Introduction
Bladder cancer (BCa) is the second most common
cancer of the genitourinary tract (1), causing numerous cancer-associated
mortalities (2). The incidence of
BCa has increased over the past decades, and specific therapeutic
strategies were developed to treat BCa, including endoscopic
resection and radical cystectomy (3). However, due to recurrence, drug
resistance and tumor metastasis, the outcomes of patients with BCa
remain poor (4). Therefore, in
order to identify better therapeutic targets and develop more
effective therapeutic methods, understanding the mechanism
underlying BCa progression is required.
The number of identified long noncoding RNAs
(lncRNAs) has been increasing (5).
lncRNAs are >200 nucleotides in length and do not possess
protein-coding potential (6).
Accumulating evidence suggests that lncRNAs serve important roles
in numerous biological processes, including immunity, embryonic
development and cancer (7–9). In humans, a previous study
demonstrated that lncRNAs serve roles in cancer progression by
regulating cell proliferation, migration, invasion and apoptosis
(10). In BCa, numerous lncRNAs
were demonstrated to be aberrantly expressed, and were associated
with BCa progression. For example, lncRNA bladder cancer-associated
transcript 2 promoted BCa metastasis (11), and lncRNA fibroblast growth factor
receptor 3 antisense transcript 1 promoted BCa growth and
metastasis (12). Additionally,
lncRNA urothelial cancer associated 1 contributed to BCa
progression and metastasis by modulating the microRNA
(miR)-143/high mobility group box 1 signaling pathway (13).
Previous studies suggested that LINC00978 promoted
the development of gastric cancer and breast cancer (14,15).
However, the role of LINC00978 in BCa has not been investigated.
The present study identified that LINC00978 was upregulated in BCa
tissues. Furthermore, knockdown of LINC00978 suppressed the
proliferation, migration and invasion of BCa cells.
Mechanistically, it was identified that LINC00978 served as a
competing endogenous RNA (ceRNA) of miR-4288. The present study
demonstrated that LINC00978 promoted BCa cell proliferation,
migration and invasion by inhibiting miR-4288. Collectively, the
present study suggested that the LINC00978/miR-4288 axis is an
important regulator of BCa progression.
Materials and methods
Clinical specimens and cell lines
A total of 60 BCa (46 males and 14 females; age
range, 37–61 years old; median age, 51 years old) and adjacent
normal tissue samples were obtained from The Tiantai People's
Hospital of Zhejiang Province (Taizhou, China) between September
2014 and November 2016. Written informed consent was obtained from
all enrolled patients. Patients treated by radiotherapy or
chemotherapy before surgery were excluded. All other patients were
included. The present study was approved by The Ethics Committee of
Tiantai People's Hospital of Zhejiang Province (approval no.
2016020342). A total of four BCa cell lines (T24, J82, UMUC3 and
5637) and a normal bladder cell line (SV-HUC-1) were purchased from
The Cell Bank of The Chinese Academy of Sciences (Shanghai, China).
Cells were cultured in the Dulbecco's modified Eagle's medium
(DMEM; Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.) and maintained at 37°C in a cell incubator with a
humidified atmosphere containing 5% CO2.
Oligonucleotide and transfection
A small interfering RNA against LINC00978
(siLINC00978; 5′-GGACAGAGCAGAAGACAAA-3′), siRNA negative control
(siNC; 5′-AAUUCUCCGAACGUGUCAC-3′), miR-4288 mimics
(5′-UUGUCUGCUGAGUUUCC-3′), miR-4288 inhibitors
(5′-GGAAACUCAGCAGACAA-3′) and controls
(5′-UCACAACCUCCUAGAAAGAGUAGA-3′) were purchased from GeneCopoeia,
Inc. (Rockville, MD, USA). To overexpress LINC00978, its coding
sequence was constructed into pcDNA3 vector (Invitrogen; Thermo
Fisher Scientific, Inc.) to generate pcDNA3-LINC00978. The siRNAs
(100 nM), mimics (100 nM), inhibitor (100 nM) r, pcDNA3-LINC00978
(1 µg) and their corresponding negative controls (100 nM or 1 µg)
were transfected into T24 cells using Lipofectamine®
2000 transfection reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. After 48 h, the
transfection efficiency was confirmed by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) as
described below.
Cell proliferation assay
Cell proliferation was measured by Cell Counting
kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan). A total of 5×103 cells were seeded in 96-well
plates and incubated for 1, 2 and 3 days. Subsequently, 10 µl CCK-8
reagent was added to the 96-well plates. Following 2 h incubation
at 37°C, the absorbance at 450 nm was measured to detect the number
of viable cells using a SUNRISE microplate reader (Tecan Group,
Ltd., Männedorf, Switzerland).
Colony formation assay
A total of 5×102 T24 cells transfected
with siLINC00978 or siNC were seeded in 6-well plates. The culture
medium was replaced every other day. Following incubation for 10
days, the colonies from three groups were fixed with ice-cold 100%
methanol for 2 h at 4°C and stained with 0.1% crystal violet for 1
h at room temperature. Subsequently, the number of colonies was
determined using a light microscope (magnification, ×100; Nikon
Corporation, Tokyo, Japan).
RT and RT-qPCR
Total RNA was extracted from cultured cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol and cDNA was
synthesized from 1 µg total RNA using a PrimerScript RT reagent kit
(Takara Bio, Inc., Otsu, Japan). The RT conditions were: Incubation
for 1 h at 42°C and inactivation at 70°C for 15 min. miRNA from
total RNA was reverse transcribed using the PrimeScript miRNA cDNA
synthesis kit (Takara Bio, Inc.). RT-qPCR was performed with the
SYBR-Green Premix Ex Taq II (Takara Bio, Inc.) using the
StepOnePlus RT PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The PCR thermocycling conditions were: 95°C for
30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 34
sec. U6 was used as the endogenous control for miRNA and LINC00978
expression analysis. The relative gene expression level was
calculated using the 2−ΔΔCq method (16). The sequences of the primers were:
LINC00978 (Forward, 5′-TAGGAGACACAGGCAGAGCC-3′ and reverse,
5′-CTGACTAGAGCTTGTCTCA-3′), miR-4288 (Forward,
5′-AACGAGACGACGACAGAC-3′ and reverse, 5′-TTGTCTGCTGAGTTTCC-3′) and
U6 (Forward, 5′-AACGAGACGACGACAGAC-3′ and reverse,
5′-GCAAATTCGTGAAGCGTTCCATA-3′).
Transwell and Matrigel assays
For Transwell migration, following transfection of
siLINC00978 or siNC, 5×103 BCa cells were plated in the
upper chamber (Corning, Inc., Corning, NY, USA) containing 200 µl
serum-free DMEM. A total of 600 µl DMEM containing 10% FBS was
added to the lower chamber. Following an incubation of 24 h, the
cells migrated into the lower chamber were fixed with 100% methanol
at 4°C and stained with 1% crystal violet for 30 min at room
temperature. Cells were imaged using a light microscope at
magnification, ×100. For the Matrigel assay, the upper chamber was
pre-coated with Matrigel (1:3; Becton, Dickinson and Company,
Franklin, Lakes, NJ, USA). The invasion assay was performed
following the protocol used for the migration assay.
Luciferase reporter assay
The potential binding site for miR-4288 in LINC00978
were predicted by the miRDB tool (http://mirdb.org/miRDB/index.html). Then the sequence
containing the wild-type or mutant binding site was constructed
into pMIR-Report vector (Promega Corporation, Madison, WI, USA).
For luciferase reporter assay, miR-4288 mimics and wild-type or
mutant LINC00978 reporter were co-transfected into the cells using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Following transfection, cells were incubated for
24 h, and luciferase activity was determined with a luciferase
assay system kit (Promega Corporation). Renilla luciferase
activity was used for normalization.
Statistical analysis
Statistical analysis was conducted using SPSS 20.0
(IBM Corp., Armonk, NY, USA). Data from three independent
experiments were expressed as mean ± standard deviation.
Significant differences were calculated using Student's t-test or
one-way analysis of variance followed by Tukey's post hoc test.
Pearson's correlation coefficient analysis was used to determine
correlation. P<0.05 was considered to indicate a statistically
significant difference.
Results
LINC00978 is upregulated in BCa
tissues
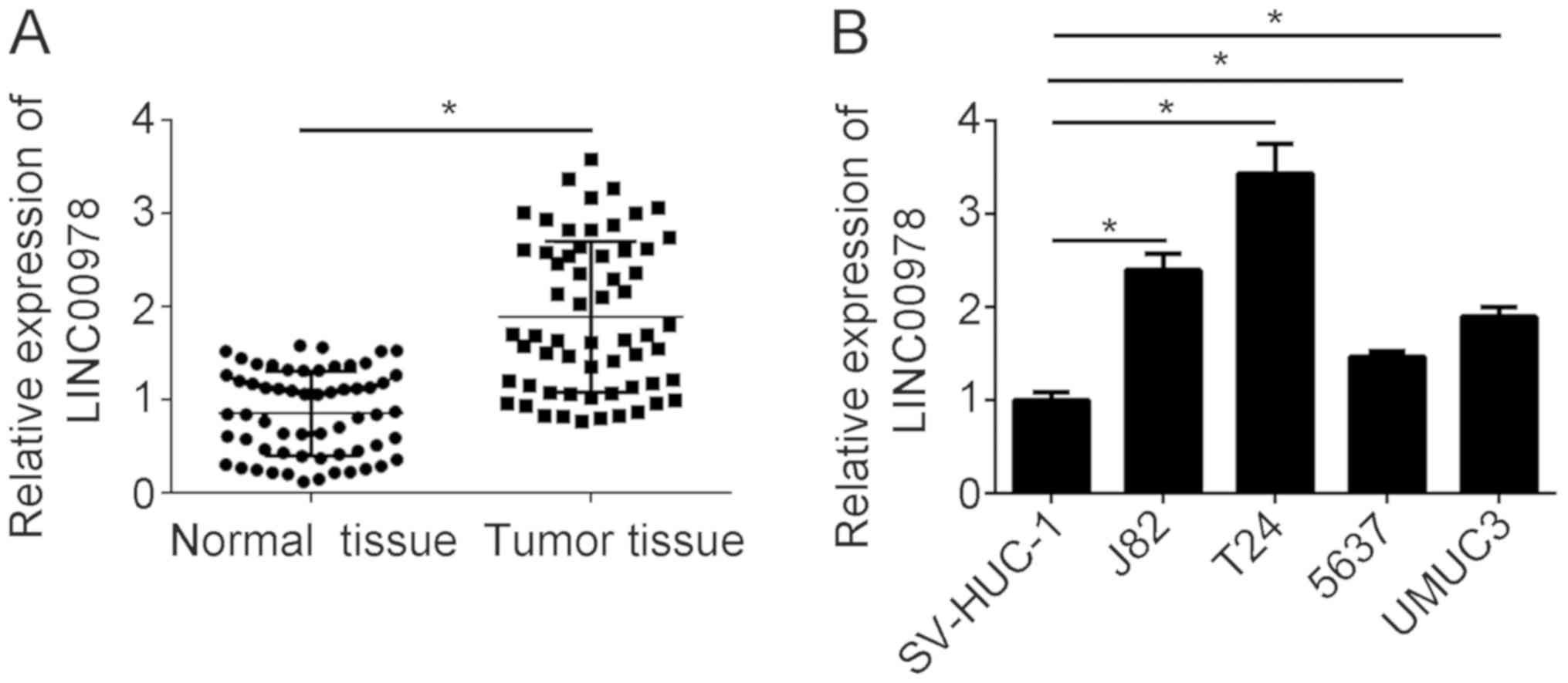
To investigate the role of LINC00978 in BCa, the
expression level of LINC00978 was analyzed by RT-qPCR. LINC00978
was significantly upregulated in BCa tissues compared with adjacent
normal tissues (Fig. 1A).
Furthermore, the expression level of LINC00978 was examined in BCa
cell lines. The results suggested that LINC00978 was significantly
upregulated in BCa cell lines, including J82, T24, 5637 and UMUC3
cells, compared with SV-HUC-1 cells (Fig. 1B). The present results suggested
that LINC00978 may be involved in BCa progression.
LINC00978 promotes BCa cell
proliferation
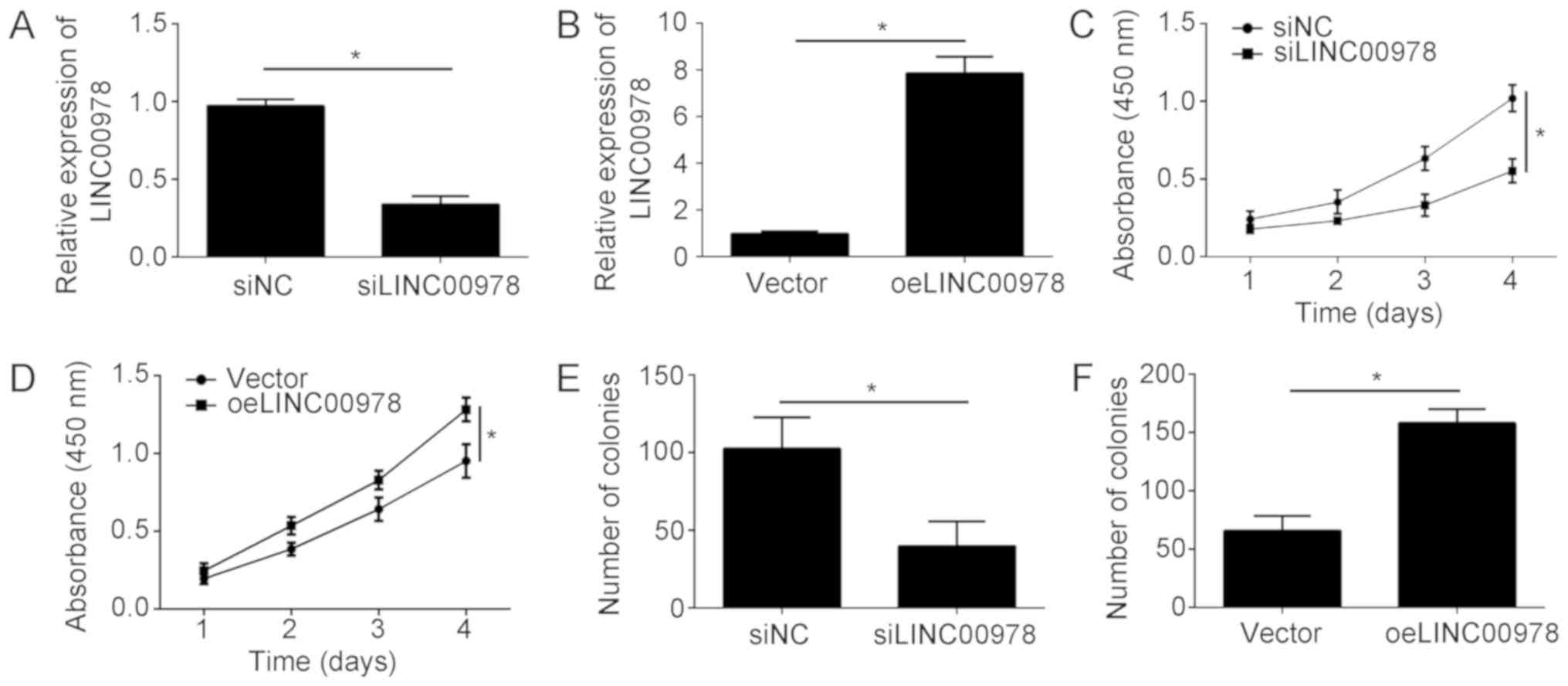
To investigate the function of LINC00978 in BCa, a
knockdown of LINC00978 was performed by transfecting cells with
siLINC00978. Additionally, overexpression of LINC00978 was
conducted by transfecting T24 cells with a plasmid expressing
LINC00978. RT-qPCR analysis demonstrated that the LINC00978
expression level was significantly decreased upon knockdown and
upregulated following overexpression (Fig. 2A and B). Subsequently, a CCK-8
assay was conducted. LINC00978 knockdown significantly inhibited
the proliferation of T24 cells at 4 days (Fig. 2C). Conversely, LINC00978
overexpression promoted cell proliferation at 4 days (Fig. 2D). To test the effect of LINC00978
on BCa cell proliferation, a colony formation assay was
additionally conducted. The results suggested that LINC00978
knockdown led to a significant decrease in the number of colonies
(Fig. 2E), whereas, LINC00978
overexpression caused an increased number of colonies (Fig. 2F). Collectively, the present data
suggested that LINC00978 promoted the proliferation of BCa
cells.
LINC00978 promotes BCa cell migration
and invasion
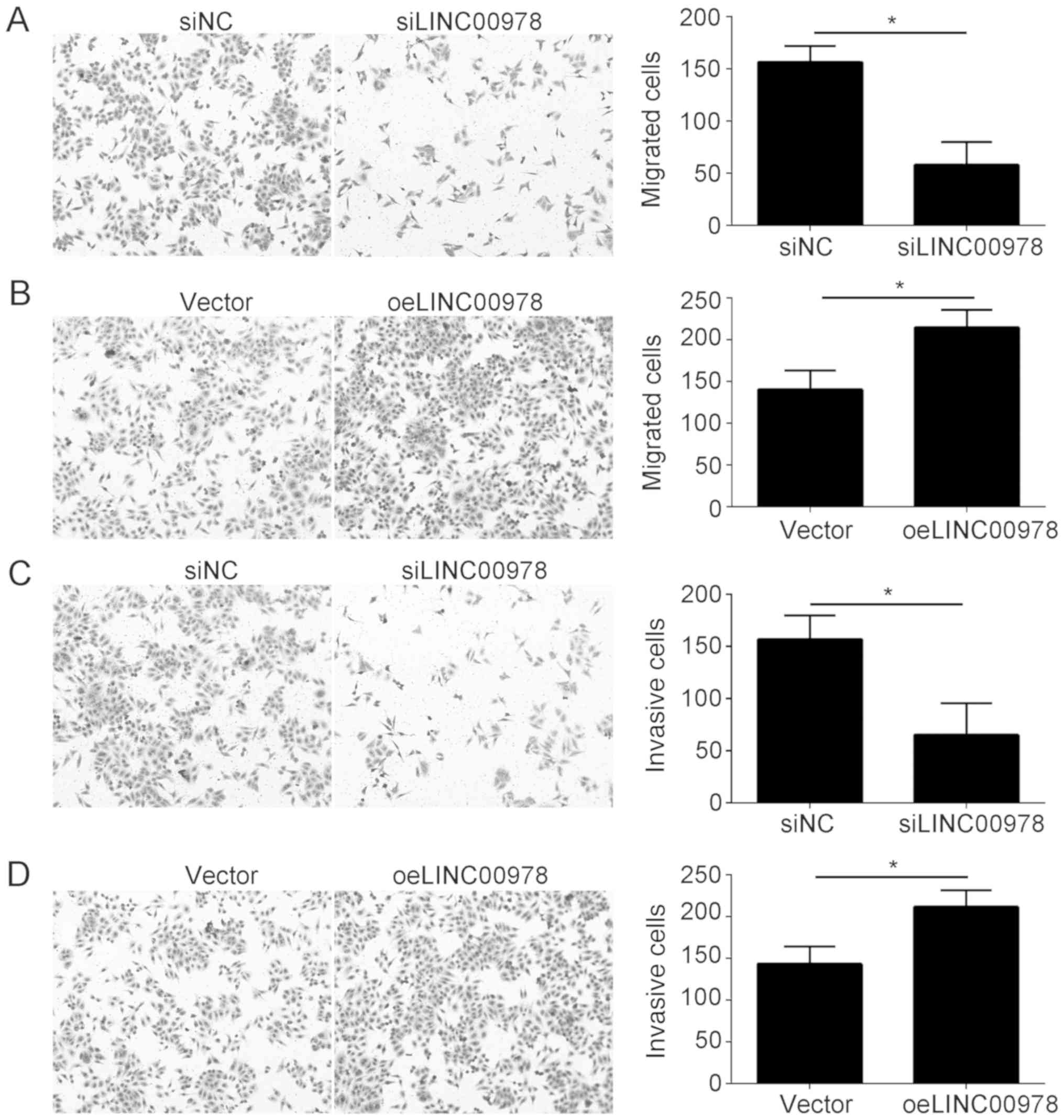
Malignancies are frequently associated with tumor
metastasis. To test whether LINC00978 had an effect on BCa
metastasis, a Transwell assay was conducted, using T24 cells.
LINC00978 knockdown significantly inhibited migration (Fig. 3A), whereas, overexpression of
LINC00978 caused the opposite effect (Fig. 3B). Similarly, LINC00978 knockdown
decreased the number of invasive cells (Fig. 3C), and overexpression of LINC00978
promoted invasion (Fig. 3D). The
present results suggested that LINC00978 contributed to the
metastasis of BCa.
LINC00978 sponges miR-4288 in BCa
cells
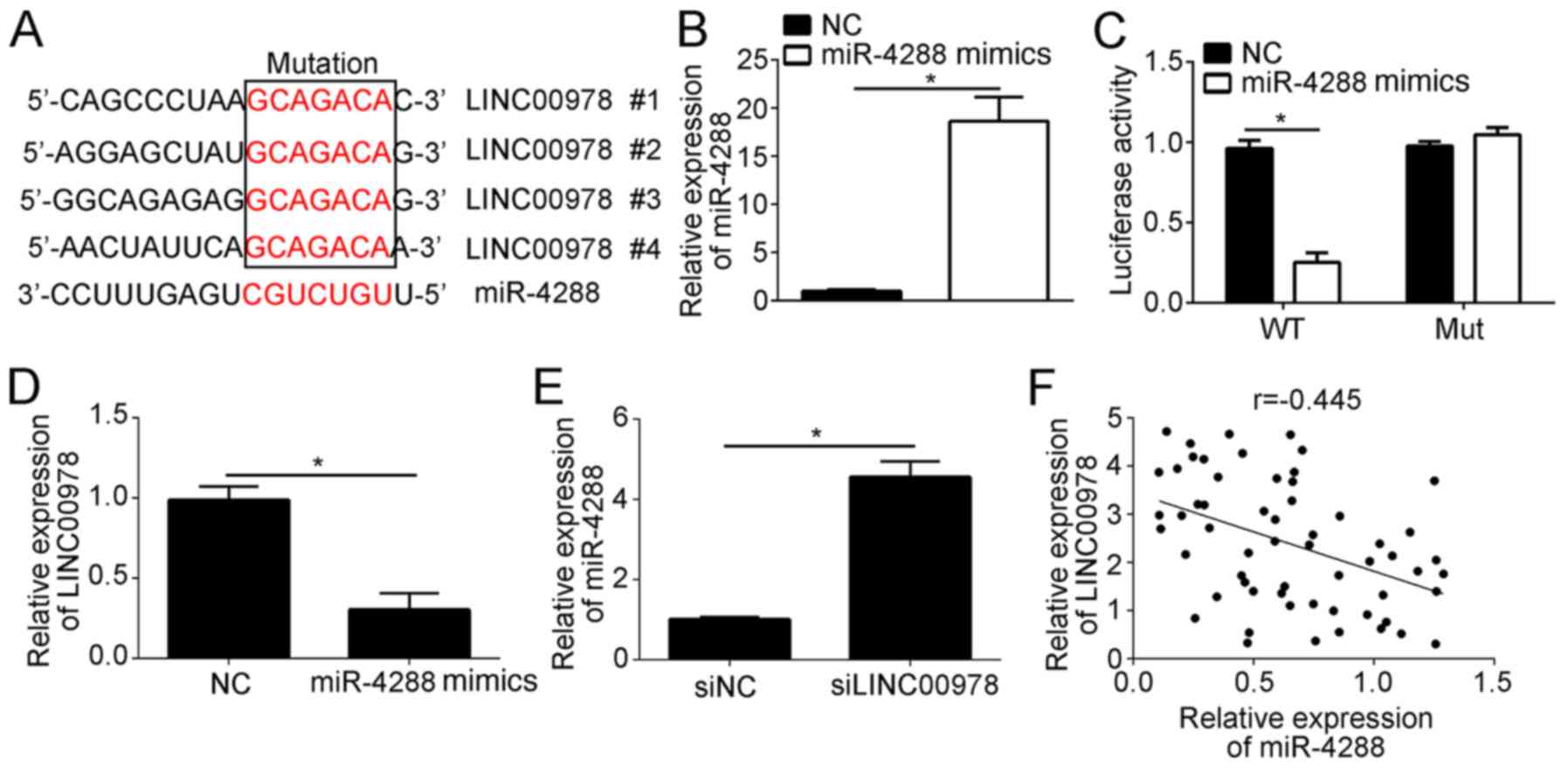
To investigate the mechanism underlying LINC00978
function in BCa, a bioinformatics analysis was performed. It was
identified that LINC00978 may serve as a ceRNA of miR-4288. In
total, four predicted binding sites of miR-4288 were identified in
LINC00978 (Fig. 4A). To test this
prediction, a luciferase reporter assay was performed, using
wild-type (WT) or mutant binding sites in a reporter plasmid. The
results demonstrated that overexpression of miR-4288 increased the
expression level of miR-4288 (Fig.
4B), and suppressed the luciferase activity of WT-LINC00978
(Fig. 4C). Notably, mutation of
the predicted binding sites abrogated this interaction (Fig. 4C). Furthermore, using RT-qPCR, it
was identified that miR-4288 overexpression significantly decreased
the expression level of LINC00978 in T24 cells (Fig. 4D). Notably, LINC00978 knockdown
increased the expression level of miR-4288 in T24 cells (Fig. 4E). Furthermore, the expression
level of LINC00978 was inversely correlated with miR-4288
expression level in BCa tissues (Fig.
4F). Collectively, the present data suggested that LINC00978
binds to miR-4288, inhibiting its expression in BCa cells.
Inhibition of miR-4288 reverses the
effects of LINC00978 knockdown on BCa cells
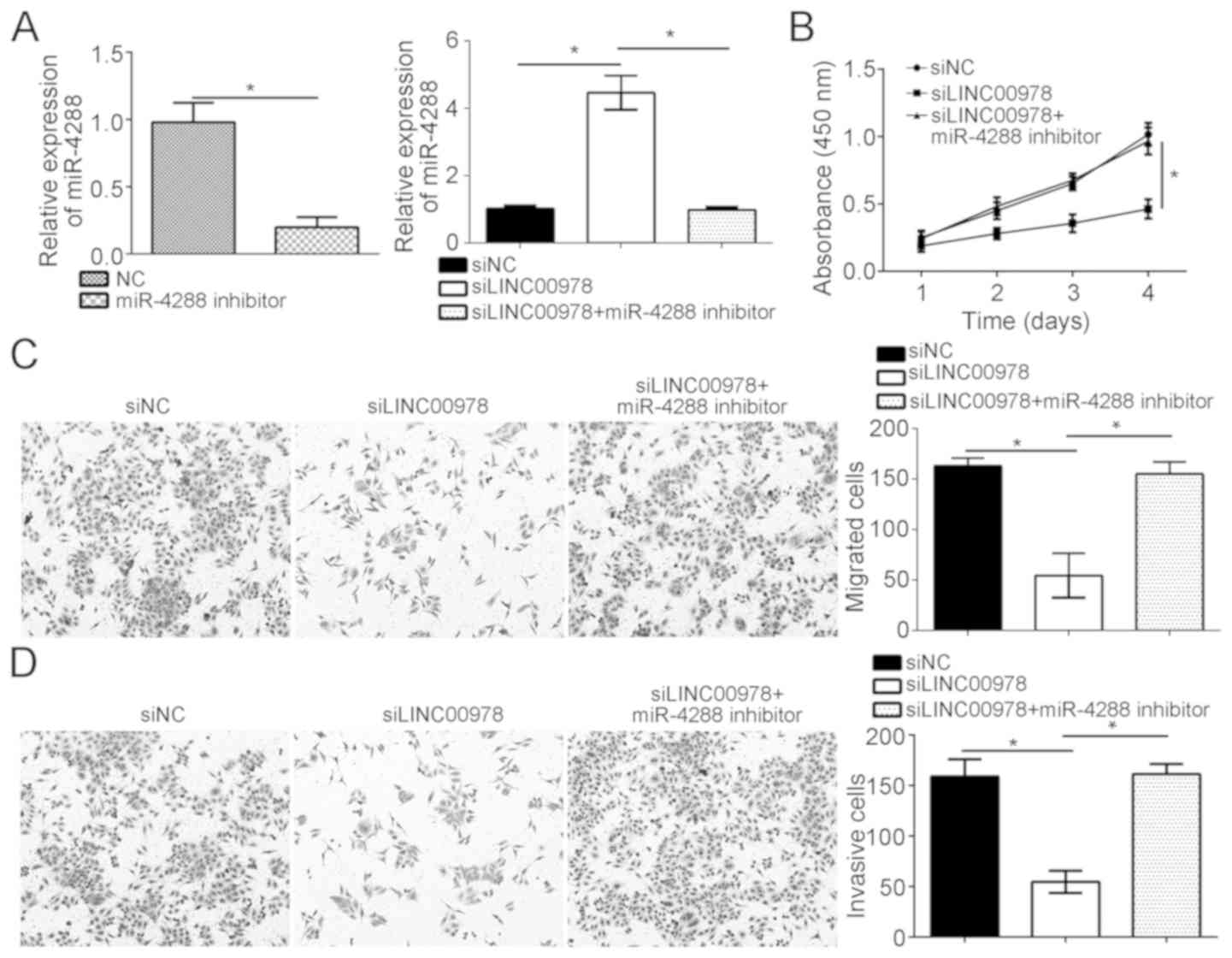
As the present study demonstrated that LINC00978
interacted with miR-4288, it was examined whether miR-4288 was
involved in regulating the function of LINC00978 in BCa cells.
miR-4288 was inhibited in LINC00978-silenced T24 cells using
miR-4288 inhibitors (Fig. 5A).
RT-qPCR analysis demonstrated that the miR-4288 expression level
was restored in LINC00978-silenced T24 cells with miR-4288
inhibitors (Fig. 5A).
Subsequently, CCK-8, Transwell and Matrigel assays were conducted.
LINC00978 knockdown inhibited the proliferation, migration and
invasion of T24 cells, whereas, inhibition of miR-4288 was able to
abrogate the effects of LINC00978 knockdown (Fig. 5B-D). Collectively, the present data
demonstrated that LINC00978 promoted the proliferation, migration
and invasion of BCa cells by inhibiting miR-4288.
Discussion
BCa is one of the most common types of cancer,
leading to numerous mortalities every year (17). BCa is divided into
non-muscle-invasive BCa (~80%) and muscle-invasive BCa (~20%),
based on the severity of tumor infiltration into the musculature of
the bladder (18). BCa is a public
health problem worldwide (2).
However, the underlying mechanism of BCa development and
progression remains poorly understood. In the present study, the
function of LINC00978 in BCa progression was investigated. The
present study demonstrated that the LINC00978 expression level was
significantly upregulated in BCa tissues and cell lines compared
with adjacent normal tissues and a normal cell line, respectively.
Knockdown of LINC00978 led to decreased proliferation of BCa cells
in vitro, and overexpression of LINC00978 led to the
opposite effect. Additionally, it was demonstrated that LINC00978
knockdown suppressed the migratory and invasive potential of BCa
cells. The present study demonstrated a novel function of LINC00978
in BCa. Collectively, the present findings suggested that LINC00978
serves an oncogenic role in BCa and may represent a potential
therapeutic target for the treatment of BCa.
A previous study suggests that lncRNAs may serve as
ceRNAs to sponge miRs and regulate gene expression (19). For example, small nucleolar RNA
host gene 16 contributed to the tumorigenesis of cervical cancer
cells by sponging miR-216-5p, which increased the expression level
of zinc finger E-box binding homeobox 1 (20). lncRNA urothelial cancer associated
1 promoted proliferation and conferred cisplatin resistance in oral
squamous cell carcinoma by suppressing miR-184 (21). lncRNA-ATB promoted
epithelial-mesenchymal transition in a model of silica-induced
pulmonary fibrosis by binding to miR-200c (22). X inactive specific transcript
promoted gastric cancer progression by targeting miR-185,
upregulating transforming growth factor β 1 expression level
(23). A previous study identified
that LINC00978 may serve as an oncogene in gastric and breast
cancer (14,15). Furthermore, LINC00978 upregulation
was associated with poor prognosis in gastric and breast cancer
(14,15). However, the mechanism underlying
LINC00978 regulation of cancer progression remains unknown.
In the present study, to examine the mechanism of
LINC00978, a bioinformatics analysis was performed. The present
results suggested that miR-4288 may represent a target of
LINC00978. A luciferase reporter assay suggested a direct
interaction between LINC00978 and miR-4288 in BCa cells.
Furthermore, it was identified that overexpression of miR-4288
inhibited LINC00978 expression in BCa cells, whereas, knockdown of
LINC00978 promoted miR-4288 expression. The expression of LINC00978
was inversely correlated with miR-4288 in BCa tissues.
Collectively, the present data suggested that LINC00978 may serve
as a ceRNA of miR-4288. To investigate whether LINC00978 serves as
an oncogene by sponging miR-4288, a rescue assay was performed. The
present results suggested that miR-4288 inhibition reversed the
effects of LINC00978 knockdown on BCa cell proliferation, migration
and invasion. Therefore, LINC00978 contributed to BCa progression
in vitro by inhibiting miR-4288.
A previous study suggests that miRNAs are able to
regulate gene expression and function, by serving as tumor
suppressors or oncogenes (24).
Although the present data suggested that miR-4288 served as a tumor
suppressor in BCa, the function of miR-4288 has not been previously
investigated, to the best of the authors' knowledge, and additional
studies are required to understand the signaling pathways
downstream of miR-4288 in BCa. Collectively, the present study
investigated the role of LINC00978 in BCa progression and suggested
that the LINC00978/miR-4288 axis may represent a potential
therapeutic target for treating BCa.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Funds of
Science Technology Department of Zhejiang Province (grant no.
LGF18H160027; China).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
WW contributed to the conception and design of the
present study. In addition, WW analyzed and interpreted the
results, and wrote the manuscript. ZX, JW and RC performed the
experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Tiantai People's Hospital of Zhejiang Province
(approval no. 2016020342; Taizhou, China). Written informed consent
was obtained from all enrolled patients.
Patient consent for publication
All patients within the present study provided
consent for the publication of their data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang M, Guo C, Wang L, Luo G, Huang C, Li
Y, Liu D, Zeng F, Jiang G and Xiao X: Long noncoding RNA GAS5
promotes bladder cancer cells apoptosis through inhibiting EZH2
transcription. Cell Death Dis. 9:2382018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tran MN, Goodwin Jinesh G, McConkey DJ and
Kamat AM: Bladder cancer stem cells. Curr Stem Cell Res Ther.
5:387–395. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Feng Z and Wang B: Long non-coding RNA
HNF1A-AS1 promotes cell viability and migration in human bladder
cancer. Oncol Lett. 15:4535–4540. 2018.PubMed/NCBI
|
|
6
|
Liao Z, Zhao J and Yang Y: Downregulation
of lncRNA H19 inhibits the migration and invasion of melanoma cells
by inactivating the NF-κB and PI3K/Akt signaling pathways. Mol Med
Rep. 17:7313–7318. 2018.PubMed/NCBI
|
|
7
|
Liu B, Ye B, Yang L, Zhu X, Huang G, Zhu
P, Du Y, Wu J, Qin X, Chen R, et al: Long noncoding RNA lncKdm2b is
required for ILC3 maintenance by initiation of Zfp292 expression.
Nat Immunol. 18:499–508. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ye B, Liu B, Yang L, Zhu X, Zhang D, Wu W,
Zhu P, Wang Y, Wang S, Xia P, et al: LncKdm2b controls self-renewal
of embryonic stem cells via activating expression of transcription
factor Zbtb3. EMBO J. 37(pii): e971742018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhao R, Wang X, Wang H, Yu T, Wang Q, Yang
X and Sun J: Inhibition of long noncoding RNA BDNF-AS rescues cell
death and apoptosis in hypoxia/reoxygenation damaged murine
cardiomyocyte. Biochimie. 138:43–49. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zheng R, Lin S, Guan L, Yuan H, Liu K, Liu
C, Ye W, Liao Y, Jia J and Zhang R: Long non-coding RNA XIST
inhibited breast cancer cell growth, migration, and invasion via
miR-155/CDX1 axis. Biochem Biophys Res Commun. 498:1002–1008. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He W, Zhong G, Jiang N, Wang B, Fan X,
Chen C, Chen X, Huang J and Lin T: Long noncoding RNA BLACAT2
promotes bladder cancer-associated lymphangiogenesis and lymphatic
metastasis. J Clin Invest. 128:861–875. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liao X, Chen J, Liu Y, He A, Wu J, Cheng
J, Zhang X, Lv Z, Wang F and Mei H: Knockdown of long noncoding RNA
FGFR3- AS1 induces cell proliferation inhibition, apoptosis and
motility reduction in bladder cancer. Cancer Biomark. 21:277–285.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo J, Chen J, Li H, Yang Y, Yun H, Yang S
and Mao X: LncRNA UCA1 promotes the invasion and EMT of bladder
cancer cells by regulating the miR-143/HMGB1 pathway. Oncol Lett.
14:5556–5562. 2017.PubMed/NCBI
|
|
14
|
Fu M, Huang Z, Zang X, Pan L, Liang W,
Chen J, Qian H, Xu W, Jiang P and Zhang X: Long noncoding RNA
LINC00978 promotes cancer growth and acts as a diagnostic biomarker
in gastric cancer. Cell Prolif. 51:2018. View Article : Google Scholar :
|
|
15
|
Deng LL, Chi YY, Liu L, Huang NS, Wang L
and Wu J: LINC00978 predicts poor prognosis in breast cancer
patients. Sci Rep. 6:379362016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(T)(-Delta DeltaC) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ploeg M, Aben KK and Kiemeney LA: The
present and future burden of urinary bladder cancer in the world.
World J Urol. 27:289–293. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Millan-Rodriguez F, Chechile-Toniolo G,
Salvador-Bayarri J, Palou J, Algaba F and Vicente-Rodriguez J:
Primary superficial bladder cancer risk groups according to
progression, mortality and recurrence. J Urol. 164:680–684. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li HJ, Sun XM, Li ZK, Yin QW, Pang H, Pan
JJ, Li X and Chen W: LncRNA UCA1 promotes mitochondrial function of
bladder cancer via the MiR-195/ARL2 signaling pathway. Cell Physiol
Biochem. 43:2548–2561. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu H, Zeng Y, Zhou CC and Ye W:
SNHG16/miR-216-5p/ZEB1 signal pathway contributes to the
tumorigenesis of cervical cancer cells. Arch Biochem Biophys.
637:1–8. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fang Z, Zhao J, Xie W, Sun Q, Wang H and
Qiao B: LncRNA UCA1 promotes proliferation and cisplatin resistance
of oral squamous cell carcinoma by sunppressing miR-184 expression.
Cancer Med. 6:2897–2908. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu Y, Li Y, Xu Q, Yao W, Wu Q, Yuan J,
Yan W, Xu T, Ji X and Ni C: Long non-coding RNA-ATB promotes EMT
during silica-induced pulmonary fibrosis by competitively binding
miR-200c. Biochim Biophys Acta Mol Basis Dis. 1864:420–431. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang Q, Chen B, Liu P and Yang J: XIST
promotes gastric cancer (GC) progression through TGF-β1 via
targeting miR-185. J Cell Biochem. 119:2787–2796. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tonouchi E, Gen Y, Muramatsu T, Hiramoto
H, Tanimoto K, Inoue J and Inazawa J: miR-3140 suppresses tumor
cell growth by targeting BRD4 via its coding sequence and
downregulates the BRD4-NUT fusion oncoprotein. Sci Rep. 8:44822018.
View Article : Google Scholar : PubMed/NCBI
|