Introduction
Cerebral ischemic stroke (IS), also known as
cerebral infarction (CI), is an ischemic event caused by
insufficient blood supply to brain tissues. Following an ischemic
event, brain function may decrease within 60–90 sec, and cerebral
ischemia may cause irreversible damage if untreated (1). Understanding of the pathogenesis of
IS is required to develop multifactorial prevention strategies and
novel therapeutic treatments in order to decrease the incidence and
recurrence rates of IS.
Epigenetic alterations have been identified to be
among the principal regulators involved in the pathogenesis of
various pathological conditions, including cardiovascular diseases
and cancer (2).
Cytosine-phosphate-guanine (CpG) islands in promoter regions may
exhibit an abnormal methylation status in various diseases;
hypermethylation may lead to repression of gene expression, whereas
hypomethylation may cause chromosomal instability and loss of DNA
imprinting (3). Accumulating
evidence demonstrates that DNA methylation may serve a principal
role in IS (4). The mRNA
expression level of tumor protein p53 (TP53) was previously
identified to be regulated by epigenetic mechanisms, and
alterations in the methylation status of TP53 promoter may affect
its expression level in pathological conditions, including cancer
and atherosclerosis (5). The TP53
gene is located on the short arm of chromosome 17 (17p13.1) and it
consists of ~20,000 base pairs (bp). TP53 is a tumor suppressor
gene that was previously identified to promote apoptosis, affect
gene stability and inhibit tumor formation (6). Notably, TP53 hypermethylation may
result in the silencing of TP53 (7). Although a previous study demonstrated
that the circulating levels of TP53 were increased in patients with
IS (8), to the best of the
authors' knowledge, no studies have investigated the association
between methylation of the TP53 promoter and IS.
In the present study, the methylation levels of the
TP53 promoter were examined using DNA extracted from the peripheral
blood of patients with IS. Methylation-specific polymerase chain
reaction (MSP) and bisulfite sequencing polymerase chain reaction
(BSP) analyses were used to investigate the methylation status of
the promoter of TP53 in patients with IS. Additionally, the
association between the methylation level of the TP53 promoter and
the incidence of CI was investigated.
Materials and methods
Subjects
In total, 78 patients with IS (female to male ratio,
19:20; age, 61.54±9.90 years) were enrolled in the present study.
All patients were hospitalized in The Second Hospital of Shandong
University (Jinan, China) between October 2010 and February 2011.
All patients were diagnosed according to the diagnostic criteria
established by The Fourth National Conference on Cerebrovascular
Diseases in 1995 (9). The samples
were collected within 48 h following IS. Transcranial magnetic
resonance diffusion-weighted imaging identified acute infarction in
conscious patients. Patients with tumors, connective tissue
disease, severe liver kidney disease, neurological diseases and
atrial fibrillation were excluded from the present study.
In the control group, 86 healthy subjects (female to
male ratio 37:49; age, 62.12±7.06 years) were enrolled between
October 2010 and February 2011. Healthy subjects did not exhibit
evidence of IS in their medical history and no signs of CI were
identified following physical examination. In a subset of healthy
individuals, the diagnosis was confirmed by computed tomography and
magnetic resonance imaging. Subjects exhibiting a history of
transient ischemic attacks, cerebral hemorrhage, blood diseases,
severe liver and kidney dysfunctions, immune system diseases and
atrial fibrillation were excluded from the present study. All
clinical studies were approved by The Local Ethics Committee of The
Second Hospital of Shandong University and informed consent was
obtained from all patients.
Blood sample collection and
processing
In total, 2 ml of blood was collected from each
subject by venipuncture and was placed into serum separator tubes
(BD Biosciences, San Jose, CA, USA). After 10–60 min, samples were
centrifuged at 1,500 × g for 10 min at 4°C. Serum was subsequently
divided in 0.5 ml aliquots and stored at −80°C.
DNA extraction and bisulfite
modification
Genomic DNA was extracted from serum using a DNA
extraction kit (cat. no. 69504; Qiagen GmbH, Hilden, Germany). A
total of 2 µg genomic DNA was denatured using 0.3 M NaOH for 15 min
at 37°C in a final volume of 20 µl, and purified according to the
manufacturer's protocol (Wizard® DNA clean-up Resin;
cat. no. A7280; Promega Corporation, Madison, WI, USA). DNA samples
were treated with sodium bisulfate to convert unmethylated cytosine
to uracil without affecting methylated cytosine using an EpiTect
Fast Bisulfite kit (Qiagen GmbH; cat. no. 59802), according to the
manufacturer's protocol.
BSP and MSP
MethPrimer (http://www.urogene.org/methprimer/) was used to design
primers for the first CpG island downstream of the transcription
start site of TP53 (10–12). The primers used in the present
study were the following: Unmethylated forward (F),
5′-GTAGTTTGAATGTTTTTATTTTGGT-3′ and unmethylated reverse (R),
5′-CCTACTACACCCTCTACAAACA-3′; methylated F,
5′-GTAGTTTGAACGTTTTTATTTTGGC-3′ and methylated R,
5′-CCTACTACGCCCTCTACAAACG-3′. BSP primers were
5′-AAGATTTTCGGGAGGAGAGG-3′ and
5′-CCTAAATACCTATATCAATACTAAATAACAA-3′. A hot start Taq polymerase
(cat. no. 14966001; Invitrogen, Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) was used to amplify the regions of interest by
MSP and BSP. PCR thermocycling conditions were as follows: Initial
denaturation at 95°C for 5 min, 40 cycles of 30 sec at 95°C, 30 sec
at 60°C and 45 sec at 72°C, and a final extension step at 72°C for
10 min. PCR products were loaded on a 2% agarose gel and the bands
were excised and purified using the Illustra GFX PCR DNA and Gel
Band Purification kit (cat. no. 28-9034-70; GE Healthcare Life
Sciences, Little Chalfont, UK). Directly sequencing PCR products
may lead to inconclusive results, due to variability in the
conversion efficiency and in the quantity of methylated sites. In
order to obtain clear sequencing results, PCR products were cloned
into plasmids prior to sequencing. Purified PCR products were
ligated into pGEM-T easy plasmids (Promega Corporation) for 2 h on
ice. DH5α Escherichia coli competent cells (cat. no.
18265017; Invitrogen; Thermo Fisher Scientific, Inc.) were
transformed by heat shock and incubated overnight at 37°C on agar
plates containing 100 µg/ml ampicillin (cat. no. BP021;
Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) (13). In total, ≥10 colonies for each
treatment were selected and expanded in lysogeny broth medium
containing 50 µg/ml ampicillin. On the following day, plasmids were
isolated using the PureLink plasmid miniprep kit (cat. no. K210010;
Invitrogen; Thermo Fisher Scientific, Inc.). The sequences of
plasmids containing the bisulfite-modified DNA were analyzed by
Sanger sequencing (Laboratory Services, University of Guelph,
Guelph, ON, Canada) and BiQ Analyzer 2.0 software (14) was used to analyze methylation
patterns and draw diagrams. Human methylated DNA following
bisulfite modification and unmethylated DNA (cat. no. 59568; Qiagen
GmbH) were used as controls for bisulfite conversion efficiency and
accuracy, respectively.
Carotid intima-media thickness (CIMT)
measurement
The CIMT of 49 patients with IS was examined by
carotid color doppler ultrasonography (Philips iE33; Philips
Medical Systems, Inc., Bothell, WA, USA). Two certified
sonographers who were blinded to all clinical information performed
carotid arterial scanning. Patients were placed in the supine
position with slight hyperextension, and rotation of the neck to
the contralateral side. CIMT measurements were collected at 10 mm
intervals of the far wall of the right common carotid arteries. The
CIMT was measured by manually examining the thickness of every free
plaque lesion. The Crouse plaque score was used to calculate the
severity of carotid artery atherosclerosis (15).
Identification of transcription factor
binding site
TFSearch viewer 1.0 (Parallel Application TRC
laboratory, RWCP, Japan) was used to identify the predicted
transcription factor binding sites in the CpG islands of the TP53
promoter (16).
Homocysteine (Hcy) concentration
detection
Hcy concentration was detected using an Hcy assay
kit (cat. no. ab228559; Abcam, Cambridge, UK) according to the
manufacturer's protocol.
Statistical analysis
The pathophysiological characteristics of the
patients were examined using a χ2 test and Student's
t-test. Associations between the methylation levels of TP53 and IS
were analyzed using Student's t-test. Multivariate logistic
regression analysis was used to adjust for age. SPSS software for
Windows (version 17.0; SPSS, Inc., Chicago, IL, USA) was used to
perform all statistical analyses. Two-tailed P<0.05 was
considered to indicate a statistically significant difference.
Results
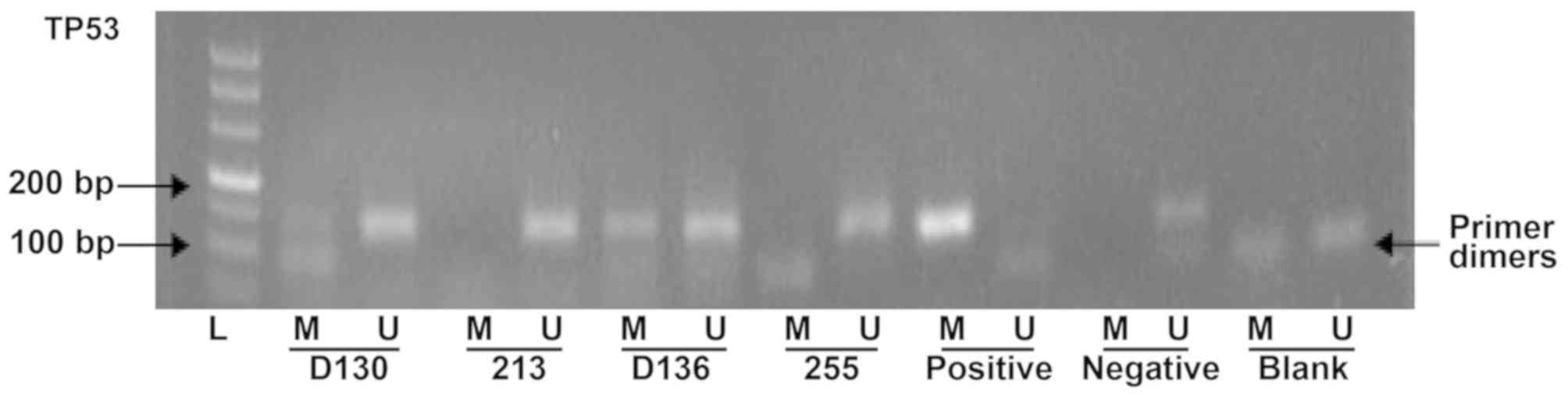
TP53 promoter methylation in IS group
and control groups
The methylation levels of TP53 promoter were
determined by MSP assay (Fig 1).
In total, 25 out of 78 (32.1%) patients with IS exhibited promoter
methylation. By contrast, 14 out of 86 (16.3%) healthy subjects
exhibited methylated CpGs. The methylation rate of the TP53
promoter in patients with IS was significantly increased compared
with the control group (Table
I).
 | Table I.Tumor protein p53 promoter methylation
in IS group and control groups. |
Table I.
Tumor protein p53 promoter methylation
in IS group and control groups.
| Group | n | Methylated promoter,
n | Unmethylated
promoter, n | P-value |
|---|
| IS | 78 | 25 (32.1%) | 53 (67.9%) | <0.001 |
| Control | 86 | 14 (16.3%) | 72 (83.7%) |
|
Association between TP53 promoter
methylation and age
Subjects were divided into two groups according to
their age: i) Elderly subjects (≥60 years); and ii) younger
subjects (<60 years). In total, 10 out of 37 (27%) young
patients and 15 out of 41 (36.6%) elderly patients with IS
exhibited methylated TP53 promoters. The rate of methylation
increased with age; however, the difference was not statistically
significant (Table II). In total,
5 out of 34 (14.7%) young patients and 9 out of 52 (17.3%) elderly
patients in the control group exhibited methylated TP53
promoters.
 | Table II.Association between tumor protein p53
promoter methylation and age. |
Table II.
Association between tumor protein p53
promoter methylation and age.
| A, Patients with
ischemic stroke |
|---|
|
|---|
| Age, years | Methylated
promoter, n | Unmethylated
promoter, n | P-value |
|---|
| <60 | 10 (27.0%) | 27 (73.0%) | 0.509 |
| ≥60 | 15 (36.6%) | 26 (63.4%) |
|
|
| B, Healthy
control subjects |
|
| Age,
years | Methylated
promoter, n | Unmethylated
promoter, n | P-value |
|
| <60 | 5
(14.7%) | 29 (85.3%) | 0.983 |
| ≥60 | 9
(17.3%) | 43 (82.7%) |
|
Association between TP53 promoter
methylation and sex
The association between the methylation level of the
TP53 promoter and the sex of the subjects was investigated. The
percentage of male and female patients with IS exhibiting a
methylated TP53 promoter was 35% (14/40) and 28.9% (11/38),
respectively. The percentage of healthy male and female subjects
with methylated TP53 promoters was 16.3% (8/49) and 16.2% (6/37),
respectively. The difference was not statistically significant in
the IS or control groups. The present results suggested that TP53
promoter methylation was not associated with sex (Table III).
 | Table III.Association between tumor protein p53
promoter methylation and sex. |
Table III.
Association between tumor protein p53
promoter methylation and sex.
| A, Patients with
ischemic stroke |
|---|
|
|---|
| Sex | Methylated
promoter, n | Unmethylated
promoter, n | P-value |
|---|
| Male | 14 (35.0%) | 26 (65.0%) | 0.632 |
| Female | 11 (28.9%) | 27 (71.1%) |
|
|
| B, Healthy
control subjects |
|
| Sex | Methylated
promoter, n | Unmethylated
promoter, n | P-value |
|
| Male | 8 (16.3%) | 41 (83.7%) | 0.999 |
| Female | 6 (16.2%) | 31 (83.8%) |
|
Association between TP53 promoter
methylation and Hcy levels in patients with IS
The circulating levels of Hcy were investigated in
patients with IS exhibiting methylated and unmethylated TP53
promoters. In total, 25 patients presenting a methylated promoter
of TP3 exhibited a circulating level of Hcy of 33.91±23.81 µM. The
concentration of Hcy in 53 patients exhibiting unmethylated TP53
promoters was 23.19±5.67 µM, significantly decreased compared with
the methylated group. Therefore, the methylation status of TP53 was
identified to be significantly associated with the circulating
levels of Hcy (Table IV).
 | Table IV.Association between tumor protein p53
promoter methylation and Hcy levels in patients with ischemic
stroke. |
Table IV.
Association between tumor protein p53
promoter methylation and Hcy levels in patients with ischemic
stroke.
| Clinical
characteristic | Methylated promoter
(n=25) | Unmethylated
promoter (n=53) | P-value |
|---|
| Hcy, µM | 33.91±23.81 | 23.19±5.67 | 0.004a |
Association between TP53 promoter
methylation and carotid intima-media thickness (CIMT) in patients
with IS
The CIMTs of 49 patients with IS were examined by
carotid color doppler ultrasonography (15). The CIMT of 19 patients exhibiting
methylated promoters of TP53 was 1.06±0.35 mm. By contrast, the
CIMT of patients exhibiting unmethylated promoters was 0.87±0.19
mm. Notably, the difference was statistically significant (Table V). Additionally, the Crouse score
was used to calculate the severity of carotid artery
atherosclerosis (15), and a total
of 19 patients in the methylated group presented a Crouse score of
4.97±4.40. In contrast, patients with IS in the unmethylated group
presented a Crouse score of 2.82±2.80, significantly decreased
compared with that of the methylated group (Table V).
 | Table V.Association between tumor protein p53
promoter methylation and CIMT in patients with ischemic stroke. |
Table V.
Association between tumor protein p53
promoter methylation and CIMT in patients with ischemic stroke.
| Clinical
characteristic | Methylated promoter
(n=19) | Unmethylated
promoter (n=30) | P-value |
|---|
| CIMT, mm | 1.06±0.35 | 0.87±0.19 | 0.018a |
| Crouse score | 4.97±4.40 | 2.82±2.80 | 0.041a |
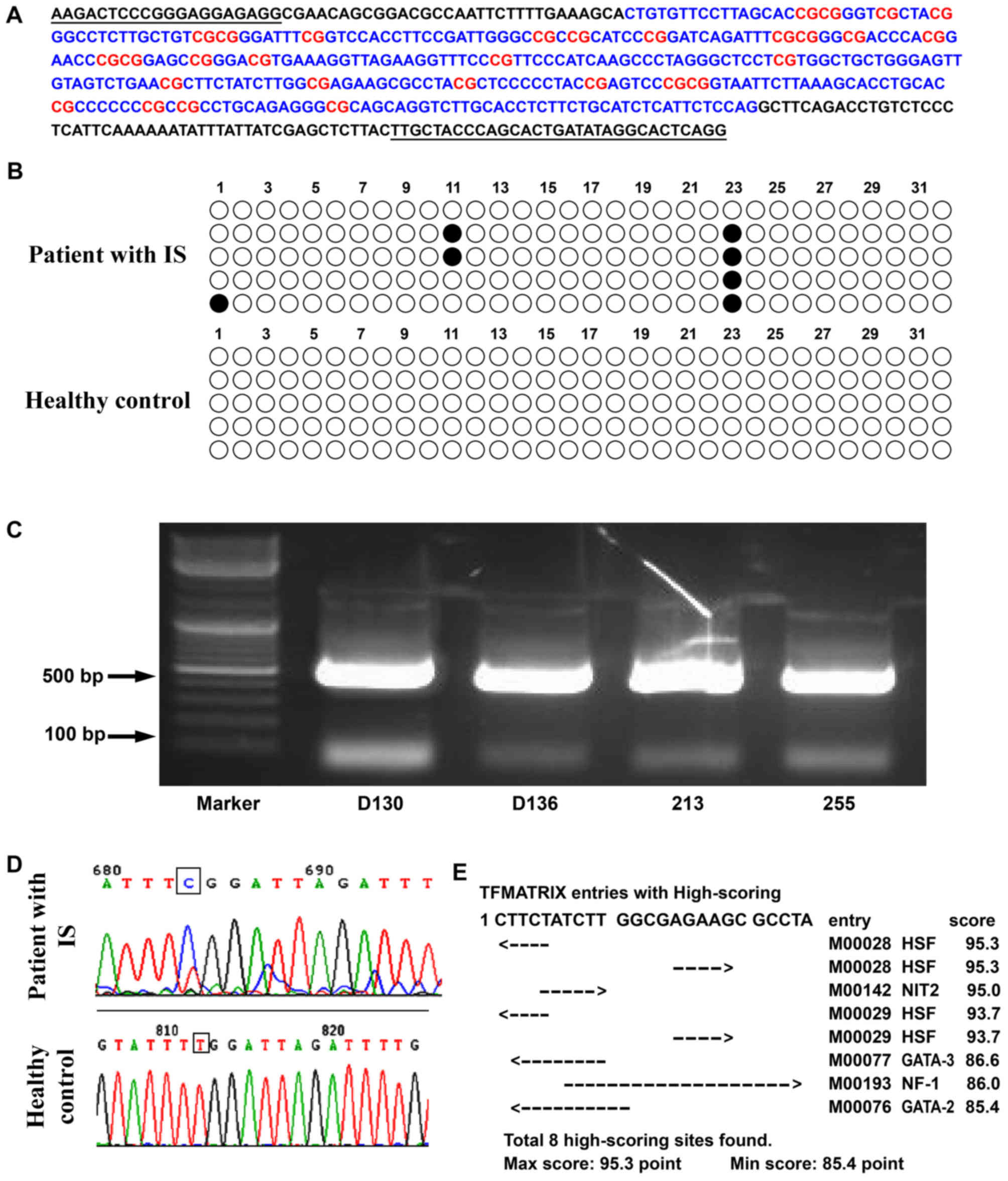
BSP analysis
The methylation status of a DNA fragment containing
32 CpG sites in the promoter region of TP53 was assessed by BSP
analysis. The methylation level of patients with IS in the
methylated group was confirmed to be increased. In particular, the
23rd CpG site was methylated in 80% of patients with IS. By
contrast, the 23rd CpG site was unmethylated in the control group
(Fig. 2). A bioinformatics
analysis was performed to identify the predicted transcription
factor binding sites in the CpG islands of the TP53 promoter using
TFSearch viewer 1.0 and the 23rd CpG island was identified to
contain a nuclear factor I (NFI) binding site.
Discussion
Epigenetic mechanisms are important physiological
mediators of development and cellular functions. Epigenetic
alterations are able to regulate gene expression without affecting
the DNA sequence. The mechanisms underlying epigenetic alterations
include DNA methylation, histone methylation, acetylation and
non-coding RNA interference (4,17,18).
Previous studies identified that whole genome hypomethylation,
epigenetic regulation of gene expression and hypermethylation of
CpG islands are common in tumors and in pathological conditions,
and the association between DNA methylation and tumor development
has attracted considerable attention (19–21).
An increasing number of studies have investigated epigenetic
events, and DNA methylation was identified to be associated with
tumors and with various diseases including atherosclerosis,
diabetes, Alzheimer's disease and autoimmune diseases (22). In the present study, the
methylation of TP53 promoter was investigated in patients with IS
and in healthy control subjects. Additionally, the association
between the methylation status of TP53 promoter and IS was
investigated.
A previous study demonstrated that TP53 may serve
its roles primarily by regulating the expression levels of its
downstream genes involved in arresting the cell cycle, promoting
cell apoptosis and maintaining gene stability (23). Additionally, a previous study
demonstrated that TP53 may serve an important role in the
occurrence and development of tumors, atherosclerosis and ischemic
injury (24). Crumrine et
al (25) observed that
knockdown of TP53 expression may protect against focal ischemic
damage in transgenic mice. The protein expression level of TP53 was
previously identified to be upregulated following ischemic brain
injury, and TP53 was able to induce apoptosis and endothelial
injury, aggravating brain injury (26). Li et al (27) suggested that TP53-immunoreactive
protein and TP53 mRNA expression levels were upregulated following
IS, and TP53 may affect the cellular response following IS. In the
present study, the methylated status of the TP53 promoter was
significantly increased in patients with IS compared with the
control group, suggesting that the hypermethylation of the TP53
promoter in peripheral blood may be associated with the incidence
of IS and carotid atherosclerosis. The present results suggested
that the alteration in the methylation status of the TP53 promoter
may be important in the occurrence of carotid atherosclerosis,
promoting the pathogenesis of IS. Therefore, it was hypothesized
that the increased methylation level of the TP53 promoter may
promote the occurrence of IS. Additionally, the increased
methylation level of the TP53 promoter may be a predictive factor
of IS.
Carotid atherosclerosis is an important risk factor
for the development of IS. Accumulating evidence demonstrates that
DNA methylation may serve a role in the development of
atherosclerosis (28). A previous
study investigating a model of atherosclerotic lesions demonstrated
that the inactivation of TP53 following promoter methylation may
promote the development of atherosclerosis (29). Newman (30) demonstrated that in vivo
atherosclerosis models exhibited whole genome hypomethylation.
Turunen et al (31)
observed that the hypermethylation of the TP53 promoter was
associated with the development of atherosclerosis. Therefore, in
the present study, it was hypothesized that the expression level of
TP53 may be involved in regulating cell proliferation in
atherosclerotic plaques. The present results suggested that the
TP53 promoter region was hypermethylated in patients with IS. TP53
promoter hypermethylation may decrease the expression level of
TP53, promoting the occurrence of atherosclerosis, thus serving a
role in the pathogenesis of IS.
Hcy, an amino acid containing sulfur, is an
intermediate product of methionine metabolism (32). A previous study demonstrated that
hyperhomocysteinemia may be a risk factor for the occurrence of
cardiovascular and cerebrovascular diseases (33). The present results suggested that
the circulating levels of Hcy were significantly increased in
patients presenting hypermethylation of the TP53 promoter.
Additionally, the results suggested that lower levels of folate may
increase the concentration of Hcy in samples with methylated TP53
promoter (34). BSP analysis
identified 32 CpG sites in the TP53 promoter, and MSP analysis
identified that these CpG sites were methylated in 32.1% (25/78) of
patients with IS and in 16.3% (14/86) of control subjects. In
particular, the 23rd CpG island in the TP53 promoter was methylated
in 80% of the samples in the methylated group. The present results
suggested that hypermethylation of the TP53 promoter may be
associated with the occurrence of IS. Moreover, a bioinformatics
analysis identified that the 23rd CpG island contained a nuclear
NFI binding site. Therefore, methylation of this CpG island may
affect the binding between NFI and the promoter of TP53, thus
decreasing the expression level of TP53.
In conclusion, the present study suggested that
methylation of TP53 promoter was significantly increased in
patients with IS compared with healthy subjects. Additionally, the
methylation level of the TP53 promoter was identified to be
associated with the degree of carotid atherosclerosis and the Hcy
concentration in peripheral blood. However, the protein expression
level of TP53 in patients with IS requires further investigation.
Additionally, further studies are required to investigate the
protein expression level of TP53 and the methylation level of TP53
promoter in brain tissues. Collectively, the development of novel
strategies aimed to alter the expression level and the activity of
TP53 in patients with IS may improve the treatment and prevention
of IS.
Acknowledgements
Not applicable.
Funding
The present work was supported by The National
Natural Science Foundation of China (grant no. 81571052), Shandong
Provincial Key Research and Development Program (grant no.
2017GSF218036), The Fundamental Research Funds of Shandong
University (grant no. 2016JC022) and The Youth Foundation of The
Second Hospital of Shandong University (grant no. Y2015010021).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
YWe and ZZ conceived and designed experiments. YWe,
ZS, ZX and YWa performed experiments. ZX and SX provided new tools
and reagents. YWe, YX, JB, SX and XZ analyzed data. YWe, JB and ZZ
wrote and made manuscript revisions. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All clinical studies were approved by The Local
Ethics Committee of The Second Hospital of Shandong University
(Jinan, China) and informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
IS
|
cerebral ischemic stroke
|
|
CI
|
cerebral infarction
|
|
MSP
|
methylation-specific polymerase chain
reaction
|
|
Hcy
|
homocysteine
|
|
CIMT
|
carotid intima-media thickness
|
References
|
1
|
Deb P, Sharma S and Hassan KM:
Pathophysiologic mechanisms of acute ischemic stroke: An overview
with emphasis on therapeutic significance beyond thrombolysis.
Pathophysiology. 17:197–218. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fattahi S, Pilehchian Langroudi M and
Akhavan-Niaki H: Hedgehog signaling pathway: Epigenetic regulation
and role in disease and cancer development. J Cell Physiol.
233:5726–5735. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Matsubara N: Epigenetic regulation and
colorectal cancer. Dis Colon Rectum. 55:96–104. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Krupinski J, Carrera C, Muino E, Torres N,
Al-Baradie R, Cullell N and Fernandez-Cadenas I: DNA methylation in
stroke. update of latest advances. Comput Struct Biotechnol J.
16:1–5. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hsue SS, Wang WC, Chen YK and Lin LM:
Expression of inhibitors of apoptosis family protein in
7,12-dimethylbenz[a]anthracene-induced hamster buccal-pouch
squamous-cell carcinogenesis is associated with mutant p53
accumulation and epigenetic changes. Int J Exp Pathol. 89:309–320.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bykov VJN, Eriksson SE, Bianchi J and
Wiman KG: Targeting mutant p53 for efficient cancer therapy. Nat
Rev Cancer. 18:89–102. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mayr U, Mayr M, Li C, Wernig F, Dietrich
H, Hu Y and Xu Q: Loss of p53 accelerates neointimal lesions of
vein bypass grafts in mice. Circ Res. 90:197–204. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mendrysa SM, Ghassemifar S and Malek R:
p53 in the CNS: Perspectives on development, stem cells, and
cancer. Genes Cancer. 2:431–442. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X: Diagnostic essentials of various
cerebrovascular diseases. Chin J Neurol. 29:379–381. 1996.
|
|
10
|
Li LC and Dahiya R: MethPrimer: Designing
primers for methylation PCRs. Bioinformatics. 18:1427–1431. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kang JH, Kim SJ, Noh DY, Park IA, Choe KJ,
Yoo OJ and Kang HS: Methylation in the p53 promoter is a
supplementary route to breast carcinogenesis: Correlation between
CpG methylation in the p53 promoter and the mutation of the p53
gene in the progression from ductal carcinoma in situ to invasive
ductal carcinoma. Lab Invest. 81:573–579. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou Y, Zheng X, Lu J, Chen W, Li X and
Zhao L: Ginsenoside 20(S)-Rg3 inhibits the warburg effect via
modulating DNMT3A/MiR-532-3p/HK2 pathway in ovarian cancer cells.
Cell Physiol Biochem. 45:2548–2559. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cohen SN, Chang AC and Hsu L:
Nonchromosomal antibiotic resistance in bacteria: Genetic
transformation of escherichia coli by R-factor DNA. Proc Natl Acad
Sci USA. 69:2110–2114. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bock C, Reither S, Mikeska T, Paulsen M,
Walter J and Lengauer T: BiQ analyzer: Visualization and quality
control for DNA methylation data from bisulfite sequencing.
Bioinformatics. 21:4067–4068. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kim GH, Youn HJ, Choi YS, Jung HO, Chung
WS and Kim CM: Carotid artery evaluation and coronary calcium
score: Which is better for the diagnosis and prevention of
atherosclerotic cardiovascular disease? Int J Clin Exp Med.
8:18591–18600. 2015.PubMed/NCBI
|
|
16
|
Lee C and Huang CH: Searching for
transcription factor binding sites in vector spaces. BMC
Bioinformatics. 13:2152012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Aslani S, Sobhani S, Gharibdoost F,
Jamshidi A and Mahmoudi M: Epigenetics and pathogenesis of systemic
sclerosis; the ins and outs. Hum Immunol. 79:178–187. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Masser DR, Hadad N, Porter H, Stout MB,
Unnikrishnan A, Stanford DR and Freeman WM: Analysis of DNA
modifications in aging research. Geroscience. 40:11–29. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lyu H, Huang J, He Z and Liu B: Epigenetic
mechanism of survivin dysregulation in human cancer. Sci China Life
Sci. 61:808–814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
El Bairi K, Tariq K, Himri I, Jaafari A,
Smaili W, Kandhro AH, Gouri A and Ghazi B: Decoding colorectal
cancer epigenomics. Cancer Genet. 220:49–76. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
James E and Jenkins TG: Epigenetics,
infertility, and cancer: Future directions. Fertil Steril.
109:27–32. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mayer W, Niveleau A, Walter J, Fundele R
and Haaf T: Demethylation of the zygotic paternal genome. Nature.
403:501–502. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hong LZ, Zhao XY and Zhang HL:
p53-mediated neuronal cell death in ischemic brain injury. Neurosci
Bull. 26:232–240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Crow MT: Revisiting p53 and its effectors
in ischemic heart injury. Cardiovasc Res. 70:401–403. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Crumrine RC, Thomas AL and Morgan PF:
Attenuation of p53 expression protects against focal ischemic
damage in transgenic mice. J Cereb Blood Flow Metab. 14:887–891.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li J, Chen G, Gao X, Shen C, Zhou P, Wu X,
Che X and Xie R: p53 participates in the protective effects of
ischemic post-conditioning against OGD-reperfusion injury in
primary cultured spinal cord neurons. Neurosci Lett. 638:129–134.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li Y, Chopp M, Zhang ZG, Zaloga C,
Niewenhuis L and Gautam S: p53-immunoreactive protein and p53 mRNA
expression after transient middle cerebral artery occlusion in
rats. Stroke. 25:849–856. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hai Z and Zuo W: Aberrant DNA methylation
in the pathogenesis of atherosclerosis. Clin Chim Acta. 456:69–74.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hiltunen MO, Turunen MP, Hakkinen TP,
Rutanen J, Hedman M, Mäkinen K, Turunen AM, Aalto-Setälä K and
Ylä-Herttuala S: DNA hypomethylation and methyltransferase
expression in atherosclerotic lesions. Vasc Med. 7:5–11. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Newman PE: Can reduced folic acid and
vitamin B12 levels cause deficient DNA methylation producing
mutations which initiate atherosclerosis? Med Hypotheses.
53:421–424. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Turunen MP, Aavik E and Yla-Herttuala S:
Epigenetics and atherosclerosis. Biochim Biophys Acta.
1790:886–891. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Durand P, Prost M, Loreau N, Lussier-Cacan
S and Blache D: Impaired homocysteine metabolism and
atherothrombotic disease. Lab Invest. 81:645–672. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mok V and Kim JS: Prevention and
management of cerebral small vessel disease. J Stroke. 17:111–122.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Joseph J and Loscalzo J: Methoxistasis:
Integrating the roles of homocysteine and folic acid in
cardiovascular pathobiology. Nutrients. 5:3235–3256. 2013.
View Article : Google Scholar : PubMed/NCBI
|