance the efficacy of docetaxel by increasing
drug-induced apoptosis in breast cancer models (56). Abubaker et al (53) reported that activation of the
PI3K/Akt pathway stimulates cell growth in CRC. Song et al
(55) demonstrated that miR-532
attenuates PI3K/Akt signaling to suppress the progression of CRC
(55). Jia et al (57) confirmed that miR-182 and miR-135b
suppress cell proliferation and motility through inhibiting the
PI3K/Akt pathway in CRC. Therefore, in this study, the effects of
miR-214 on the PI3K/Akt pathway were examined in LoVo cells via
western blotting. The results confirmed that miR-214 suppressed
activation of PI3K/Akt signaling. Furthermore, activation of
PI3K/Akt reversed the effects of miR-214 on the expression levels
of MMP-9 and TIMP-2. Therefore, these findings suggested that
inhibition of the PI3K/Akt pathway may be associated with the
antitumor effect of miR-214.
In conclusion, the present results demonstrated that
TGM2 was a target gene for mi-214, and that a negative correlation
existed between miR-214 and TGM2 expression in CRC. miR-214
markedly suppressed the viability, migration and invasion of CRC
cells, which was associated with downregulation of the PI3K/Akt
signaling pathway. These findings indicated that miR-214 may be
considered a novel target for CRC therapy.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HS wrote the manuscript. HS, XFZ and CJC performed
the experiments and data analysis. HS and CJC designed the study
and revised the manuscript. All authors reviewed the
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Affiliated Dongtai Hospital of Nantong University.
Patients provided written informed consent permitting their tissues
to be used.
Patient consent for publication
Informed consent was obtained from all participants
for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Peng BJ, Cao CY, Li W, Zhou YJ, Zhang Y,
Nie YQ, Cao YW and Li YY: Diagnostic performance of intestinal
fusobacterium nucleatum in colorectal cancer: A meta-analysis. Chin
Med J (Engl). 131:1349–1356. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM, Bray F, Ferlay J and Pisani P:
Global cancer statistics, 2002. CA Cancer J Clini. 55:74–108. 2005.
View Article : Google Scholar
|
|
3
|
Liao Y, Li S, Chen C, He X, Lin F, Wang J,
Yang Z and Lan P: Screening for colorectal cancer in Tianhe,
Guangzhou: Results of combining fecal immunochemical tests and risk
factors for selecting patients requiring colonoscopy. Gastroenterol
Rep (Oxf). 6:132–136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun J, Hu J, Wang G, Yang Z, Zhao C, Zhang
X and Wang J: LncRNA TUG1 promoted KIAA1199 expression via miR-600
to accelerate cell metastasis and epithelial-mesenchymal transition
in colorectal cancer. J Exp Clini Cancer Res. 37:1062018.
View Article : Google Scholar
|
|
5
|
Wang H, Yao L, Gong Y and Zhang B: TRIM31
regulates chronic inflammation via NF-κB signal pathway to promote
invasion and metastasis in colorectal cancer. Am J Transl Res.
10:1247–1259. 2018.PubMed/NCBI
|
|
6
|
Wang JL, Guo CR, Su WY, Chen YX, Xu J and
Fang JY: CD24 overexpression related to lymph node invasion and
poor prognosis of colorectal cancer. Clin Lab. 64:497–505. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hua Q, Mi B and Huang G: The emerging
co-regulatory role of long noncoding RNAs in epithelial-mesenchymal
transition and the Warburg effect in aggressive tumors. Crit Rev
Oncol Hematol. 126:112–120. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lamouille S, Xu J and Derynck R: Molecular
mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell
Biol. 15:178–196. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cai Z, Cao Y, Luo Y, Hu H and Ling H:
Signalling mechanism(s) of epithelial-mesenchymal transition and
cancer stem cells in tumour therapeutic resistance. Clin Chim Acta.
483:156–163. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maeda S and Nakagawa H: Roles of
E-cadherin in Hepatocarcinogenesis. Innovative Medicine: Basic
research and development. Nakao K, Minato N and Uemoto S: Springer;
Tokyo: pp. 71–77. 2015, View Article : Google Scholar
|
|
11
|
Chi Y, Huang Q, Lin Y, Ye G, Zhu H and
Dong S: Epithelial-mesenchymal transition effect of fine
particulate matter from the Yangtze River Delta region in China on
human bronchial epithelial cells. J Environ Sci (China).
66:155–164. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
He J, Tang F, Liu L, Chen L, Li J, Ou D,
Zhang L, Li Z, Feng D, Li W and Sun LQ: Positive regulation of TAZ
expression by EBV-LMP1 contributes to cell proliferation and
epithelial-mesenchymal transition in nasopharyngeal carcinoma.
Oncotarget. 8:52333–52344. 2016.PubMed/NCBI
|
|
13
|
Zhou WJ, Chen J, Feng Y, Fan YP, Li Q, Fu
J and Zhang P: Inhibition of cigarettes smoke-induced epithelial to
mesenchymal transition by the SMO inhibitor PF-5274857 in Beas-2b
epithelial cells. Sichuan Da Xue Xue Bao Yi Xue Ban. 47:485–490.
2016.(In Chinese). PubMed/NCBI
|
|
14
|
Haider MT and Taipaleenmäki H: Targeting
the metastatic bone microenvironment by MicroRNAs. Front Endocrinol
(Lausanne). 9:2022018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Amarilyo G, Pillar N, Ben-Zvi I,
Weissglas-Volkov D, Zalcman J, Harel L, Livneh A and Shomron N:
Analysis of microRNAs in familial mediterranean fever. PLoS One.
13:e01978292018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
De Los Reyes-Garcia AM, Arroyo AB,
Teruel-Montoya R, Vicente V, Lozano ML, González-Conejero R and
Martinez C: MicroRNAs as potential regulators of platelet function
and bleeding diatheses. Platelets. 1–6. 2018.(Epub ahead of print).
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Duo Y, Bi J, Zeng X, Mei L, Bao S,
He L, Shan A, Zhang Y and Yu X: Targeted delivery of anti-miR-155
by functionalized mesoporous silica nanoparticles for colorectal
cancer therapy. Int J Nanomedicine. 13:1241–1256. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin Y, Chen F, Shen L, Tang X, Du C, Sun
Z, Ding H, Chen J and Shen B: Biomarker microRNAs for prostate
cancer metastasis: Screened with a network vulnerability analysis
model. J Transl Med. 16:1342018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tu J and Zhang YS: Advance on microRNA and
prostate cancer. Zhonghua Bing Li Xue Za Zhi. 47:388–390. 2018.(In
Chinese). PubMed/NCBI
|
|
20
|
Vera O, Jimenez J, Pernia O,
Rodriguez-Antolin C, Rodriguez C, Sanchez Cabo F, Soto J, Rosas R,
Lopez-Magallon S, Esteban Rodriguez I, et al: DNA methylation of
miR-7 is a mechanism involved in platinum response through MAFG
overexpression in cancer cells. Theranostics. 7:4118–4134. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Hong-Yuan W and Xiao-Ping C: miR-338-3p
suppresses epithelial-mesenchymal transition and metastasis in
human nonsmall cell lung cancer. Indian J Cancer. 52 (Suppl
3):E168–E171. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Y, An H, Pang J, Huang L, Li J and Liu
L: MicroRNA profiling identifies miR-129-5p as a regulator of EMT
in tubular epithelial cells. Int J Clin Exp Med. 8:20610–20616.
2015.PubMed/NCBI
|
|
23
|
Lili LN, Huang AD, Zhang M, Wang L,
McDonald LD, Matyunina LV, Satpathy M and McDonald JF: Time-course
analysis of microRNA-induced mesenchymal-to-epithelial transition
underscores the complexity of the underlying molecular processes.
Cancer Lett. 428:184–191. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Penna E, Orso F and Taverna D: miR-214 as
a key hub that controls cancer networks: Small player, multiple
functions. J Invest Dermatol. 135:960–969. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Alonso L, Okada H, Pasolli HA, Wakeham A,
You-Ten AI, Mak TW and Fuchs E: Sgk3 links growth factor signaling
to maintenance of progenitor cells in the hair follicle. J Cell
Biol. 170:559–570. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu Y, Zhou H, Ma L, Hou Y, Pan J, Sun C,
Yang Y and Zhang J: MiR-214 suppressed ovarian cancer and
negatively regulated semaphorin 4D. Tumour Biol. 37:8239–8248.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yang TS, Yang XH, Wang XD, Wang YL, Zhou B
and Song ZS: MiR-214 regulate gastric cancer cell proliferation,
migration and invasion by targeting PTEN. Cancer Cell Int.
13:682013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang X, Chen J, Li F, Lin Y, Zhang X, Lv Z
and Jiang J: MiR-214 inhibits cell growth in hepatocellular
carcinoma through suppression of beta-catenin. Biochem Biophys Res
Commun. 428:525–531. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Penna E, Orso F, Cimino D, Tenaglia E,
Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De
Pitta C, et al: microRNA-214 contributes to melanoma tumour
progression through suppression of TFAP2C. EMBO J. 30:1990–2007.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang H, Kong W, He L, Zhao JJ, O'Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, et al:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang XJ, Ye H, Zeng CW, He B, Zhang H and
Chen YQ: Dysregulation of miR-15a and miR-214 in human pancreatic
cancer. J Hematol Oncol. 3:462010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chandrasekaran KS, Sathyanarayanan A and
Karunagaran D: miR-214 activates TP53 but suppresses the expression
of RELA, CTNNB1, and STAT3 in human cervical and colorectal cancer
cells. Cell Biochem Funct. 35:464–471. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Huang SD, Yuan Y, Zhuang CW, Li BL, Gong
DJ, Wang SG, Zeng ZY and Cheng HZ: MicroRNA-98 and microRNA-214
post-transcriptionally regulate enhancer of zeste homolog 2 and
inhibit migration and invasion in human esophageal squamous cell
carcinoma. Mol Cancer. 11:512012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Schwarzenbach H, Milde-Langosch K,
Steinbach B, Müller V and Pantel K: Diagnostic potential of
PTEN-targeting miR-214 in the blood of breast cancer patients.
Breast Cancer Res Treat. 134:933–941. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Shih TC, Tien YJ, Wen CJ, Yeh TS, Yu MC,
Huang CH, Lee YS, Yen TC and Hsieh SY: MicroRNA-214 downregulation
contributes to tumor angiogenesis by inducing secretion of the
hepatoma-derived growth factor in human hepatoma. J Hepatol.
57:584–591. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yang Z, Chen S, Luan X, Li Y, Liu M, Li X,
Liu T and Tang H: MicroRNA-214 is aberrantly expressed in cervical
cancers and inhibits the growth of HeLa cells. IUBMB Life.
61:1075–1082. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Chandrasekaran KS, Sathyanarayanan A and
Karunagaran D: MicroRNA-214 suppresses growth, migration and
invasion through a novel target, high mobility group AT-hook 1, in
human cervical and colorectal cancer cells. Br J Cancer.
115:741–751. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lorand L and Graham RM: Transglutaminases:
Crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell
Biol. 4:140–156. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kang S, Oh SC, Min BW and Lee DH:
Transglutaminase 2 regulates self-renewal and stem cell marker of
human colorectal cancer stem cells. Anticancer Res. 38:787–794.
2018.PubMed/NCBI
|
|
41
|
Yamaguchi H, Kuroda K, Sugitani M,
Takayama T, Hasegawa K and Esumi M: Transglutaminase 2 is
upregulated in primary hepatocellular carcinoma with early
recurrence as determined by proteomic profiles. Int J Oncol.
50:1749–1759. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yin J, Oh YT, Kim JY, Kim SS, Choi E, Kim
TH, Hong JH, Chang N, Cho HJ, Sa JK, et al: Transglutaminase 2
Inhibition reverses mesenchymal transdifferentiation of glioma stem
cells by regulating C/EBPbeta signaling. Cancer Res. 77:4973–4984.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zonca S, Pinton G, Wang Z, Soluri MF,
Tavian D, Griffin M, Sblattero D and Moro L: Tissue
transglutaminase (TG2) enables survival of human malignant pleural
mesothelioma cells in hypoxia. Cell Death Dis. 8:e25922017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fang Q, Yao S, Luo G and Zhang X:
Identification of differentially expressed genes in human breast
cancer cells induced by 4-hydroxyltamoxifen and elucidation of
their pathophysiological relevance and mechanisms. Oncotarget.
9:2475–2501. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Miyoshi N, Ishii H, Mimori K, Tanaka F,
Hitora T, Tei M, Sekimoto M, Doki Y and Mori M: TGM2 is a novel
marker for prognosis and therapeutic target in colorectal cancer.
Ann Surg Oncol. 17:967–972. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Park KS, Kim HK, Lee JH, Choi YB, Park SY,
Yang SH, Kim SY and Hong KM: Transglutaminase 2 as a cisplatin
resistance marker in non-small cell lung cancer. J Cancer Res Clin
Oncol. 36:493–502. 2010. View Article : Google Scholar
|
|
47
|
Sodek KL, Ringuette MJ and Brown TJ:
Compact spheroid formation by ovarian cancer cells is associated
with contractile behavior and an invasive phenotype. Int J Cancer.
124:2060–2070. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lu Q, Xu L, Li C, Yuan Y, Huang S and Chen
H: miR-214 inhibits invasion and migration via downregulating
GALNT7 in esophageal squamous cell cancer. Tumour Biol.
37:14605–14614. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao X, Lu C, Chu W, Zhang Y, Zhang B,
Zeng Q, Wang R, Li Z, Lv B and Liu J: microRNA-214 governs lung
cancer growth and metastasis by targeting carboxypeptidase-D. DNA
Cell Biol. 35:715–721. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Li B, Han Q, Zhu Y, Yu Y, Wang J and Jiang
X: Down-regulation of miR-214 contributes to intrahepatic
cholangiocarcinoma metastasis by targeting Twist. FEBS J.
279:2393–2398. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Cai HK, Chen X, Tang YH and Deng YC:
MicroRNA-194 modulates epithelial-mesenchymal transition in human
colorectal cancer metastasis. OncoTargets Ther. 10:1269–1278. 2017.
View Article : Google Scholar
|
|
52
|
Yan MD, Yao CJ, Chow JM, Chang CL, Hwang
PA, Chuang SE, Whang-Peng J and Lai GM: Fucoidan elevates
MicroRNA-29b to regulate DNMT3B-MTSS1 axis and inhibit EMT in human
hepatocellular carcinoma cells. Mar Drugs. 13:6099–6116. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Abubaker J, Bavi P, Al-Harbi S, Ibrahim M,
Siraj AK, Al-Sanea N, Abduljabbar A, Ashari LH, Alhomoud S,
Al-Dayel F, et al: Clinicopathological analysis of colorectal
cancers with PIK3CA mutations in Middle Eastern population.
Oncogene. 27:3539–3545. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Qin Y, Huo Z, Song X, Chen X, Tian X and
Wang X: mir-106a regulates cell proliferation and apoptosis of
colon cancer cells through targeting the PTEN/PI3K/AKT signaling
pathway. Oncol Lett. 15:3197–3201. 2018.PubMed/NCBI
|
|
55
|
Song Y, Zhao Y, Ding X and Wang X:
microRNA-532 suppresses the PI3K/Akt signaling pathway to inhibit
colorectal cancer progression by directly targeting IGF-1R. Am J
Cancer Res. 8:435–449. 2018.PubMed/NCBI
|
|
56
|
Wallin JJ, Guan J, Prior WW, Lee LB, Berry
L, Belmont LD, Koeppen H, Belvin M, Friedman LS and Sampath D:
GDC-0941, a novel class I selective PI3K inhibitor, enhances the
efficacy of docetaxel in human breast cancer models by increasing
cell death in vitro and in vivo. Clin Cancer Res. 18:3901–3911.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Jia L, Luo S, Ren X, Li Y, Hu J, Liu B,
Zhao L, Shan Y and Zhou H: miR-182 and miR-135b mediate the
tumorigenesis and invasiveness of colorectal cancer cells via
targeting ST6GALNAC2 and PI3K/AKT pathway. Dig Dis Sc.
62:3447–3459. 2017. View Article : Google Scholar
|