Introduction
Microglia represent immune cells of the central
nervous system (CNS) and are the main cells responsible for the
intrinsic brain immune system (1,2).
Under normal conditions, microglia monitor the surroundings
continuously to identify the presence of CNS injury or damage, and
they seem to preserve homeostasis through several interactions with
neurons. However, continuous microglial activation increases the
release of proinflammatory cytokines (1), which may cause detrimental
inflammatory effects (3). This
chronic inflammatory process enlists effector cells and can create
a feedback loop, perpetuating inflammation and finally damaging the
neurons (4,5). Microglial production of cytokines
occurs as a response to injury, trauma or infection (6). Thus, the chronic activation of
microglia is a critically important step in the development of
neurodegeneration, leading to neuronal malfunction or complete
neuron disability (1).
The knowledge of the detrimental effects caused by
inflammation in the development of neurodegeneration shows that
reducing inflammation may assist in delaying disease advancement.
Compounds that can modulate microglial activity and decrease
inflammation to controllable levels could be beneficial for
neuronal survival (7). Moreover,
the role of neuroinflammation in Alzheimer's disease (AD)
progression was previously reported, showing the effects of NSAIDs
on AD progression (8,9). Furthermore, several AD models were
used to demonstrate the neuroprotective effects of different
compounds with anti-inflammatory potential (10). Consequently, attenuating
inflammatory mediators can help reduce or retard the advancement of
AD.
Various compounds extracted from plants have shown
strong anti-inflammatory activity. The polyphenol pentagalloyl
glucose (1,2,3,4,6-penta-O-galloyl-β-D-glucose, PGG) is found in
many medicinal plants, including Rhus chinensis Mill and
Paeonia suffruticosa (5,11).
This compound has been reported in several in vivo and in
vitro studies showing its potential in the therapy and
prevention of several inflammatory diseases. Our earlier work
showed that PGG inhibited the release of MCP-5 (monocyte
chemoattractant protein-5) and pro-MMP-9 (pro-metalloproteinase-9)
in activated BV-2 cells (12). PGG
also modulated genes and proteins involved in the nuclear-factor-κB
(NF-κB) and mitogen-activated protein kinase (MAPK) signaling
pathways, which may affect the MAPK cascade, NF-κB activation, and
the subsequent release of MCP-5 and pro-MMP-9 (13). The current work investigated the
effect of PGG on the expression of proteins that may be involved in
the pathogenesis of neuroinflammation and neurodegenerative
diseases using an LPS/IFNγ-activated BV-2 microglial cell
model.
Materials and methods
Chemicals and reagents
Polyphenol pentagalloyl glucose
(1,2,3,4,6-penta-O-galloyl-β-D-glucose-purity 96.8%), bovine serum
albumin (BSA), dimethyl sulfoxide (DMSO), lipopolysaccharide from
Salmonella enterica (LPS), interferon γ (IFNγ), urea,
Tris/HCl, iodoacetamide, trypsin, NaCl, trifluoracetic acid
(CF3COOH), ammonium bicarbonate
(NH4HCO3), and general chemicals were
purchased from Sigma-Aldrich Co. and VWR International. Dulbecco's
modified Eagle's medium-high glucose medium (DMEM),
penicillin/streptomycin, fetal bovine serum heat inactivated
(FBS-HI), trypsin-EDTA, and Hank's Balanced Salt Solution (HBSS)
were purchased from Genesee Scientific. All reagents and plates for
western blot assays were purchased from ProteinSimple. Bradford
reagent, PCR primers, and reagents were from Bio-Rad, and primary
antibodies were from Thermo Fisher Scientific (Table I).
 | Table I.List of primary antibodies. |
Table I.
List of primary antibodies.
| Antibody | Catalog no. | Type | Species
reactivity | Host/Isotype |
|---|
| Septin-7 | PA5-56181 | Polyclonal | human, mouse,
rat | Rabbit/IgG |
| Ataxin-2 | PA5-54336 | Polyclonal | human, mouse,
rat | Rabbit/IgG |
| ADSS | PA5-55070 | Polyclonal | human, mouse,
rat | Rabbit/IgG |
Cell culture and treatments
The immortalized murine microglial BV-2 cell line
was provided by Dr Elizabetta Blasi (14). Cells were grown in T-75 flasks
using DMEM supplemented with 10% heat-inactivated FBS and 1%
penicillin (100 U/ml)/streptomycin (0.1 mg/ml) in an incubator set
for 5% CO2. at 37°C. For the experimental media, only
2.5% of heat-inactivated FBS was added. LPS and IFNγ were stored at
−20°C at concentrations of 1 mg/ml and 200 ng/ml, respectively. The
working concentrations of 0.2 µg/ml and ٠.٢ ng/ml were then used in
the experiments. DMSO was used to dissolve PGG, and the final
concentration of DMSO did not exceed 0.1% (15). The selected concentration of PGG
was based on the cell viability results from our prior study
(12), which showed >80% of
viable cells after treatment with 25 µM PGG. The count of cells in
all the experiments was ٥x١٠5/ml, and the treatments
consisted of the control (DMSO), PGG, LPS/IFNγ, and PGG followed by
the combination of LPS/IFNγ after 1 h.
Proteomic assay
In this experiment, BV-2 cells were plated in
experimental media and incubated overnight. The next day, the cells
were treated as described before. After 24 h, cells were harvested
using 0.25% trypsin-EDTA by first aspirating the medium,
centrifuging, and washing the cells twice with PBS. Once the pellet
was ready, using the Filter Aided Sample Preparation (FASP)
protocol, samples were prepared by mixing 30 µl of cell lysate with
200 µl of UA (8 M urea in 0.1 M Tris/HCl pH 8.5) in the filter unit
and centrifuged twice. The flow through from the collection tube
was discarded. Then, 100 µl of IAA (0.05 M iodoacetamide in UA)
solution was added and mixed at 600 rpm in a thermomixer for 1 min
and incubated without mixing for 20 min. After centrifuging, 100 µl
of UA was added to the filter unit and centrifuged. Next, 100 µl of
ABC (0.05 M NH4HCO3 in water) was added to
the filter unit and centrifuged. Then, 40 µl ABC with trypsin
(enzyme to protein ratio 1:100) was added and mixed at 600 rpm in a
thermomixer for 1 min. The units were incubated at 37°C for 18 h,
and the filter units were transferred to new collection tubes.
Then, 50 µl of NaCl (٠.٥ M) was pipetted into the filter and
centrifuged. Using CF3COOH, the filtrate was acidified,
desalted and sent to the Translational Science Laboratory for
Orbitrap/QExactive Proteomic LC-MS/MS (liquid chromatography-mass
spectrometry) for complex mixture analysis. Using ‘Scaffold version
4.4’ software, the results were then analyzed, and the fold change
of each protein of interest was calculated.
Protein assay
Cells were appropriately treated and kept in the
incubator for 24 h. The next day, cells were harvested, washed
twice with PBS, and lysed with buffer containing protease inhibitor
cocktail. Using Bradford reagent, the concentration of protein was
measured. Five µl of standards in concentrations of ٠ to ٢ mg/ml or
5 µl of samples and ٢٠٠ µl of protein assay reagent were added to
the ٩٦-well plate. Using a Synergy HTX Multi-Reader (BioTek), the
concentration of proteins was measured at 595 nm wavelength.
Capillary electrophoresis western blot
analysis
Total protein expression was determined using
western blot analysis (Wes, ProteinSimple). The protein
concentration was determined as described above. ProteinSimple
provided the reagents and protocol for the assay. The
concentrations of antibody and protein for the experiments were
first optimized by being tested at 3 different concentrations. The
best concentration was then selected for further tests. Briefly,
the protein samples (concentration: 0.2 mg/ml) were mixed with
sample buffer, fluorescent molecular weight markers, and
dithiothreitol and left in a heat block at 95°C for 5 min. The
microplate provided with the kit was then loaded with blocking
buffer, antibodies (in dilutions ranging from 1:5 to 1:25),
secondary antibody, chemiluminescent substrate, separation and
stacking matrices. The microplate was then placed in the instrument
and through the capillary system; the electrophoresis and
immunodetection occurred. The reaction identifies specific proteins
by using primary and secondary antibodies and a chemiluminescent
substrate. The Wes instrument provided us with real-time results of
the experiment. By using a charge-coupled device camera, the
chemiluminescence reaction and the digital image were analyzed by
the software (ProteinSimple Compass).
Reverse transcriptase-polymerase chain
reaction (RT-qPCR)
RNA extraction and cDNA synthesis
Cell pellets from the different treatments were
initially lysed with 1 ml of TRIzol reagent. Chloroform (0.2 ml)
was added, vortexed, incubated at 15–30°C for 2–3 min, and
centrifuged at 10,000 g for 15 min at 2–8°C. The aqueous phase of
the lysed samples was transferred to a fresh tube, and the RNA was
precipitated by mixing with isopropyl alcohol (0.5 ml). After
centrifugation, 75% ethanol was used to wash the RNA pellet. After
centrifugation at 7,500 g for 5 min at 2–8°C, the pellet was left
to dry for 15 min and then dissolved in water (RNase free). The
purity and RNA concentration were measured using a NanoDrop (Thermo
Fischer Scientific). Using iScript advanced reverse transcriptase
from Bio-Rad, the cDNA strands were synthesized based on the mRNA.
The solutions were loaded into 0.2 ml tubes and included 5X iScript
advanced reaction mix (4 µl) (containing primers), reverse
transcriptase (١ µl), sample (١.٥ µg/٧.٥ µl), and water (٧.٥ µl) in
a total of ٢٠ µl. The thermal cycling for reverse transcription
steps included ٣٠ min at 42°C and 5 min at 85°C.
RT-PCR
PCR amplification followed the Bio-Rad protocol. A 1
µl aliquot of the sample (٢٠٠ ng cDNA/reaction), 10 µl of
SsoAdvanced™ Universal SYBR®-Green Supermix (Bio-Rad), 1
µl of primer, and ٨ µl of water were pipetted into each well. Using
the CFX٩٦ Real-Time System from Bio-Rad, the protocol provided by
the company was followed: Initial 95°C for 2 min (hold step) and
95°C for 15 sec (denaturation), followed by 40 cycles of 60°C for
30 sec (annealing/extension) and 65–95°C for ٥ sec/step (melting
curve). Primers were selected according to the specific genes of
interest. The identification of each primer was provided by the
manufacturer (Bio-Rad).
– Septin-7-UniqueAssayID:
qMmuCID0017359
– Ataxin-2-UniqueAssayID:
qMmuCID0025396
– ADSS-UniqueAssayID:
qMmuCID0005512
Data analysis
Statistical analysis
Statistical analysis of the obtained results
included a minimum of 3 biological replicates and 3 technical
replicates for each one of the assays. For the proteomics assay,
data analysis was performed using Scaffold software, and the
significance of the difference between the groups was assessed
using Student's t-test (P<0.05). The results from the western
blot assay were analyzed using ProteinSimple Compass software from
the manufacturer. For the RT-PCR assays, Bio-Rad CFX manager
software was used. The software automatically calculates the
differences in mRNA expression for each of the treatments compared
to the reference gene (GAPDH). All data are expressed as the mean ±
standard error of the mean, and the significance of the difference
between the groups was assessed using a one-way analysis of
variance, followed by Dunnett's multiple comparison post hoc tests.
P<0.05 was considered to indicate a statistically significant
difference.
Bioinformatics analysis
The NCBI (National Center for Biotechnology
Information-ncbi.nlm.nih.gov)
(16) and PANTHER (Protein
Analysis Through Evolutionary Relationships-pantherdb.org) (17) databases were used to identify
biological and molecular functions, as well as signaling pathways
associated with the proteins.
Results
A proteomic approach was used to profile the effect
of PGG on global protein expression in LPS/IFNγ-activated BV-2
microglia cells. A total of 1895 proteins were identified as being
differentially expressed after cells were treated with PGG,
LPS/IFNγ or the combinations of PGG + LPS/IFNγ. Among those,
specific proteins were selected according to the fold change.
Proteins with at least a 7-fold change in inhibition after PGG
treatment were chosen, resulting in a total of 24 proteins. Then,
statistical analysis was used to select those that showed
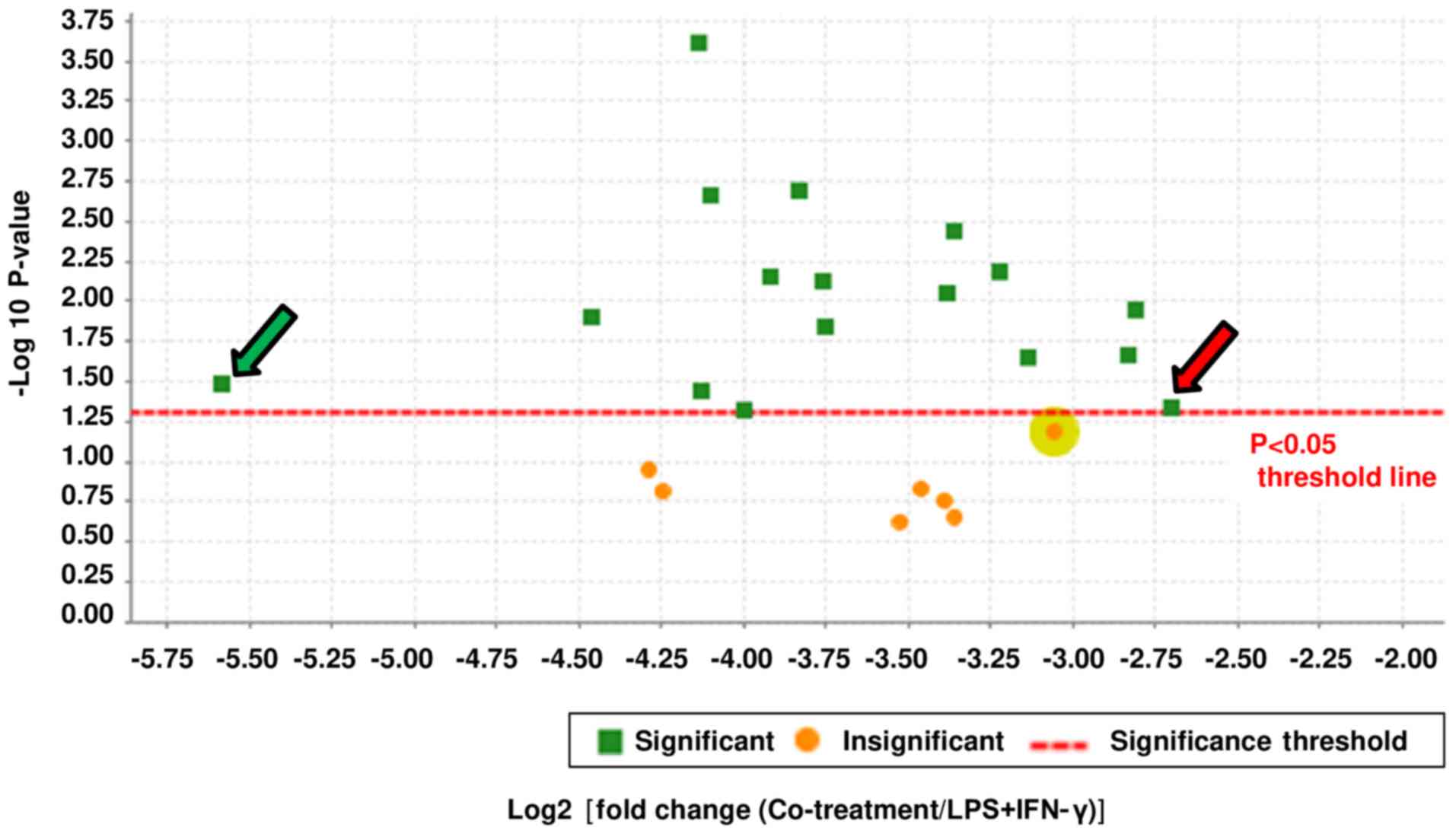
significant inhibition after PGG treatment. A total of 17 proteins
were identified for further studies, as shown in the volcano plot
(Fig. 1). The fold change was
calculated by comparing the control vs. LPS/IFNγ groups (Table II) and the LPS/IFNγ vs. PGG +
LPS/IFNγ groups (Table III). The
data obtained show that even for those proteins where LPS/IFNγ did
not induce an augmentation in protein expression, PGG pretreatment
downregulated their expression up to 53-fold (Tables II and III). Some of these highly downregulated
proteins are known to be involved in cancer progression.
 | Table II.Effect of LPS/IFNγ on the protein
expression of BV-2 cells. |
Table II.
Effect of LPS/IFNγ on the protein
expression of BV-2 cells.
| Protein name | Protein ID | Molecular weight,
kDa | P-value
(t-test) | Fold change |
|---|
| Ataxin-2-like
protein | ATX2L | 111 | 0.047 | +3 |
| Septin-7 | SEPT7 | 51 | 0.011 | +2 |
| SNW
domain-containing protein | SNW1 | 61 | 0.048 | +5 |
|
WAS/WAS/L-interacting protein | WIPF1 | 50 | 0.050 | 1 |
| Thymidylate
synthase | TYSY | 35 | <0.00010 | 1 |
| Acidic leucine-rich
nuclear phosphoprotein 32 family member | AN32B | 31 | 0.011 | 1 |
| 5′-3′
exoribonuclease 2 | XRN2 | 109 | 0.0018 | 1 |
|
Protein-L-isoaspartate (D-aspartate)
O-methyltransferase | PIMT | 25 | 0.0036 | 1 |
| Puromycin-sensitive
aminopeptidase | PSA | 103 | <0.00014 | +3.4 |
| Coronin-1B | COR1B | 54 | 0.017 | 1 |
| Bleomycin
hydrolase | BLMH | 53 | 0.040 | +1.6 |
| Emerin | EMD | 29 | 0.013 | +1.7 |
| Nuclear transport
factor 2 | NTF2 | 14 | <0.00095 | 1 |
| Treacle
protein | TCOF | 135 | <0.00098 | 1 |
| Adenylosuccinate
synthetase isozyme 2 | PURA2 | 50 | <0.00010 | 1 |
| Protein
phosphatase | PPM1G | 59 | <0.00010 | 1 |
| Glyoxalase
domain-containing protein 4 | GLOD4 | 33 | <0.00010 | +3.2 |
 | Table III.Effect of PGG on LPS/IFNγ-activated
BV-2 Cells |
Table III.
Effect of PGG on LPS/IFNγ-activated
BV-2 Cells
| Protein name | Protein ID | Molecular weight,
kDa | P-value
(t-Test) | Fold change |
|---|
| Ataxin-2-like
protein | ATX2L | 111 | 0.015 | −7.2 |
| Septin-7 | SEPT7 | 51 | 0.017 | −8 |
| SNW
domain-containing protein | SNW1 | 61 | 0.048 | −8.4 |
|
WAS/WAS/L-interacting protein | WIPF1 | 50 | 0.022 | −9.4 |
| Thymidylate
synthase | TYSY | 35 | 0.01 | −9.7 |
| Acidic leucine-rich
nuclearphosphoprotein 32 family member | AN32B | 31 | 0.025 | −10 |
| 5′-3′
exoribonuclease 2 | XRN2 | 109 | 0.015 | −14 |
|
Protein-L-isoaspartate (D-aspartate)
O-methyltransferase | PIMT | 25 | 0.0013 | −14 |
| Puromycin-sensitive
aminopeptidase | PSA | 103 | 0.008 | −15 |
| Coronin-1B | COR1B | 54 | 0.029 | −16 |
| Bleomycin
hydrolase | BLMH | 53 | 0.0089 | −17 |
| Emerin | EMD | 29 | 0.046 | −18 |
| Nuclear transport
factor 2 | NTF2 | 14 | 0.0075 | −20 |
| Treacle
protein | TCOF | 135 | 0.012 | −23 |
| Adenylosuccinate
synthetase isozyme 2 | PURA2 | 50 | 0.0028 | −23 |
| Protein
phosphatase | PPM1G | 59 | 0.041 | −30 |
| Glyoxalase
domain-containing protein 4 | GLOD4 | 33 | 0.022 | −53 |
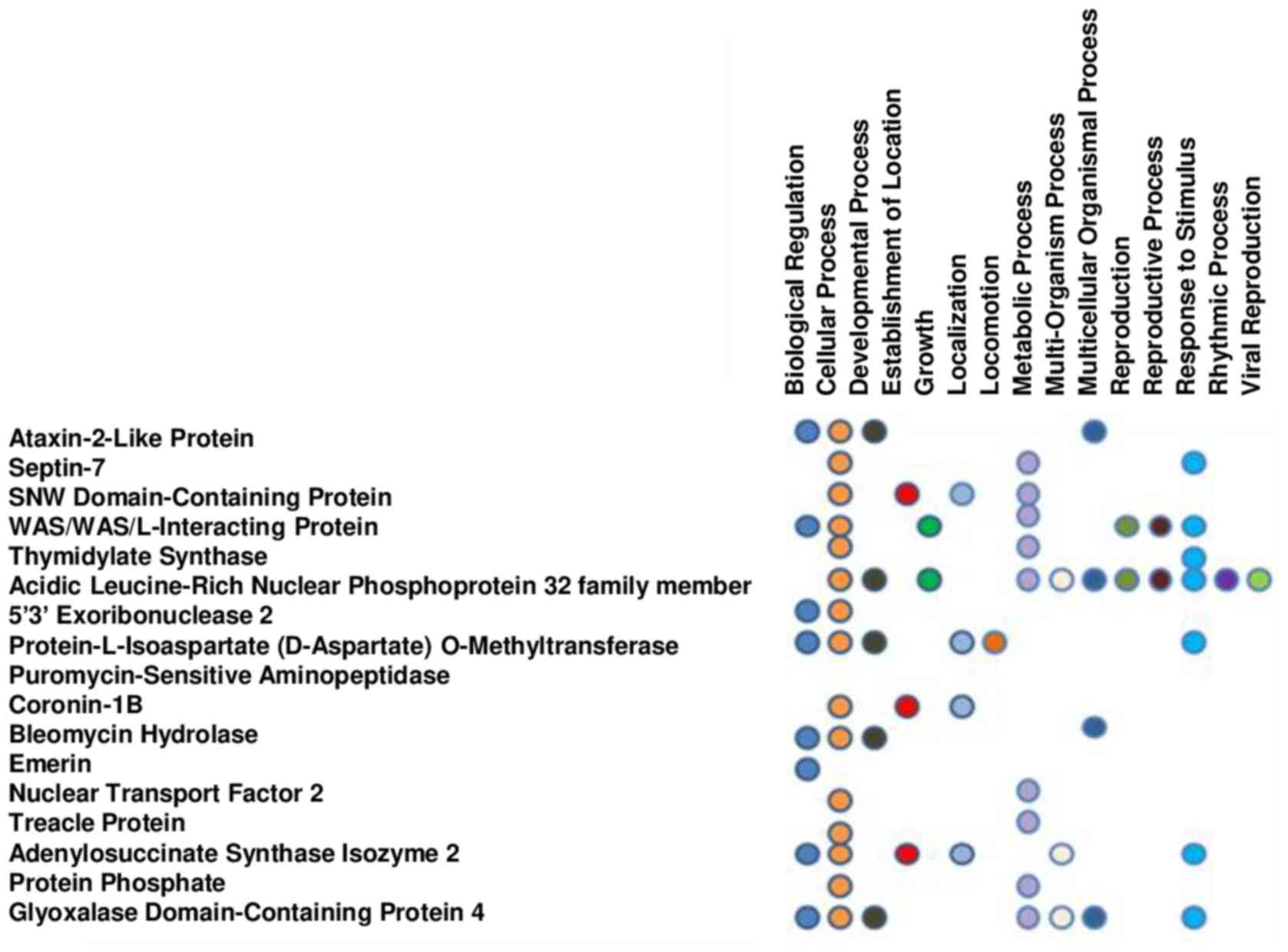
Annotation of the function of the different proteins
was performed using the NCBI data file to identify the biological
relevance of the differentially expressed proteins in response to
PGG treatment. The proteins were placed into 15 biological
processes (biological regulations, cellular and developmental
processes, establishment of localization, growth, localization,
locomotion, metabolic process, multi-organism and multicellular
organism processes, reproduction, reproductive process, response to
stimulus, rhythmic process and viral reproduction) (Fig. 2) and 5 molecular functions
(binding, catalytic activity, molecular function, transcription
regulator activity, and transporter activity).
The PANTHER classification system was used to
classify the proteins according to the signaling pathway that they
are associated with. The database found that 3 proteins
(adenylosuccinate synthase isozyme 2 (ADSS), thymidylate synthase,
and 5′-3′ exoribonuclease) were involved in 6 signaling pathways:
DNA replication, de novo purine biosynthesis, de novo pyrimidine
deoxyribonucleotide biosynthesis, formyltetrahydrofolate
biosynthesis, tetrahydroformate biosynthesis, and the Wnt signaling
pathway.
Among the 17 proteins significantly downregulated by
PGG, septin-7, ataxin-2 and ADSS were of special interest because
these proteins were previously described as being highly expressed
in neurodegenerative diseases and/or involved in the development of
neuronal circuits and control of AD and Parkinson's disease (PD)
pathogenesis. To validate the proteomic results, we performed
western blot assays with specific antibodies against Septin-7,
Ataxin-2 and ADSS. The results showed that LPS/IFNγ induced the
expression of these proteins in BV-2 microglial cells and
significantly reduced their expression when activated BV-2
microglial cells were pretreated with PGG (Figs. 3–5).
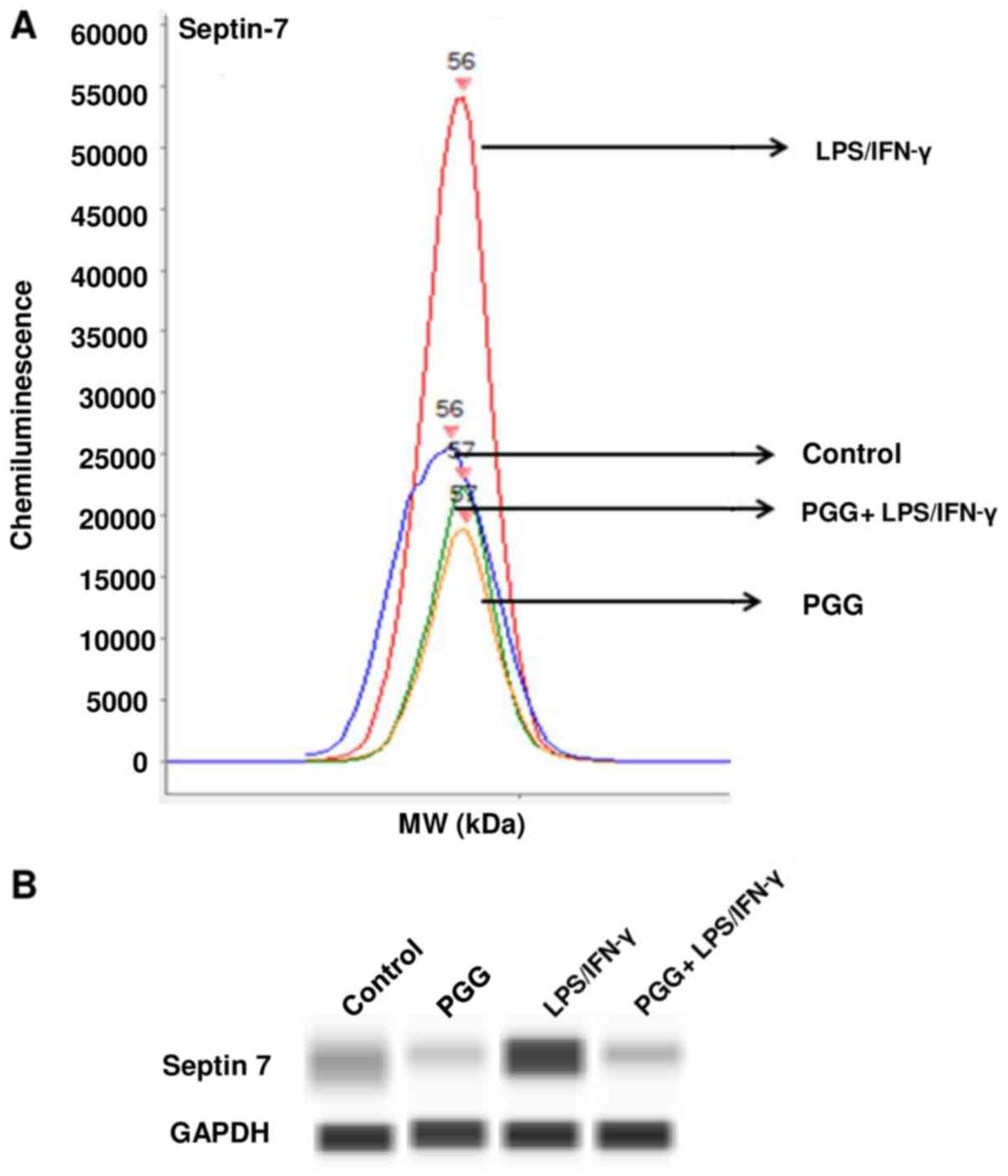 | Figure 3.Effect of PGG on the expression of
septin-7 as determined by western analysis with automated capillary
electrophoresis. (A) Graph of the amount of chemiluminescence
measured in the different treatments. (B) Bands representing the
protein expression in control (DMSO), PGG (25 µM), LPS/IFNγ, and
pretreated cells (PGG + LPS/IFNγ) after 24 h of treatment (n=3).
PPG, polyphenol pentagalloyl glucose
(1,2,3,4,6-penta-O-galloyl-β-D-glucose); LPS, lipopolysaccharide;
IFNγ, interferon γ. |
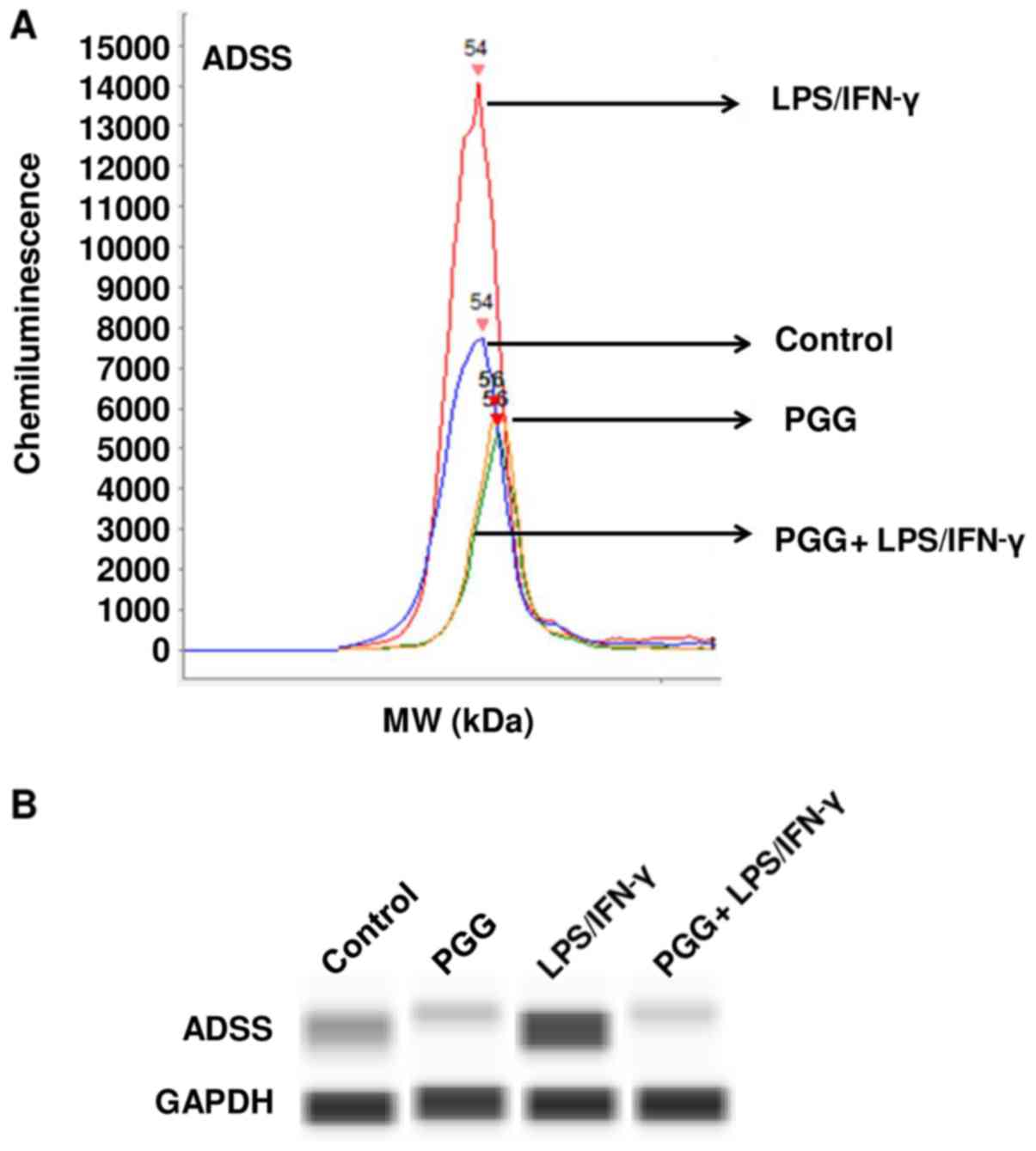 | Figure 5.Effect of PGG on the expression of
ADSS as determined by western analysis with automated capillary
electrophoresis. (A) Graph presenting the chemiluminescence levels
measured in the different treatments. (B) shows bands representing
the protein expression in control (DMSO), PGG (25 µM), LPS/IFNγ,
and pretreated cells (PGG + LPS/IFNγ) after 24 h of treatment
(n=3). ADSS, adenylosuccinate synthetase isozyme 2; PPG, polyphenol
pentagalloyl glucose (1,2,3,4,6-penta-O-galloyl-β-D-glucose); LPS,
lipopolysaccharide; IFNγ, interferon γ; MW, molecular weight. |
In this investigation, to confirm whether the
changes observed in protein expression were the result of
transcriptional regulation, we used RT-PCR to determine the mRNA
levels of the three proteins studied. The RT-PCR assay results were
consistent with the results obtained from the proteomics and
western blot assays. PGG significantly suppressed the expression of
the ataxin-2, septin-7 and ADSS genes (Fig. 6A-C). Therefore, these results
confirmed the inhibitory effect of PGG on related proteins and
genes involved in neurodegeneration.
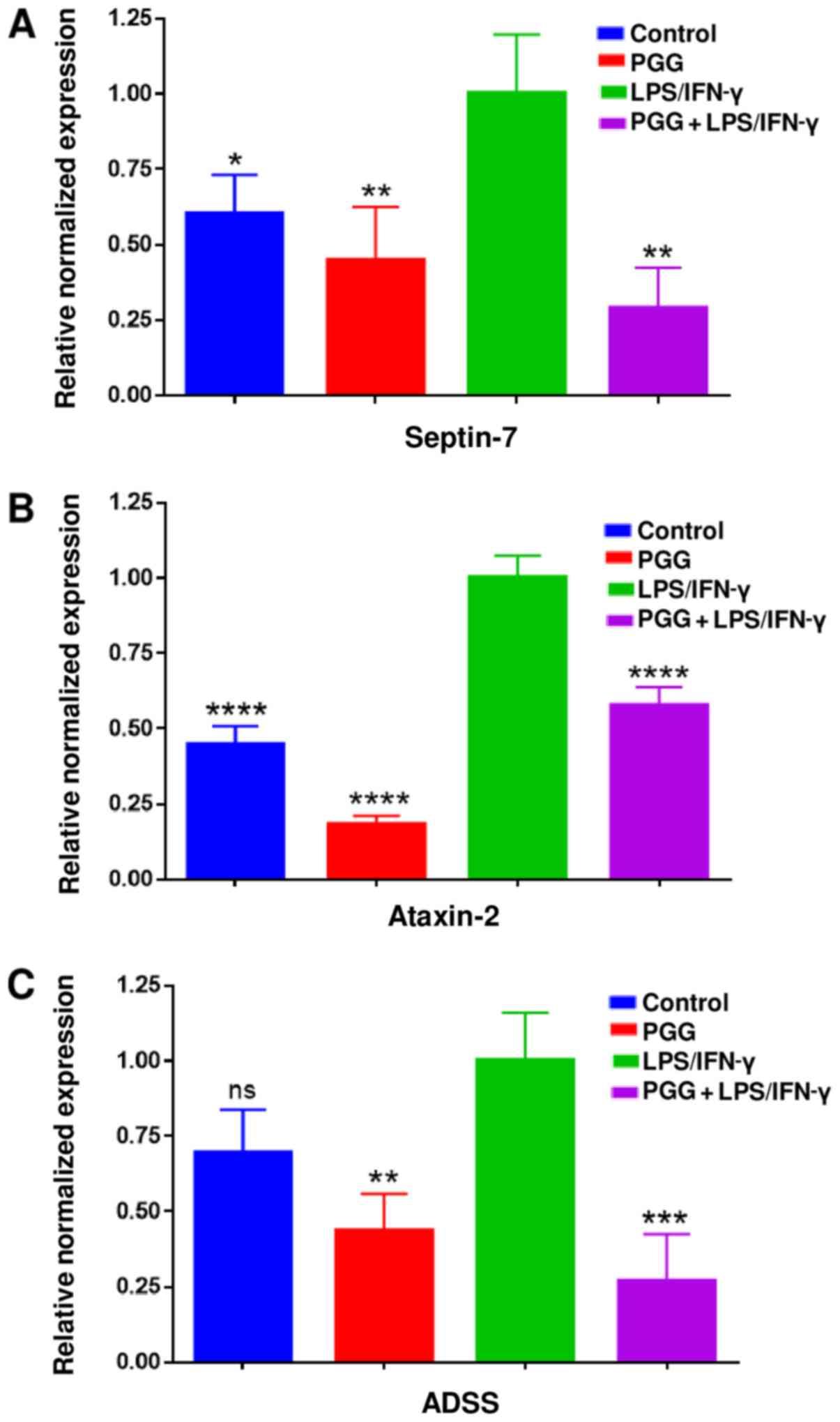 | Figure 6.Effect of PGG on mRNA expression of
(A) septin-7, (B) ataxin-2 and (C) ADSS using RT-PCR assays. Graphs
presenting the changes in the mRNA levels after the different
treatments, including control, PGG (25 µM), LPS/IFNγ, and cotreated
cells (PGG + LPS/IFNγ). Data represent gene expression presented as
the mean ± standard error of the mean (n=3). *P<0.05,
**P<0.01, ***P<0.001 and ****P<0.0001 vs. LPS/IFNγ. ADSS,
adenylosuccinate synthetase isozyme 2; PPG, polyphenol pentagalloyl
glucose (1,2,3,4,6-penta-O-galloyl-β-D-glucose); LPS,
lipopolysaccharide; IFNγ, interferon γ; ns, not significant. |
Discussion
Neuroinflammation is a prominent trait common to
many neurodegenerative diseases (7), such as AD, PD, and several other
disorders, such as amyotrophic lateral sclerosis, multiple
sclerosis, spinal cord injury, and traumatic brain (18). Persistent activation of innate
immunity, including responses mediated by microglial activation, is
a common association among these diseases, causing activation of
neurotoxic pathways that may lead to progressive neurodegeneration
(18).
The current study indicates that PGG has profound
effects on the proteome, causing up to a 53-fold inhibition of
protein expression. PGG has proven to be a very potent antioxidant
agent (19,20). While gallic acid (GA) is the
primary product of tannic acid hydrolysis (21), PGG showed higher antioxidant
activity (22). Moreover, oral PGG
was found to reduce the accumulation of human amyloid β (Aβ)
protein in the brains of transgenic mice overexpressing this
protein (23). PGG inhibited
accumulation at doses more than 10 times lower than that of the
plant extract. The study showed that PGG suppresses Aβ fibril
accumulation in treated animals, implying that PGG treatment could
offer a therapeutic aid for AD patients with moderate cognitive
injury (23). Additionally, by
applying the combined technology of ion mobility associated with
mass spectrometry, transmission electron microscopy, and molecular
kinetics, studies indicated that PGG interacts with the N-terminal
metal binding segment and the first central hydrophobic core,
interfering with Aβ1-40 and Aβ1-42 amyloid
assembly. These results showed that PGG is a potential therapeutic
drug to mitigate Aβ oligomer neurotoxicity (24).
Currently, there are many cell models used to
examine neuroinflammation, including primary microglial cultures
and immortalized microglial cell cultures (25). In the present work, we used BV-2
microglial cells that showed similar properties to primary
microglia (2,25). Henn et al (2) showed that in response to LPS, 90% of
genes induced in BV-2 cells were also induced in primary microglia;
however, the upregulation of genes in BV-2 was less pronounced.
Although BV-2 cells have an inflammatory response that is not
identical to primary cultures, they are still an important and
widely used cell model for neuroinflammation. Our prior studies
showed that PGG inhibited the expression of MCP-5 and pro-MMP-9 in
LPS/IFNγ-activated BV-2 microglial cells. These proinflammatory
cytokines may have a connection with the development of
neurofibrillary tangles and senile plaques in AD patients. Our
earlier studies also showed that PGG can modulate genes that
participate in NF-κB and MAPK signaling, which are key components
in the process of neuroinflammation. PGG modulated the expression
of CDK2, CHUK, IRAK1 and NF-κB1 at the transcriptional and protein
levels, which may help to explain how PGG downregulates the release
of MCP-5 and pro-MMP-9 in stimulated BV-2 microglial cells
(12,13).
In the present study, the data obtained indicate
that PGG was very effective at inhibiting the expression of
proteins and genes that are critical to neurodegeneration,
including septin-7, ataxin-2 and ADSS. These proteins have been
found to be involved in the pathogenesis of neurodegenerative
diseases. Kinoshita and colleagues reported that septins were
concentrated in the brains of AD patients, specifically in the
intracellular neurofibrillary tangles, dystrophic neurites in
senile plaques, and neuropil threads (26). Septins were shown to be involved in
neurodegeneration and neurobehavioral conditions. Reports have
shown that the septin family is found in postsynaptic densities
when analyzed by mass spectrometry (27,28),
indicating a possible involvement in neurodegenerative diseases and
cognitive impairment. Septins may aid in the development of
cellular aggregates, although their function in neurodegeneration
is still unknown. Additionally, the septin filaments that maintain
the structure and shape of the cells become unfolded, which may
facilitate the formation of aggregates that interrupt cell
function, leading to cell death (29). The septin family provides
attractive candidates that may be involved in the essential
mechanisms of synaptic dysfunction and neurodegeneration in
neurodegenerative diseases. Several reports have described the
association of septins with many diseases, including AD, PD,
Huntington's disease, frontotemporal lobar degeneration, and Down's
syndrome (26,29,30–35),
indicating that these proteins are involved in the pathogenic
mechanism of neurodegeneration.
Another critical protein whose expression was
attenuated by PGG in this study is ataxin 2, which is an
RNA-binding protein found in the body with multiple roles in RNA
metabolism and is broadly expressed in the mammalian nervous system
(36). Ataxin 2 was found to be
involved in neurodegenerative diseases, such as amyotrophic lateral
sclerosis (ALS) and spinocerebellar ataxia type 2 (SCA2). In mouse
models, reduced ataxin 2 levels improved motor performance of the
animals in both diseases. In addition, this protein could alter the
toxicity produced by the RNA-binding protein TDP43 (encoded by
TARDBP), which participates in the formation of aggregates in the
brain and spinal cords of ALS patients. A decrease in disease
progression was observed when TARDBP transgenic mice were crossed
with Atxn2-knockout mice, increasing the median lifespan in 80% of
the animals studied. This work suggests that neurodegenerative
conditions could be improved by targeting ataxin-2 (36,37).
Furthermore, adenylosuccinate synthetase isozyme 2
(ADSS) was another protein of interest, which was inhibited by PGG
in the current study. ADSS is an enzyme that participates in the de
novo and salvage pathways for purine and nucleotide biosynthesis,
both of which play a role in the formation of nucleotide inosine
monophosphate. Vertebrates present two isozymes, one basic (ADSS1)
and the other acidic (ADSS2), which respond in a different way to
inhibitors, indicating that they differ in their regulation
(٣٨-41). The regulation of ADSS is also implicated in the
maintenance of AMP/GMP ratios in the cell (٣٨). ADSS was identified
as one of the altered proteins in MPTP
(1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine)-treated animal
models of PD (42). Since
mitochondrial dysfunction is likely to be one of the mechanisms
that leads to PD, an investigation using quantitative proteomic
analysis for mitochondrial protein showed that MPTP-treated animals
also presented an increase in the expression of ADSS, from 50 to
100% (43). The present study,
together with previous proteomic studies in the literature,
revealed that proteins such as ADSS may be connected to PD
pathogenesis and could be regulated by PGG, indicating that this
compound may have potential use in targeted therapy.
Since ataxin-2, septin-7 and ADSS were found to be
concentrated in neurofibrillary tangles and/or involved in synapse
impairment induced by Aβ protein (26,44),
these proteins are attractive candidates that may be associated
with the fundamental mechanisms of synaptic damage in
neurodegenerative diseases. Thus, by inhibiting the expression of
these proteins, PGG may provide a useful target compound for
neurodegenerative disease therapies.
The proteomic results not only revealed the
inhibitory effect of PGG on proteins overexpressed in
neurodegenerative diseases but also showed that PGG modulates many
proteins involved in different cellular functions, which
participate in several signaling pathways. PGG exhibited a
significant fold change inhibition in the expression of
WAS/WASL-interacting protein family member 1 (WIPF1) (−9.4-fold↓),
thymidylate synthase (−9.7-fold↓), and glyoxalase (−53-fold↓),
whose overexpression has been shown to be associated with cancer
development and progression. The expression of WIPF1 had a critical
role in thyroid cancer cell migration and invasion, suggesting
WIPF1 as a novel therapeutic target for thyroid cancer (45). Additionally, high levels of
thymidylate synthase expression have been correlated with poor
prognosis in breast cancer (46),
gastric cancer and colorectal cancers (47,48).
In the present study, the most remarkable effect of
PGG was on the expression of glyoxalase 4 (GLOD4), with a 53-fold
inhibition. The overexpression of glyoxalases has been reported in
numerous types of malignant tumors, such as colon, breast, ovarian,
and prostate cancers (49–54). Furthermore, glyoxalases I and II
are involved in the detoxification of methylglyoxal, a cytotoxic
product of glycolysis, whose overexpression was observed in 79% of
tumors (55). Methylglyoxal and
glycation end-products are associated with an increase in
proinflammatory markers and affect AD, Aβ peptide depositions and
neurofibrillary tangles (56–58).
In tumor cells, increased cellular levels of toxic methylglyoxal
and S-d-lactoylglutathione metabolites are present due to a
higher metabolism compared to normal cells. In response to the
increase in the production of these cytotoxic products, tumor cells
augment the activity of the detoxifying glyoxalase system to
minimize the intracellular concentrations of toxic metabolites
(59). Methylglyoxal also
accumulates with age, and it is associated with age-associated
pathologies, such as diabetic complications and neurodegenerative
disorders (60). Previous studies
using mutant GLOD-4/GLO1-animals (Caenorhabditis elegans
model) showed that animals quickly exhibited several pathogenic
phenotypes reminiscent of α-dicarbonyl stress-induced
age-associated disorders (61).
Therefore, glyoxalase inhibitors, such as PGG, may have potential
as antitumor agents as well as regulatory mediators in
neurodegenerative diseases. While the role of glyoxalase 4 is not
well established in humans, our results suggest that PGG may
regulate the expression of distinct but overlapping genes and
proteins involved in neurodegeneration and cancer progression.
Although the current study did not directly confirm
the effect of PGG on the expression of CDK2, CHUK, IRAK1, and
NF-κB1 observed in our previous manuscript (13), our current results indirectly
showed that there may be an association among all the proteins that
were downregulated by PGG. CDK2, CHUK, IRAK1 and NF-κB1 are
proteins involved in the NF-κB and MAPK signaling pathways, which
are both activated by pathogenic and noxious stimuli. Evidence
suggests that MAPKs can participate in the regulation of NF-κB
transcriptional activity, demonstrating that both the c-Jun
N-terminal kinase and the p38 pathways are implicated in the
activation of NF-κB in the cytoplasm, as well as in modulation of
its transactivation potential in the nucleus, confirming the
association between these signaling pathways (62). Moreover, Septin 7 was identified as
a novel ERK3-interacting protein by using a Ras recruitment system,
and evidence showed that Sept7, ERK3 and MK5 exist in the same
complex and, together with Kal7, are of physiological relevance in
neuronal plasticity by regulating dendritic spine formation
(63). Furthermore, Ataxin-2 was
described as a positive regulator of Notch signaling (64), which may maintain NF-κB activity by
direct interaction with p50/c-Rel in the nucleus (65). Thus, we can conclude that there is
an indirect association with the current and previous results due
to the overlapping functions and interactions of multiple proteins
involved in the NF-κB and MAPK signaling pathways.
The results of this work provide evidence that the
polyphenolic compound PGG can modulate the expression of several
proteins and transcripts whose expression is increased in
LPS/IFNγ-stimulated BV-2 microglial cells. Among the proteins, PGG
significantly inhibited the expression of ataxin-2, septin-7 and
ADSS, which play a critical role in synapse impairment and
pathogenesis of neurodegenerative diseases. Therefore, this study
suggests that PGG may have therapeutic potential for
neuroinflammation and neurodegeneration by targeting ataxin-2,
septin-7 and ADSS.
Acknowledgements
Not applicable.
Funding
The present study was supported by NIH NIMHD (grant
nos. G12 MD007582 and P20 MD 006738).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
PM and KFAS conceived and designed the study. PM and
ET performed the experiments. PM analyzed and interpreted the data,
and wrote the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Aβ
|
amyloid-β
|
|
AD
|
Alzheimer's disease
|
|
ADSS
|
adenylosuccinate synthetase isozyme
2
|
|
ALS
|
amyotrophic lateral sclerosis
|
|
CNS
|
central nervous system
|
|
IFNγ
|
interferon γ
|
|
FASP
|
Filter Aided Sample Preparation
|
|
GA
|
gallic acid
|
|
GLOD4
|
Glyoxalase 4
|
|
LPS
|
lipopolysaccharide
|
|
MAPK
|
mitogen-activated protein kinase
|
|
MCP-5
|
monocyte chemoattractant protein-5
|
|
NCBI
|
National Center for Biotechnology
Information
|
|
NF-κB
|
nuclear factor-κB
|
|
PANTHER
|
Protein Analysis Through Evolutionary
Relationships
|
|
PD
|
Parkinson's disease
|
|
PGG
|
1,2,3,4,6-penta-O-galloyl-β-D-glucose
|
|
Pro MMP-9
|
pro-metalloproteinase-9
|
|
SCA2
|
spinocerebellar ataxia type 2
|
|
TLR4
|
Toll-like receptor 4
|
|
WIPF1
|
WAS/WASL-interacting protein family
member 1
|
References
|
1
|
Street WJ, Mark RE and Griffin WS:
Microglia and neuroinflammation: A pathological perspective. J
Neuroinflammation. 1:142004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Henn A, Lund S, Hedtjärn M, Schrattenholz
A, Pörzgen P and Leist M: The suitability of BV-2 cells as an
alternative model system for primary microglia cultures for animal
experiments examining brain inflammation. ALTEX. 26:83–94. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schwartz M and Baruch K: The resolution of
neuroinflammation in neurodegeneration: Leukocyte recruitment via
the choroid plexus. EMBO J. 33:7–22. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gendelman HE: Neural immunity: Friend or
foe? J Neurovirol. 8:474–479. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Oh GS, Pae HO, Choi BM, Lee HS, Kim IK,
Yun YG, Kim JD and Chung HT: Penta-O-galloyl-Beta-D-glucose
inhibits phorbol myristate acetate-induced interleukin-8
[correction of interleukin-8] gene expression in human monocytic
U937 cells through its inactivation of nuclear factor-kappa B. Int
Immunopharmacol. 4:377–386. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hanisch UK: Microglia as a source and
target of cytokines. Glia. 40:140–155. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gao HM and Hong JS: Why neurodegenerative
diseases are progressive: Uncontrolled inflammation drives disease
progression. Trends Immunol. 29:357–365. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Etminan M, Gill S and Samii A: Effect of
non-steroidal anti-inflammatory drugs on risk of Alzheimer's
disease: Systematic review and meta-analysis of observational
studies. BMJ. 327:1282003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
McGeer PL and McGeer EG: The amyloid
cascade-inflammatory hypothesis of Alzheimer disease: Implications
for therapy. Acta Neuropathol. 126:479–497. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Heneka MT, Kummer MP, Weggen S, Bulic B,
Multhaup G, Münter L, Hüll M, Pflanzner T and Pietrzik CU:
Molecular mechanisms and therapeutic application of NSAIDs and
derived compounds in Alzheimer's disease. Curr Alzheimer Res.
8:115–131. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cao Y, Himmeldirk KB, Qian Y, Ren Y, Malki
A and Chen X: Biological and biomedical functions of
Penta-O-Galloyl-D-Glucose and its derivatives. J Nat Med.
68:465–472. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mendonca P, Taka E, Bauer D,
Cobourne-Duval M and Soliman KF: The attenuating effects of
1,2,3,4,6 Penta-O-Galloyl-β-D-glucose on inflammatory cytokines
release from activated BV-2 microglial cells. J Neuroimmunol.
305:9–15. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mendonca P, Taka E, Bauer D, Reams RR and
Soliman KFA: The attenuating effects of 1,2,3,4,6
penta-O-galloyl-β-D-glucose on pro-inflammatory responses of
LPS/IFNγ-activated BV-2 microglial cells through NFκB and MAPK
signaling pathways. J Neuroimmunol. 324:43–53. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Blasi E, Barluzzi R, Bocchini V, Mazzolla
R and Bistoni F: Immortalization of murine microglial cells by a
v-raf/v-myc carrying retrovirus. J Neuroimmunol. 27:229–237. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Elsisi N, Darling-Reed S, Lee EY, Oriaku
ET and Soliman KF: Ibuprofen and apigenin induce apoptosis and cell
cycle arrest in activated microglia. Neurosci Lett. 375:91–96.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
NCBI Resource Coordinators: Database
resources of the National Center for Biotechnology Information.
Nucleic Acids Res. 44:D7–D19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thomas PD, Campbell MJ, Kejariwal A, Mi H,
Karlak B, Daverman R, Diemer K, Muruganujan A and Narechania A:
PANTHER: A library of protein families and subfamilies indexed by
function. Genome Res. 13:2129–2141. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Amor S, Peferoen LA, Vogel DY, Breur M,
van der Valk P, Baker D and van Noort JM: Inflammation in
neurodegenerative diseases-an update. Immunology. 142:151–166.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Abdelwahed A, Bouhlel I, Skandrani I,
Valenti K, Kadri M, Guiraud P, Steinman R, Mariotte AM, Ghedira K,
Laporte F, et al: Study of antimutagenic and antioxidant activities
of Gallic acid and 1,2,3,4,6 penta galloyl glucose from Pistacia
lentiscus. Confirmation by microarray expression profiling. Chem
Biol Interact. 165:1–13. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim BH, Choi MS, Lee HG, Lee SH, Noh KH,
Know S, Jeong AJ, Lee H, Yi EH, Park JY, et al: The photoprotective
potential of penta-O-galloyl-β-D-glucose by targeting NF-κB and
MAPK signaling in UVB radiation-induced human dermal fibroblasts
and mouse skin. Mol Cells. 38:982–990. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Aithal M and Belur P: Enhancement of
propyl gallate yield in a nonaqueous medium using novel
cell-associated tannase of Bacillus massiliensis. Prep Biochem
Biotechnol. 43:445–455. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang KJ, Yang CR and Zhang YJ: Phenolic
antioxidants from Chinese toon (fresh young leaves and shoots of
Toona sinensis). Food Chemistry. 101:365–371. 2006. View Article : Google Scholar
|
|
23
|
Fujiwara H, Tabuchi M, Yamaguchi T,
Iwasaki K, Furukawa K, Sekiguchi K, Ikarashi Y, Kudo Y, Higuchi M,
Saido TC, et al: Traditional medicinal herb Paeonia suffruticosa
and its active constituent
1,2,3,4,6-Penta-O-Galloyl-Beta-D-Glucopyranose have potent
anti-aggregation effects on Alzheimer's amyloid beta proteins in
vitro and in vivo. J Neurochem. 109:1648–1657. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
De Almeida NEC, Do TD, LaPointe NE, Tro M,
Feinstein SC, Shea JE and Bowers MT:
1,2,3,4,6-Penta-O-Galloyl-β-D-glucopyranose binds to the N-terminal
metal binding region to inhibit amyloid β-protein oligomer and
fibril formation. Int J Mass Spectrom. 420:24–34. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Stansley B, Post J and Hensley K: A
comparative review of cell culture systems for the study of
microglial biology in Alzheimer's disease. J Neuroinflammation.
9:1152012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kinoshita A, Kinoshita M, Akiyama H,
Tomimoto H, Akiguchi I, Kumar S, Noda M and Kimura J:
Identification of septins in neurofibrillary tangles in Alzheimer's
disease. Am J Pathol. 153:1551–1560. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Tada T, Simonetta A, Batterton M,
Kinoshita M, Edbauer D and Sheng M: Role of septin cytoskeleton in
spine morphogenesis and dendrite development in neurons. Curr Biol.
17:1752–1758. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Walikonis RS, Jensen ON, Mann M, Provance
DW Jr, Mercer JA and Kennedy MB: Identification of proteins in the
postsynaptic density fraction by mass spectrometry. J Neurosci.
20:4069–4080. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Barr AM, Young CE, Sawada K, Trimble WS,
Phillips AG and Honer WG: Abnormalities of presynaptic protein
CDCrel-1 in the striatum of rats reared in social isolation:
Relevance to neural connectivity in schizophrenia. Eur J Neurosci.
20:303–307. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ageta-Ishihara N, Yamakado H, Morita T,
Hattori S, Takao K, Miyakawa T, Takahashi R and Kinoshita M:
Chronic overload of SEPT4, a parkin substrate that aggregates in
Parkinson's disease, causes behavioral alterations but not
neurodegeneration in mice. Mol Brain. 6:352013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong Z, Ferger B, Paterna JC, Vogel D,
Furler S, Osinde M, Feldon J and Büeler H: Dopamine-dependent
neurodegeneration in rats induced by viral vector-mediated
overexpression of the parkin target protein, CDCrel-1. Proc Natl
Acad Sci USA. 100:12438–12443. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gozal YM, Seyfried NT, Gearing M, Glass
JD, Heilman CJ, Wuu J, Duong DM, Cheng D, Xia Q, Rees HD, et al:
Aberrant septin 11 is associated with sporadic frontotemporal lobar
degeneration. Mol Neurodegener. 6:822011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ihara M, Yamasaki N, Hagiwara A, Tanigaki
A, Kitano A, Hikawa R, Tomimoto H, Noda M, Takanashi M, Mori H, et
al: Sept4, a component of presynaptic scaffold and Lewy bodies, is
required for the suppression of alpha-synuclein neurotoxicity.
Neuron. 53:519–533. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Pissuti Damalio JC, Garcia W, Alves Macedo
JN, De Almeida Marques I, Andreu JM, Giraldo R, Garratt RC and
Ulian Araújo AP: Self-assembly of human septin 2 into amyloid
filaments. Biochimie. 94:628–636. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang Y, Gao J, Chung KK, Huang H, Dawson
VL and Dawson TM: Parkin functions as an E2-dependent
ubiquitin-protein ligase and promotes the degradation of the
synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci
USA. 97:13354–13359. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Crunkhorn S: Neurodegenerative disorders:
Ataxin 2 reduction rescues motor defects. Nat Rev Drug Discov.
16:384–385. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schmidt EE, Pelz O, Buhlmann S, Kerr G,
Horn T and Boutros M: RNAi: A database for cell-based and in vivo
RNAi phenotypes, 2013 update. Nucleic Acids Res. 41:D1021–D1026.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Stayton MM, Rudolph FB and Fromm HJ:
Regulation, genetics, and properties of adenylosuccinate
synthetase. Curr Top Cell Regul. 22:103–141. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Borza T, Iancu CV, Pike E, Honzatko RB and
Fromm HJ: Variations in the response of mouse isozymes of
adenylosuccinate synthetase to inhibitors of physiological
relevance. J Biol Chem. 278:6673–6679. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Oshchepkova-Nedosekina EA and Likhoshvai
VA: A mathematical model for the adenylosuccinate synthetase
reaction involved in purine biosynthesis. Theor Biol Med Model.
4:112007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Nomura Y, Nozawa A and Tozawa Y:
Biochemical analyses of ppGpp effect on adenylosuccinate
synthetases, key enzymes in purine biosynthesis in rice. Biosci
Biotechnol Biochem. 78:1022–1025. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Sowell RA, Owen JB and Butterfield DA:
Proteomics in animal models of Alzheimer's and Parkinson's
diseases. Ageing Res Rev. 8:1–17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Jin J, Meredith GE, Chen L, Zhou Y, Xu J,
Shie FS, Lockhart P and Zhang J: Quantitative proteomic analysis of
mitochondrial proteins: Relevance to Lewy body formation and
Parkinson's disease. Brain Res. Mol. Brain Res. 134:119–138.
2005.
|
|
44
|
Wan W, Xia S, Kalionis B, Liu L and Li Y:
The role of Wnt signaling in the development of Alzheimer's
disease: A potential therapeutic target? Biomed Res Int.
2014:3015752014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Zhang T, Shen X, Liu R, Zhu G, Bishop J
and Xing M: Epigenetically upregulated WIPF1 plays a major role in
BRAF V600E-promoted papillary thyroid cancer aggressiveness.
Oncotarget. 8:900–914. 2017.PubMed/NCBI
|
|
46
|
Pestalozzi BC, Peterson HF, Gelber RD,
Goldhirsch A, Gusterson BA, Trihia H, Lindtner J, Cortés-Funes H,
Simmoncini E, Byrne MJ, et al: Prognostic importance of thymidylate
synthase expression in early breast cancer. J Clin Oncol.
15:1923–1931. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Johnston PG, Lenz HJ, Leichman CG,
Danenberg KD, Allegra CJ, Danenberg PV and Leichman L: Thymidylate
synthase gene and protein expression correlate and are associated
with response to 5-fluorouracil in human colorectal and gastric
tumors. Cancer Res. 55:1407–1412. 1995.PubMed/NCBI
|
|
48
|
Nimmagadda S and Shields AF: The role of
DNA synthesis imaging in cancer in the era of targeted
therapeutics. Cancer Metastasis Rev. 27:575–587. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Ranganathan S and Tew KD: Analysis of
Glyoxalase-I from normal and tumor tissue from human colon. Biochim
Biophys Acta. 1182:311–316. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ranganathan S, Walsh ES and Tew KD:
Glyoxalase I in detoxification: Studies using glyoxalase I
transfectant cell line. Biochem J. 309:127–131. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Rulli A, Carli L, Romani R, Baroni T,
Giovannini E, Rosi G and Talesa V: Expression of glyoxalase I and
II in normal and breast cancer tissues. Breast Cancer Res Treat.
66:67–72. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Jones MB, Krutzsch H, Shu H, Zhao Y,
Liotta LA, Kohn EC and Petricoin EF III: Proteomic analysis and
identification of new biomarkers and therapeutic targets for
invasive ovarian cancer. Proteomics. 2:76–84. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Di Ilio C, Angelucci S, Pennelli A, Zezza
A, Tenaglia R and Sacchetta P: Glyoxalase activities in tumor and
non-tumor human urogenital tissues. Cancer Lett. 96:189–193. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Davidson SD, Cherry JP, Choudhury MS,
Tazaki H, Mallouh C and Konno S: Glyoxalase I activity in human
prostate cancer: A potential marker and importance in chemotherapy.
J Urol. 161:690–691. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fonseca-Sánchez MA, Rodríguez-Cuevas S,
Mendoza-Hernández G, Bautista-Piña V, Arechaga Ocampo E, Hidalgo
Miranda A, Quintanar Jurado V, Marchat LA, Alvarez-Sánchez E, Pérez
Plasencia C and López-Camarillo C: Breast cancer proteomics reveals
a positive correlation between glyoxalase 1 expression and high
tumor grade. Int J Oncol. 41:670–680. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Ledesma MD, Bonay P and Avila J: Tau
protein from Alzheimer's disease patients is glycated at its
tubulin-binding domain. J Neurochem. 65:1658–1664. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Vitek MP, Bhattacharya K, Glendening JM,
Stopa E, Vlassara H, Bucala R, Manogue K and Cerami A: Advanced
glycation end products contribute to amyloidosis in Alzheimer
disease. Proc Natl Acad Sci USA. 91:4766–4770. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Angeloni C, Zambonin L and Hrelia S: Role
of methylglyoxal in Alzheimer's disease. Biomed Res Int.
2014:2384852014. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Al-Balas QA, Hassan MA, Al-Shar'i NA,
Mhaidat NM, Almaaytah AM, Al-Mahasneh FM and Isawi IH: Novel
glyoxalase-I inhibitors possessing a ‘zinc-binding feature’ as
potential anticancer agents. Drug Des Devel Ther. 10:2623–2629.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Thornalley PJ: Methylglyoxal, glyoxalases
and the development of diabetic complications. Amino Acids.
6:15–23. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Chaudhuri J, Bose N, Gong J, Hall D,
Rifkind A, Bhaumik D, Peiris TH, Chamoli M, Le CH, Liu J, et al: A
Caenorhabditis elegans model elucidates a conserved role for
TRPA1-Nrf signaling in reactive α-Dicarbonyl detoxification. Curr
Biol. 26:3014–3025. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Schulze-Osthoff K, Ferrari D, Riehemann K
and Wesselborg S: Regulation of NF-kappa B activation by MAP kinase
cascades. Immunobiology. 198:35–49. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Brand F, Schumacher S, Kant S, Menon MB,
Simon R, Turgeon B, Britsch S, Meloche S, Gaestel M and Kotlyarov
A: The extracellular signal-regulated kinase 3 (mitogen-activated
protein kinase 6 [MAPK6])-MAPK-activated protein kinase 5 signaling
complex regulates septin function and dendrite morphology. Mol Cell
Biol. 32:2467–2478. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Shukla JP, Deshpande G and Shashidhara LS:
Ataxin 2-binding protein 1 is a context-specific positive regulator
of Notch signaling during neurogenesis in Drosophila melanogaster.
Development. 144:905–915. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shin HM, Minter LM, Cho OH, Gottipati S,
Fauq AH, Golde TE, Sonenshein GE and Osborne BA: Notch1 augments
NF-kappaB activity by facilitating its nuclear retention. EMBO J.
25:129–138. 2006. View Article : Google Scholar : PubMed/NCBI
|