Introduction
Hepatocellular carcinoma (HCC) is one of the most
common human cancers globally and the second leading cause of
cancer-associated mortality (1).
At present, the major treatment strategies for HCC are early
detection, surgical resection, liver implantation and gene therapy
(1). Despite major advances in the
diagnosis and treatment of HCC, the 5-year survival of patients
with HCC remains poor due to the high incidence of cancer
recurrence and metastasis (1).
Therefore, it is urgent to investigate the molecular mechanisms
underlying HCC development. A number of reports have indicated that
microRNAs (miRNAs/miRs) serve important roles in HCC carcinogenesis
(2–16).
miRNAs are ~22-nucleotide, small, noncoding RNAs
that can post-transcriptionally regulate target gene expression via
complementary base pairing to block the translation or promote the
degradation of target mRNAs (17).
miRNAs have been implicated in various physiological and
pathological processes, including cell differentiation, embryonic
development and carcinogenesis (18). Increasing evidence demonstrates
that dysregulated miRNAs are involved in the development and
progression of a variety of cancers, including HCC (2). For example, miR-199-3p, an miRNA
reported to be downregulated in HCC, can inhibit tumorigenesis of
HCC cells by targeting zinc fingers and homeoboxes protein
1/p53-upregulated modulator of apoptosis signaling, vascular
endothelial growth factor (VEGF)A, VEGF receptor (VEGFR)1, VEGFR2,
hepatocyte growth factor, matrix metalloproteinase-2,
yes-associated protein 1 and CD151, and increasing the doxorubicin
sensitivity of human HCC cells by regulating mTOR and c-Met
(3–9). Furthermore, miR-199-3p was
demonstrated to efficiently inhibit tumor growth in a transgenic
mouse model of HCC and an HCC tumor-bearing nude mouse model
(10,11). A recent report revealed that
miR-501-3p was downregulated in metastatic HCC cell lines and
tissue samples from patients with recurring and metastasized HCC,
and its expression was associated with tumor progression and poor
prognosis in patients with HCC; further mechanistic study revealed
that Lin-7 homolog A mediated the suppressive effects of miR-501-3p
on the metastasis and progression of HCCs (12). Recent studies also revealed that
HCC cell-derived exosomal miRNAs serve important roles in HCC
development, particularly in metastasis (13–16).
For example, tumor-derived exosomal miR-25-5p promoted tumor
self-seeding by enhancing the invasive and migratory abilities of
recipient HCC cells (14).
Hepatoma cell-secreted miR-103 increased vascular permeability and
promoted metastasis by directly inhibiting the expression of
VE-cadherin, p120-catenin and zonula occludens 1 (15). It has been reported that miR-425-5p
expression was dysregulated in various cancers, including gastric
cancer, HCC, cervical cancer, esophageal squamous cell carcinoma,
nasopharyngeal carcinoma, renal cell carcinoma and melanoma, and
that the altered expression of miR-425-5p is associated with the
pathogenesis and progression of cancer (19–26);
however, the roles and mechanisms of miR-425-5p in the development
of HCC are yet to be determined.
Forkhead box D3 (FOXD3) is a member of the forkhead
box family of transcription factors. The forkhead box proteins are
characterized by a distinct conserved ‘forkhead’ or ‘winged helix’
DNA-binding domain (27). FOXD3 is
an important stem cell factor expressed in various types of stem
cells and has an established role in development (28). Various studies have reported that
FOXD3 is also involved in the development of a number of cancers
(29–37); however, reports into its roles and
mechanisms in HCC are limited.
In the present study, the potential roles of
miR-425-5p in the development of HCC were investigated. The present
findings demonstrated that miR-425-5p was significantly upregulated
in HCC tissues and cells. miR-425-5p promoted the proliferation,
migration and invasion of hepatoma cells by directly targeting and
repressing FOXD3, the expression of which may suppress the
malignant behavior of HCC cells. The present findings may be useful
in understanding the molecular mechanisms underlying the
development of HCC identifying novel treatment strategies for
HCC.
Materials and methods
Clinical tissues
A total of 30 pairs of HCC tissue specimens were
collected from patients who had undergone complete surgical
resection at Henan Provincial People's Hospital. Among the 30 pairs
of HCC tissue specimens, 20 were hepatitis B virus-positive.
Patient were divided into ‘miR-425-5p high’ and ‘miR-425-5p low’
groups using a threshold of 0.5 relative expression of
miR-425-5p/U6. All tissue specimens (HCC tissues and adjacent
nontumor tissues) were immediately snap-frozen in liquid nitrogen
and stored at −80°C following surgery. Written informed consent was
obtained from each patient. The study was approved by the Ethics
Committee of Henan Provincial People's Hospital. The clinical
information are presented in Tables
I and SI.
 | Table I.Clinical characteristics of patients
with hepatocellular carcinoma and association with miRNA-425-5p
levels. |
Table I.
Clinical characteristics of patients
with hepatocellular carcinoma and association with miRNA-425-5p
levels.
| Clinical
parameters | miR-425-5p high
(n=19) | miR-425-5p low
(n=11) | P-value |
|---|
| Age (years) |
|
| 0.429 |
|
<50 | 11 | 6 |
|
|
≥50 | 8 | 5 |
|
| Sex |
|
| 0.725 |
|
Male | 15 | 9 |
|
|
Female | 4 | 2 |
|
| TNM grade |
|
| 0.022a |
|
I+II | 14 | 8 |
|
|
III+IV | 5 | 3 |
|
Cell culture and transfection
The human hepatoma cell lines HCCLM3, Hep3B and Huh7
cells were purchased from the American Type Culture Collection. The
liver cancer cell line HepG2 and liver cell line L02 were purchased
from the Chinese Academy of Sciences Cell Bank. All cells were
cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.)
containing 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) and
incubated in a humidified 37°C incubator with 5% CO2.
Cell transfection was performed using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocols.
Plasmid construction
The potential targets of miR-425-5p were predicted
using TargetScanHaman 7.2 (www.targetscan.org) and PicTar databases (pictar.mdc-berlin.de) (38,39).
The FOXD3 gene was amplified from cDNA derived from HCCLM3 cells
and cloned into pcDNA3.0 (Invitrogen; Thermo Fisher Scientific,
Inc.) Pfu DNA polymerase (Promega Corporation) was used to amplify
cDNA, and the PCR conditions were as follows: 95°C for 5 min,
followed by 30 cycles of 94°C for 30 sec, 58°C for 2 min and 72°C
for 30 sec, with a final step of 72°C for 10 min. FOXD3-specific
and negative control small interfering RNA (siRNA; si-FOXD3 and
si-control) were synthesized by Shanghai GenePharma Co., Ltd.
miR-425-5p mimics and the antisense oligonucleotide of miR-425-5p
(ASO-miR-425-5p) were commercially synthesized by Shanghai
GenePharma Co., Ltd. Negative control miRNA (miR-NC) containing a
scramble sequence of miRNA was used as the control for miR-425-5p
mimics transfection. ASO-NC containing a scramble sequence of miRNA
was used as the control for ASO-miR-425-5p. The FOXD3 3′
untranslated region (3′UTR) harboring the miR-425-5p target
sequence and the seed sequence mutated type (FOXD3 3′UTR mut) were
synthesized by Shanghai GenePharma Co., Ltd., and inserted into the
pmiR-GLO vector (Promega Corporation). The primers and
oligonucleotides used during the study are presented in Table II.
 | Table II.Primers and oligonucleotide
sequences. |
Table II.
Primers and oligonucleotide
sequences.
| Name | Sequences
(5′-3′) |
|---|
| FOXD3-plasmid | Sense:
CGGGATCCATGACCCTCTCCGGCGGCG |
|
| Antisense:
CGGAATTCCTATTGCGCCGGCCATTTGGCT |
| FOXD3-qPCR | Forward:
GCCCCCAACGCCTACCTTCC |
|
| Reverse:
ATTTCCCTCCCATCCCCACG |
| GAPDH-qPCR | Forward:
ATGACATCAAGAAGGTGGTGAAGCAGG |
|
| Reverse:
GCGTCAAAGGTGGAGGAGTGGGT |
| si-FOXD3 | Forward:
ACGGCCGAGGACGTGGACATC |
|
| Reverse:
GATGTCCACGTCCTCGGCCGT |
| miR-NC mimics | Forward:
UCACAACCUCCUAGAAAGAGUAGA |
|
| Reverse:
UCUACUCUUUCUAGGAGGUUGUGA |
| miR-425-5p
mimics | Forward:
AAUGACACGAUCACUCCCGUUGA |
|
| Reverse:
UCUUCGGGUGUGAUCGUGUCAUU |
| ASO-control |
CAGUACUUUUGUGUAGUACAA |
| ASO-miR-425-5p |
AAUGACACUUUUUGCCUUUUUGA |
| miR-425-5p-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTGCA |
|
|
CTGGATACGACTCTTCGGGT |
| U6-RT |
AACGCTTCACGAATTTGCGTG |
|
miR-425-5p-qPCR |
GCCCGCAATGACACGATCACTC |
| U6-qPCR | Forward:
GCTCGCTTCGGCAGCACA |
|
| Reverse:
GAGGTATTCGCACCAGAGGA |
| FOXD3 3′UTR wt | Forward:
UCAAAAAGGCAAAAAGUGUCAUU |
|
| Reverse:
AATGACACTTTTTGCCTTTTTGA |
| FOXD3 3′UTR
mut | Forward:
UCAAAAAGGCAAAAACACAGUAU |
|
| Reverse:
ATACTGTGTTTTTGCCTTTTTGA |
HCCLM3 cells at a density of 70% were transfected
with miR-425-5p mimics (50 nM) or ASO-miR-425-5p (100 nM), FOXD3
expressing plasmid (2 µg/well, 12-well plates; 4 µg/well, 6-well
plates) or si-FOXD3 (100 nM). In the rescue experiment, cells were
transfected with miR-425-5p mimics (50 nM) and FOXD3 (1 µg/well,
12-well plates). For reverse transcription-quantitative PCR
(RT-qPCR), RNA was isolated 48 h following transfection.
RNA isolation and RT-qPCR
Total RNA was isolated using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocols. Briefly, for tissue samples, the
tissue section was first cut into small pieces and ground in liquid
nitrogen. Then, the samples were homogenized in TRIzol and RNA was
extracted. cDNA was synthesized using the Moloney murine leukemia
virus (M-MLV) reverse transcriptase (Promega Corporation). Briefly,
2 µg RNA with primers was incubated at 70°C for 5 min, then the
M-MLV RT enzyme, M-MLV 5X Reaction Buffer, dNTPs, RNasin (Promega
Corporation) and Nuclease-Free Water were added to a final volume
of 25 µl, which was incubated for 60 min at 42°C. qPCR was
performed using an SYBR Premix Ex Taq kit (Takara Biotechnology
Co., Ltd.). qPCR was conducted as follows: 94°C for 3 min, followed
by 40 cycles of 94°C for 30 sec, 56°C for 30 sec, and 72°C for 30
sec. GAPDH and U6 were used as internal controls for mRNA and miRNA
quantification, respectively. The relative expression was
calculated using the 2−ΔΔCq method (40). The primers and oligonucleotides
used are listed in Table II.
Western blotting
At 48 h following transfection, cells were lysed
with RIPA buffer (Beyotime Institute of Biotechnology). The protein
concentrations were measured with the Bradford assay (Bio-Rad
Laboratories, Inc.). Protein samples (30 µg) were separated via 10%
SDS-PAGE and then transferred onto PVDF membranes. Membranes were
incubated with blocking buffer (5% skim milk) at room temperature
for 2 h. The membranes were then incubated overnight at 4°C with
primary antibodies against FOXD3 (1:1,000; cat. no. ab64807),
E-cadherin (1:1,000; cat. no. ab15148), Vimentin (1:1,000; cat. no.
ab92547) and GAPDH (1:500; cat. no. ab181602), and then with goat
anti-rabbit horseradish peroxidase-conjugated antibodies (1:1,000;
cat. no. ab205718) at room temperature for 2 h. All antibodies were
purchased from Abcam. GAPDH was used as an internal control.
Proteins were visualized by enhanced chemiluminescence (Applygen
Technologies, Inc.). LabWorks 4.0 image acquisition and analysis
software was used to quantify band density (UVP, LLC).
Luciferase reporter assay
HCCLM3 cells were co-transfected with miR-425-5p
mimics (50 nM), ASO-miR-425-5p (100 nM) or their respective
controls, and pMIR-GLO-FOXD3 3′UTR or pMIR-GLO-FOXD3 3′UTR mut
reporter (100 ng/well, 24-well plates) with Lipofectamine 2000. The
hRluc-neo was used as an internal control. Then, 48 h following
transfection, the luciferase activities were determined using the
Dual-Glo Luciferase Reporter Assay system (Promega Corporation).
The results were expressed as relative luciferase activity (firefly
luciferase/Renilla luciferase).
MTT assay and colony formation
assay
MTT and colony formation assays were performed as
previously described (41,42). Briefly, HCCLM3 cells were
transfected with plasmids or oligonucleotides as aforementioned.
Then, 24 h following transfection, the cells were plated in 96-well
plates at a density of 3,000 cells/well. The viability of cells at
24, 48 and 72 h was evaluated using an MTT assay at 570 nm.
Briefly, 10 µl MTT reagent (10 mg/ml) was added to each well prior
to incubation at 37°C for 4 h, following which 100 µl dimethyl
sulfoxide was added into each well. The absorbance was detected at
570 nm using a microplate reader (BioTek Instruments, Inc.). Cell
viability was expressed as the relative A570 in
experimental cells compared with in control cells [viable cells
(%)=(abssample-absblank)/(abscontrol-absblank)
×100]. For the colony formation assay, at 24 h following
transfection, cells were plated in 12-well plates at a density of
300 cells/well in triplicate. The culture medium was changed every
3 days. At ~2 weeks later, the cells were stained with crystal
violet for 20 min at room temperature, and colonies containing ≥50
cells were counted. The colony formation rate was expressed as
colony number compared to the cells plated in each well [(colony
number/cell number added ×100)].
Migration and invasion assays
HCCLM3 cells were transfected with the
aforementioned plasmids or oligonucleotides. Then, at 24 h
following transfection, 5×104 HCCLM3 cells were seeded
into the upper chamber of Transwell chamber inserts (pore size, 8
µm) without or with Matrigel (used for invasion assays). DMEM with
10% FBS was added to the upper chamber, whereas DMEM with 20% FBS
was added to the lower chamber. The chambers were maintained in 5%
CO2 at 37°C for 24 h and then stained with 0.1% crystal
violet at room temperature for 20 min. The cells on the lower side
of the filter were counted under a microscope; for each sample,
five fields was counted and then averaged. Images were captured
under a light microscope (magnification, ×40; Leica Microsystems
GmbH).
Flow cytometry analysis
HCCLM3 cells were transfected as indicated and then
seeded into 6-well plates (70% confluence) and cultured for 48 h.
Then, the cells were collected for apoptosis analysis using an
Annexin V-FITC apoptosis detection kit (BD Biosciences) according
to the manufacturer's protocols. Briefly, 5 µl of Annexin V-FITC
was added to a 100-µl cell solution (1×105 cells) and
incubated at room temperature for 15 min in the dark. Then, 5 µl PI
was added to the cell solution and incubated at room temperature
for 15 min, before cells were washed with PBS. Cell apoptosis was
evaluated using a BD FACSCalibur™ flow cytometer (BD Biosciences),
and data were analyzed using FlowJo version 10 software (FlowJo
LLC).
Statistical analysis
All data were presented as the mean ± SEM.
Statistical analyses were performed using paired Student's t-tests,
χ2 test and one-way ANOVA with Bonferroni post hoc
analysis. P<0.05 was considered to indicate a statistically
significant difference.
Results
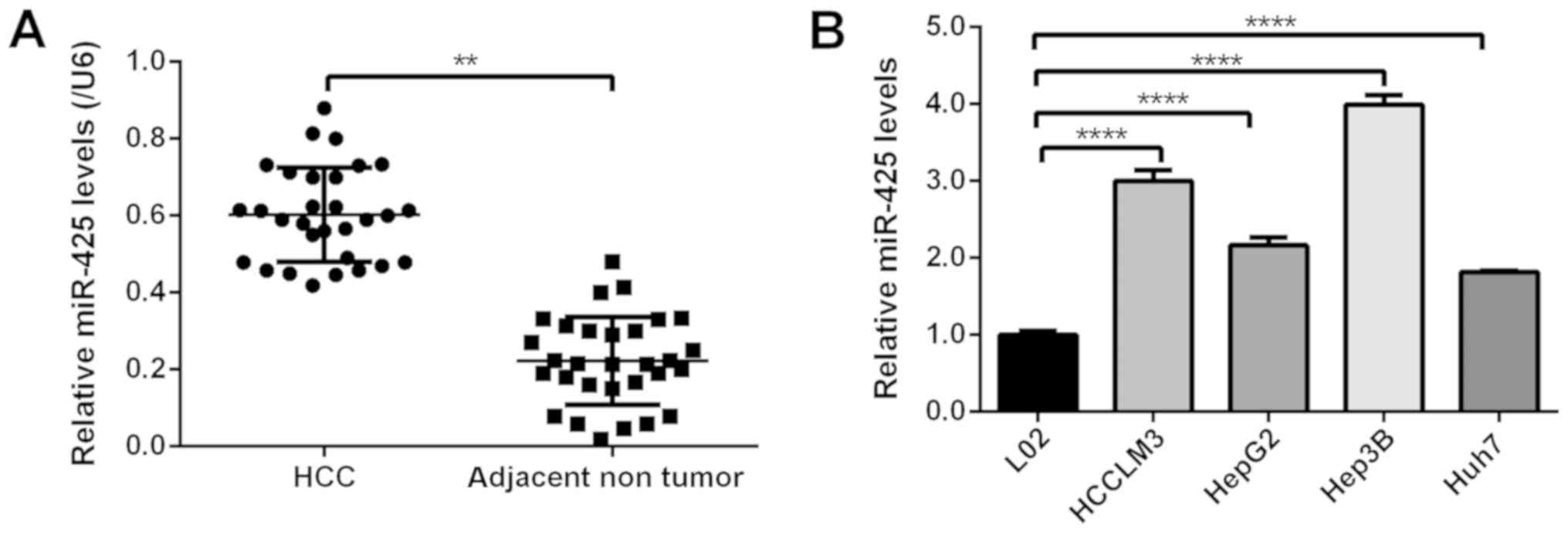
miR-425-5p is upregulated in HCC
tissues and cells
To study the expression profile of miR-425-5p in
HCC, its expression levels were determined in 30 HCC tissues and
paired adjacent nontumor tissues via RT-qPCR analysis. As presented
in Fig. 1A, the expression levels
of miR-425-5p were significantly increased in HCC tissue compared
with in adjacent nontumor tissues. The associations between
miR-425-5p expression and clinical parameters were then analyzed.
As presented in Table I, the
expression of miR-425-5p was associated with clinical stage
(P<0.01). Similarly, the levels of miR-425-5p were significantly
upregulated in HCC and liver cancer cell lines (Huh7, Hep3B, HCCLM3
and HepG2) compared with a normal liver cell line (L02; Fig. 1B). These data indicated that
miR-425-5p expression was upregulated in HCC.
miR-425-5p promotes cell
proliferation, migration and invasion in vitro
To investigate the roles of miR-425-5p in HCC
progression, its effects on the malignant behaviors of HCC cells
were determined. miR-425-5p mimics or ASO-miR-425-5p were
transfected into HCCLM3 cells. RT-qPCR analysis revealed that the
expression of miR-425-5p was significantly increased following
transfection with miR-425-5p mimics compared with miR-NC, but was
significantly decreased following ASO-miR-425-5p transfection
compared with ASO-NC (Fig. 2A).
MTT and colony formation assays revealed that the viability and
colony formation ability of HCCLM3 cells were significantly
increased when miR-425-5p was overexpressed, whereas miR-425-5p
knockdown significantly decreased viability and colony formation
(Fig. 2B and C). Additionally, to
address whether the increase in cell proliferation was associated
with cell apoptosis, flow cytometry analysis was used to evaluate
the apoptosis of HCCLM3 cells. Annexin V staining revealed that the
number of apoptotic cells was significantly decreased when HCCLM3
cells were transfected with miR-425-5p mimics, whereas transfection
of ASO-miR-425-5p induced opposing effects (Fig. 2D). Furthermore, Transwell assays
revealed that the migratory and invasive abilities of HCCLM3 cells
were promoted following miR-425-5p overexpression, but were
decreased by miR-425-5p knockdown (Fig. 2E and F). To determine the
mechanisms underlying the effects of miR-425-5p on the migration
and invasion of cells, the expression of epithelial-mesenchymal
transition markers were evaluated via western blot analysis. As
presented in Fig. 2G,
overexpression of miR-425-5p significantly increased Vimentin
expression but decreased E-cadherin expression, whereas miR-425-5p
knockdown increased E-cadherin expression, but decreased Vimentin
expression. These data demonstrated that miR-425-5p promoted the
proliferation, migration and invasion of HCC cells.
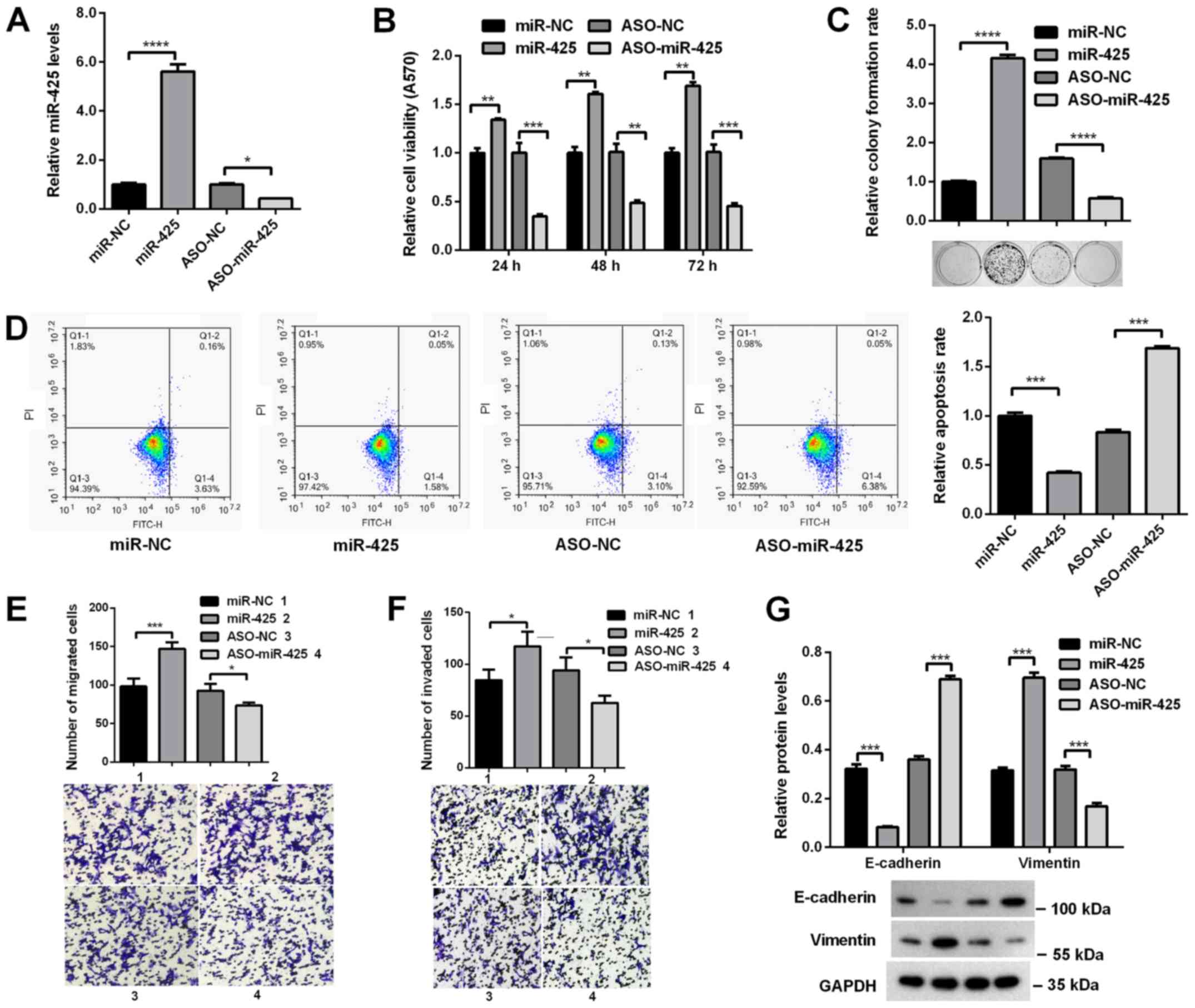 | Figure 2.miR-425-5p promotes the
proliferation, migration and invasion of hepatocellular carcinoma
cells. (A) miR-425-5p expression in HCCLM3 cells following
transfection with miR-425-5p mimics or ASO-miR-425-5p, as
determined via reverse transcription-quantitative PCR analysis. (B)
HCCLM3 cells were transfected with miR-425-5p mimics or
ASO-miR-425-5p, and cell viability was assessed using an MTT assay.
(C) Colony formation ability was evaluated using a colony formation
assay. (D) HCCLM3 cells were transfected with miR-425-5p mimics or
ASO-miR-425-5p and cell apoptosis was measured via flow cytometry.
X-axis: Annexin V-FITC; y-axis: Propidium iodide. HCCLM3 cells were
transfected with miR-425-5p mimics or ASO-miR-425-5p, and the (E)
migratory and (F) invasive abilities of cells were determined using
Transwell assays. Numbers in the associated key indicate the origin
of the Transwell images. (G) HCCLM3 cells were transfected with
miR-425-5p mimics or ASO-miR-425-5p, and the protein levels of
E-cadherin and Vimentin were analyzed via western blotting. Data
are presented as the mean ± standard error of the mean of three
independent experiments. *P<0.05, **P<0.01, ***P<0.001,
****P<0.0001. miR-425-5p, microRNA-425-5p; ASO, antisense
oligonucleotide; NC, negative control; miR-NC, miR-425-5p mimics
NC. |
FOXD3 is a target of miR-425-5p in HCC
cells
To investigate the underlying mechanisms by which
miR-425-5p promotes HCC development, the potential targets of
miR-425-5p were predicted using the TargetScan and PicTar
databases. Among the predicted targets, FOXD3 was selected, as it
has been reported as a tumor suppressor gene in various tumors,
including HCC (29,30). Its expression in HCC tissues was
determined via RT-qPCR analysis; as presented in Fig. 3A, its expression was significantly
decreased in HCC tissues compared with the adjacent nontumor
tissues. Bioinformatics analysis revealed that there was one
putative binding site in the 3′UTR of FOXD3 mRNA (Fig. 3B); this site is conserved among
vertebrates. First, a dual luciferase assay was conducted to
determine whether miR-425-5p directly targets FOXD3. As presented
in Fig. 3C, overexpression of
miR-425-5p significantly decreased the luciferase activity of FOXD3
3′UTR (by ~60%), whereas blocking miR-425-5p markedly increased the
luciferase activities of FOXD3 3′UTR (by ~50%). Conversely, when
the potential binding sites for miR-425-5p in the FOXD3 3′UTR were
mutated, miR-425-5p did not significantly affect the luciferase
activities of FOXD3 3′UTRmut (Fig.
3C). Then, the effects of miR-425-5p on endogenous FOXD3 mRNA
and protein were investigated via RT-qPCR and western blot
analyses. RT-qPCR revealed that the mRNA levels of FOXD3 were
significantly decreased when miR-425-5p was overexpressed, whereas
knockdown of miR-425-5p resulting in opposing effects (Fig. 3D). Western blot analysis also
revealed that overexpression of miR-425-5p significantly decreased,
whereas knockdown of miR-425-5p increased the protein levels of
FOXD3 (Fig. 3E). It was also
revealed that its promoter was hypermethylated in HCC tissues and
HCCLM3 cells (Table SII). These
results indicated that miR-425-5p inhibits FOXD3 expression partly
by binding directly to the FOXD3 3′UTR.
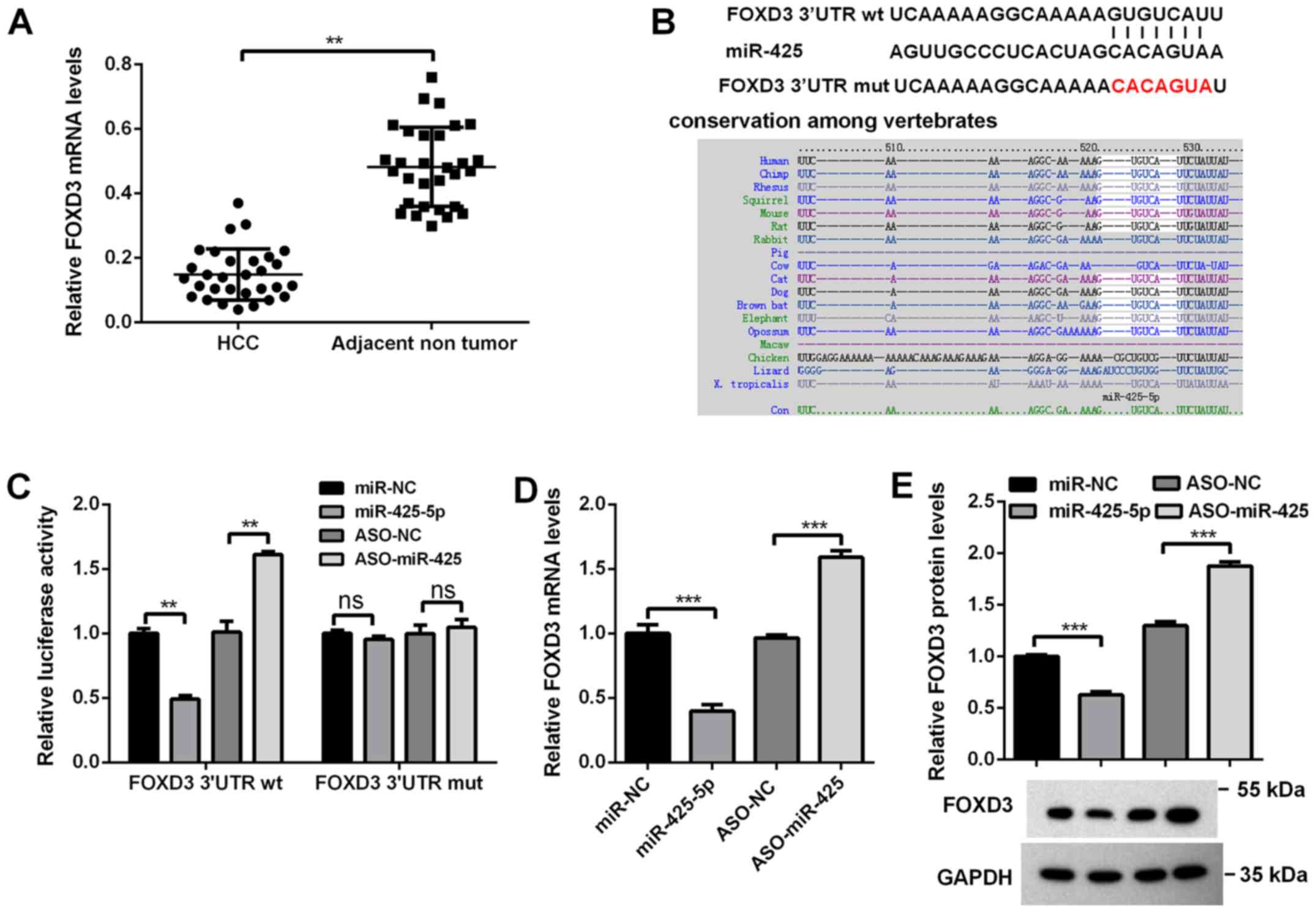 | Figure 3.FOXD3 is a direct target of
miR-425-5p. (A) mRNA levels of FOXD3 in HCC tissues and matched
tumor-adjacent tissues (n=30/group) as determined via RT-qPCR
analysis. (B) Predicted binding site of miR-425-5p in the 3′UTR of
FOXD3 mRNA and the mut sequence. (C) HCCLM3 cells were
co-transfected with the indicated reporter plasmids, and miR-425-5p
mimic or ASO-miR-425-5p, and the luciferase activity was determined
via dual-luciferase assays. HCCLM3 cells were transfected with
miR-425-5p mimic or ASO-miR-425-5p, and (D) mRNA and (E) protein
levels of FOXD3 were determined via RT-qPCR and western blot
analyses, respectively. Data are presented as the mean ± standard
error of the mean of three independent experiments. **P<0.01,
***P<0.001; ns, not significant. miR-425-5p, microRNA-425-5p;
ASO, antisense oligonucleotide; NC, negative control; miR-NC,
miR-425-5p mimics NC; FOXD3, forkhead box D3; HCC, hepatocellular
carcinoma; RT-qPCR, reverse transcription-quantitative PCR; ns, no
significance; wt, wild-type; mut, mutant; 3′UTR, 3′ untranslated
region. |
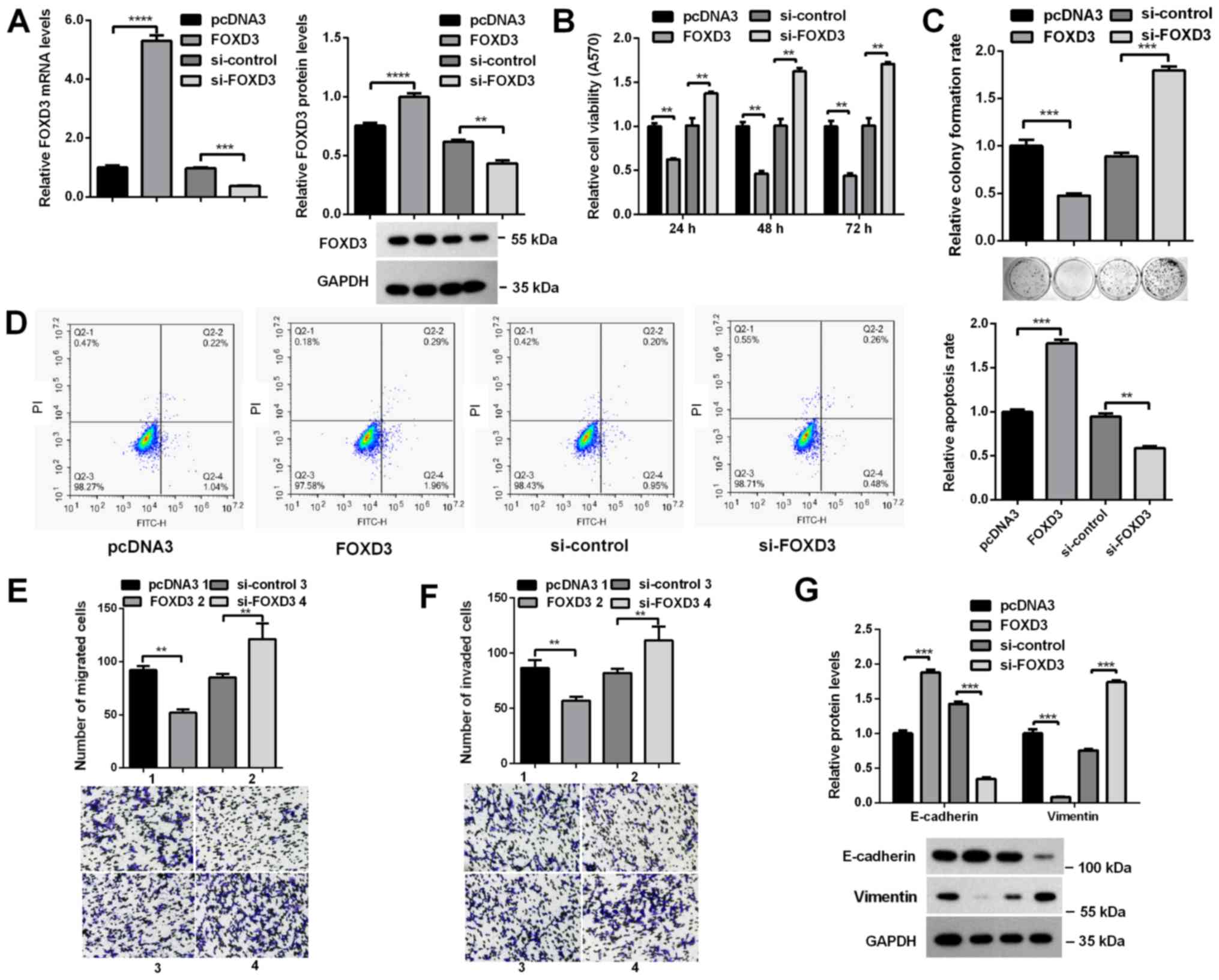
FOXD3 suppresses HCC cell
proliferation, migration and invasion in vitro
To determine whether FOXD3 was involved in the
effects of miR-425-5p on HCC cells, a FOXD3 overexpression plasmid
(pcDNA3.0/FOXD3) and si-FOX3D were constructed, and the efficiency
of these vectors was demonstrated by RT-qPCR and western blot
analyses (Fig. 4A). The effects of
FOXD3 on the malignant behaviors of HCC cells were determined. MTT
and colony formation assays revealed that overexpression of FOXD3
in HCCLM3 cells significantly decreased the viability and colony
formation ability of cells, whereas knockdown of FOXD3 induced
opposing effects (Fig. 4B and C).
Flow cytometry analysis demonstrated that the apoptosis rate of
cells was significantly increased when FOXD3 was overexpressed, but
decreased following FOXD3 knockdown (Fig. 4D). Furthermore, the migratory and
invasive abilities of HCCLM3 cells were significantly decreased
following overexpression of FOXD3, whereas FOXD3 knockdown
significantly increased the migratory and invasive abilities of
HCCLM3 cells (Fig. 4E and F).
Western blot analysis revealed that overexpression of FOXD3
significantly increased E-cadherin, but decreased Vimentin
expression, whereas FOXD3 knockdown downregulated E-cadherin and
upregulated Vimentin expression (Fig.
4G). These data indicated that FOXD3 inhibited the
proliferation, migration and invasion of HCC cells.
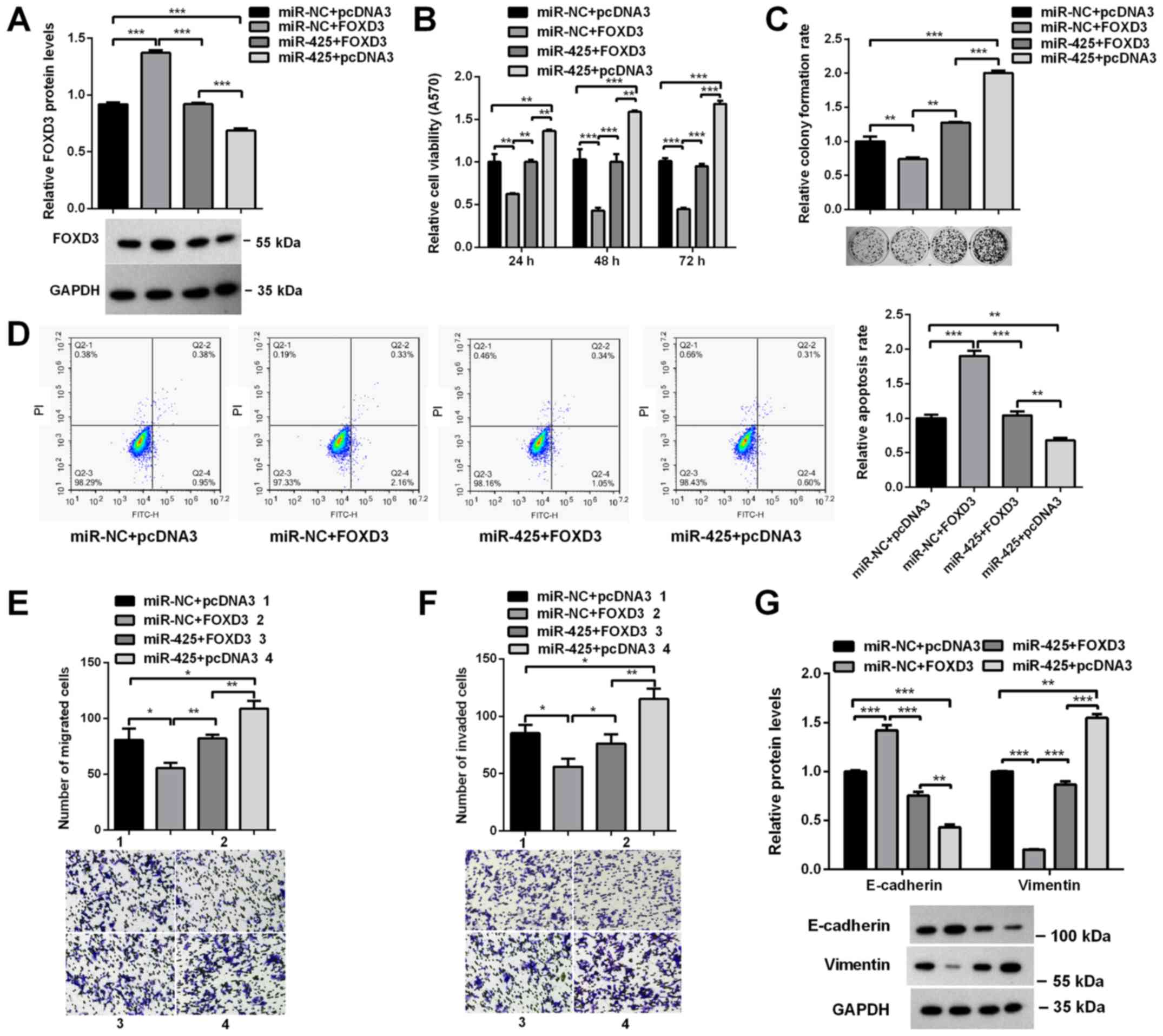
miR-425-5p promotes HCC cell
proliferation, migration and invasion via the downregulation of
FOXD3 expression
miR-425-5p represses endogenous FOXD3 mRNA
expression; however, the overexpression of FOXD3 without its 3′UTR
can recover the protein levels of FOXD3. Therefore, to investigate
whether the effects of miR-425-5p on the progression of HCC were
mediated by downregulating FOXD3, HCCLM3 cells were co-transfected
with miR-425-5p and pcDNA3.0/FOXD3, which encodes the FOXD3 coding
sequence but lacks the 3′UTR. Western blot analysis revealed that
transfection with FOXD3 significantly reversed the effects of
miR-425-5p on FOXD3 expression (Fig.
5A). MTT and colony formation assays revealed that the
restoration of FOXD3 expression significantly rescued the effects
of miR-425-5p on cell viability and colony formation ability
(Fig. 5B and C). Flow cytometry
analysis demonstrated that the inhibition of cell apoptosis by
miR-425-5p was attenuated by FOXD3 overexpression (Fig. 5D). Transwell assays revealed that
the positive effects of miR-425-5p on cell migration and invasion
were suppressed by FOXD3 overexpression (Fig. 5E and F). Similar results were
observed for the expression of E-cadherin and Vimentin (Fig. 5G). Collectively, these results
indicated that miR-425-5p promoted the proliferation, migration and
invasion of HCC cells by repressing FOXD3 expression.
Discussion
It has been reported that miR-425-5p expression is
dysregulated in various cancers (19–26);
its expression was increased in the majority of cases, with the
exception of nasopharyngeal carcinoma and melanoma. In the present
study, it was observed that miR-425-5p was upregulated in HCC,
consistent with a previous study (21); however, the specific mechanisms
responsible for its upregulation in HCC require further
exploration. Furthermore, miR-425-5p was reported to be associated
with the pathogenesis and progression of numerous cancers (19–21,24,26);
however, its role varies in different cancers. For example,
miR-425-5p promoted cell proliferation, migration and invasion by
targeting PTEN and the lysine deubiquitinase CYLD in gastric cancer
(19,20). miR-425-5p promoted the invasion and
metastasis of tumor cells via the suppressor of cancer cell
invasion protein (SCAI)-mediated dysregulation of multiple
signaling pathways in HCC (21),
whereas it acted as a tumor suppressor in nasopharyngeal carcinoma
and melanoma cells by targeting hepatoma-derived growth factor and
insulin-like growth factor 1, respectively (24,26).
In the present study, it was reported that miR-425-5p promoted HCC
cell proliferation, migration and invasion by targeting
transcription factor FOXD3, suggesting that it serves an oncogenic
role.
miRNAs are post-transcriptional regulators that
function by degrading target mRNAs or inhibiting the translation of
target mRNAs. To search for targets of miR-425-5p, TargetScan and
PicTar, two online software tools, were employed, which identified
FOXD3 as a potential target. First, its expression was decreased in
HCC tissues compared with adjacent nontumor tissues, consistent
with a previous study that reported that the downregulation of
FOXD3 in HCC due to promoter hypermethylation (30). It was further revealed that the
promoter of FOXD3 was hypermethylated in HCC tissues and HCCLM3
cells. Whether there are other mechanisms responsible for its
downregulation in HCC requires further study. Then, luciferase
assays, and RT-qPCR and western blot analyses revealed that
miR-425-5p bound directly to and suppressed the expression of
endogenous FOXD3 mRNA. Additionally, rescue experiments indicated
that the promoting effects of miR-425-5p on HCC progression in
HCCLM3 cells were mediated via the downregulation of FOXD3. As the
miR-425-5p levels are different in different HCC cells, the degree
to which malignant behaviors are rescued by FOXD3 may vary. With
previous findings that miR-425-5p promotes the invasion and
metastasis of HCC cells by targeting SCAI and regulating its
downstream signaling pathways (21), miR-425-5p may promote HCC
development via more than one target; however, whether the other
targets are involved in the effects of miR-425-5p on HCC cells
requires further investigation. Of note, in the present study, it
was demonstrated that miR-425-5p increased the viability and colony
formation ability of HCCLM3 cells; flow cytometry analysis
suggested that this may be partly due to the inhibition on
apoptosis. Conversely, in a previous study, the proliferation and
colony number of SMMC-7721 and HCCLM3 cells were not affected by
miR-425-5p (21). This difference
may be a result of different cell culture conditions, but the
specific reason requires further investigation. In addition, when
the effects of miR-425-5p on the colony formation ability of HCCLM3
cells were investigated, it was observed that ASO-NC markedly
increased colony formation compared with miR-NC. The reasons for
this are unclear, as the two oligonucleotides are scrambles of the
experimental molecules and share no homology to other host genes;
however, it may be that the molecules induced distinct cytotoxic
effects on cells, as ASO-NC is single-stranded, whereas miR-NC is
double-stranded.
It was reported that FOXD3 has an important role in
the development of numerous cancers (29–37).
In colon cancer, FOXD3 suppressed the invasive ability and
inhibited the apoptosis of cells by inhibiting the epidermal growth
factor receptor-Ras-Raf-mitogen activated protein kinase kinase
(MEK)-ERK signaling pathway, or regulating miR-214 expression and
function (32,33). FOXD3 can also suppress cell
invasion by inhibiting the expression of the short isoform 2 of
doublecortin-like kinase 1, which is a cancer stem cell marker
(34). In melanoma, FOXD3 was
shown to be involved in cancer cell proliferation and migration via
the regulation of Twist-related protein 1, C-X-C chemokine receptor
4 and paired box gene 3; its expression was controlled by MEK
activity (38–40). In HCC, FOXD3 was reported to
suppress cell proliferation, invasion and metastasis by regulating
miR-137 expression (29). The
present findings revealed similar effects on the malignant
behaviors of HCC cells; however, whether miR-425-5p functions via
an miR-425-5p/FOXD3/miR-137 axis or via other pathways requires
further investigation.
In conclusion, the present results demonstrated that
miR-425-5p was downregulated in HCC, and that miR-425 suppressed
HCC cell proliferation, migration and invasion by targeting FOXD3.
This study may improve understanding of the molecular mechanisms
underlying HCC development, providing novel insight for the
treatment of HCC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the Scientific and
Technological Project of Henan Province (grant no.
172102310098).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HW, JS, WZ and JL performed the experiments. HW, HN
and NC analyzed and interpreted the data. HW and JS drafted the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Written informed consent was obtained from each
patient. The study was approved by the Ethics Committee of Henan
Provincial People's Hospital.
Patient consent for approval
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
miRNA/miR
|
microRNA
|
|
FOXD3
|
forkhead box D3
|
|
HCC
|
hepatocellular carcinoma
|
References
|
1
|
Vasuri F, Visani M, Acquaviva G, Brand T,
Fiorentino M, Pession A, Tallini G, D'Errico A and de Biase D: Role
of microRNAs in the main molecular pathways of hepatocellular
carcinoma. World J Gastroenterol. 24:2647–2660. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Croce CM: Causes and consequences of
microRNA dysregulation in cancer. Nat Rev Genet. 10:704–714. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Giovannini C, Fornari F, Dallo R,
Gagliardi M, Nipoti E, Vasuri F, Coadă CA, Ravaioli M, Bolondi L
and Gramantieri L: MiR-199-3p replacement affects E-cadherin
expression through Notch1 targeting in hepatocellular carcinoma.
Acta Histochem. 120:95–102. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guan J, Liu Z, Xiao M, Hao F, Wang C, Chen
Y, Lu Y and Liang J: MicroRNA-199a-3p inhibits tumorigenesis of
hepatocellular carcinoma cells by targeting ZHX1/PUMA signal. Am J
Transl Res. 9:2457–2465. 2017.PubMed/NCBI
|
|
5
|
Ghosh A, Dasgupta D, Ghosh A, Roychoudhury
S, Kumar D, Gorain M, Butti R, Datta S, Agarwal S, Gupta S, et al:
MiRNA199a-3p suppresses tumor growth, migration, invasion and
angiogenesis in hepatocellular carcinoma by targeting VEGFA,
VEGFR1, VEGFR2, HGF and MMP2. Cell Death Dis. 8:e27062017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ren K, Li T, Zhang W, Ren J, Li Z and Wu
G: miR-199a-3p inhibits cell proliferation and induces apoptosis by
targeting YAP1, suppressing Jagged1-Notch signaling in human
hepatocellular carcinoma. J Biomed Sci. 23:792016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kim JH, Badawi M, Park JK, Jiang J, Mo X,
Roberts LR and Schmittgen TD: Anti-invasion and anti-migration
effects of miR-199a-3p in hepatocellular carcinoma are due in part
to targeting CD151. Int J Oncol. 49:2037–2045. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Henry JC, Park JK, Jiang J, Kim JH,
Nagorney DM, Roberts LR, Banerjee S and Schmittgen TD: miR-199a-3p
targets CD44 and reduces proliferation of CD44 positive
hepatocellular carcinoma cell lines. Biochem Biophys Res Commun.
403:120–125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fornari F, Milazzo M, Chieco P, Negrini M,
Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L and Gramantieri
L: MiR-199a-3p regulates mTOR and c-Met to influence the
doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res.
70:5184–5193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Callegari E, D'Abundo L, Guerriero P,
Simioni C, Elamin BK, Russo M, Cani A, Bassi C, Zagatti B,
Giacomelli L, et al: miR-199a-3p modulates MTOR and PAK4 pathways
and inhibits tumor growth in a hepatocellular carcinoma transgenic
mouse model. Mol Ther Nucleic Acids. 11:485–493. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Varshney A, Panda JJ, Singh AK, Yadav N,
Bihari C, Biswas S, Sarin SK and Chauhan VS: Targeted delivery of
microRNA-199a-3p using self-assembled dipeptide nanoparticles
efficiently reduces hepatocellular carcinoma in mice. Hepatology.
67:1392–1407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Luo C, Yin D, Zhan H, Borjigin U, Li C,
Zhou Z, Hu Z, Wang P, Sun Q, Fan J, et al: microRNA-501-3p
suppresses metastasis and progression of hepatocellular carcinoma
through targeting LIN7A. Cell Death Dis. 9:5352018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu H, Chen W, Zhi X, Chen EJ, Wei T,
Zhang J, Shen J, Hu LQ, Zhao B, Feng XH, et al: Tumor-derived
exosomes promote tumor self-seeding in hepatocellular carcinoma by
transferring miRNA-25-5p to enhance cell motility. Oncogene.
37:4964–4978. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lin XJ, Fang JH, Yang XJ, Zhang C, Yuan Y,
Zheng L and Zhuang SM: Hepatocellular carcinoma cell-secreted
exosomal MicroRNA-210 promotes angiogenesis in vitro and in vivo.
Mol Ther Nucleic Acids. 11:243–252. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Fang JH, Zhang ZJ, Shang LR, Luo YW, Lin
YF, Yuan Y and Zhuang SM: Hepatoma cell-secreted exosomal
microRNA-103 increases vascular permeability and promotes
metastasis by targeting junction proteins. Hepatology.
68:1459–1475. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fang T, Lv H, Lv G, Li T, Wang C, Han Q,
Yu L, Su B, Guo L, Huang S, et al: Tumor-derived exosomal
miR-1247-3p induces cancer-associated fibroblast activation to
foster lung metastasis of liver cancer. Nat Commun. 9:1912018.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lee YS and Dutta A: MicroRNAs in cancer.
Annu Rev Pathol. 4:199–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yan YF, Gong FM, Wang BS and Zheng W:
MiR-425-5p promotes tumor progression via modulation of CYLD in
gastric cancer. Eur Rev Med Pharmacol Sci. 21:2130–2136.
2017.PubMed/NCBI
|
|
20
|
Zhang Z, Li Y, Fan L, Zhao Q, Tan B, Li Z
and Zang A: microRNA-425-5p is upregulated in human gastric cancer
and contributes to invasion and metastasis in vitro and in vivo.
Exp Ther Med. 9:1617–1622. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fang F, Song T, Zhang T, Cui Y, Zhang G
and Xiong Q: MiR-425-5p promotes invasion and metastasis of
hepatocellular carcinoma cells through SCAI-mediated dysregulation
of multiple signaling pathways. Oncotarget. 8:31745–31757.
2017.PubMed/NCBI
|
|
22
|
Sun L, Jiang R, Li J, Wang B, Ma C, Lv Y
and Mu N: MicoRNA-425-5p is a potential prognostic biomarker for
cervical cancer. Ann Clin Biochem. 54:127–133. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu L, Zhao Z, Zhou W, Fan X, Zhan Q and
Song Y: Enhanced expression of miR-425 promotes esophageal squamous
cell carcinoma tumorigenesis by targeting SMAD2. J Genet Genomics.
42:601–611. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu W, Ma Y, Zhuang X and Jin X:
MicroRNA-425 is downregulated in nasopharyngeal carcinoma and
regulates tumor cell viability and invasion by targeting
hepatoma-derived growth factor. Oncol Lett. 15:6345–6351.
2018.PubMed/NCBI
|
|
25
|
Quan J, Li Y, Pan X, Lai Y, He T, Lin C,
Zhou L, Zhao L, Sun S, Ding Y, et al: Oncogenic miR-425-5p is
associated with cellular migration, proliferation and apoptosis in
renal cell carcinoma. Oncol Lett. 16:2175–2184. 2018.PubMed/NCBI
|
|
26
|
Liu P, Hu Y, Ma L, Du M, Xia L and Hu Z:
miR-425 inhibits melanoma metastasis through repression of PI3K-Akt
pathway by targeting IGF-1. Biomed Pharmacother. 75:51–57. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Weigel D and Jäckle H: The fork head
domain: A novel DNA binding motif of eukaryotic transcription
factors? Cell. 63:455–456. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yong JS, Intriago-Baldeón DP and Lam EW:
FOXD3 controls pluripotency through modulating enhancer activity.
Stem Cell Investig. 3:172016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liu LL, Lu SX, Li M, Li LZ, Fu J, Hu W,
Yang YZ, Luo RZ, Zhang CZ and Yun JP: FoxD3-regulated microRNA-137
suppresses tumour growth and metastasis in human hepatocellular
carcinoma by targeting AKT2. Oncotarget. 5:5113–5124. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
He G, Hu S, Zhang D, Wu P, Zhu X, Xin S,
Lu G, Ding Y and Liang L: Hypermethylation of FOXD3 suppresses cell
proliferation, invasion and metastasis in hepatocellular carcinoma.
Exp Mol Pathol. 99:374–382. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yin H, Meng T, Zhou L, Zhao F, Li X, Li Y,
Hu M, Chen H and Song D: FOXD3 regulates anaplastic thyroid cancer
progression. Oncotarget. 8:33644–33651. 2017.PubMed/NCBI
|
|
32
|
He GY, Hu JL, Zhou L, Zhu XH, Xin SN,
Zhang D, Lu GF, Liao WT, Ding YQ and Liang L: The
FOXD3/miR-214/MED19 axis suppresses tumour growth and metastasis in
human colorectal cancer. Br J Cancer. 115:1367–1378. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li K, Guo Q, Yang J, Chen H, Hu K, Zhao J,
Zheng S, Pang X, Zhou S, Dang Y and Li L: FOXD3 is a tumor
suppressor of colon cancer by inhibiting EGFR-Ras-Raf-MEK-ERK
signal pathway. Oncotarget. 8:5048–5056. 2017.PubMed/NCBI
|
|
34
|
Sarkar S, O'Connell MR, Okugawa Y, Lee BS,
Toiyama Y, Kusunoki M, Daboval RD, Goel A and Singh P: FOXD3
regulates CSC marker, DCLK1-S, and invasive potential: Prognostic
implications in colon cancer. Mol Cancer Res. 15:1678–1691. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kubic JD, Little EC, Kaiser RS, Young KP
and Lang D: FOXD3 promotes PAX3 expression in melanoma cells. J
Cell Biochem. 117:533–541. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kubic JD, Lui JW, Little EC, Ludvik AE,
Konda S, Salgia R, Aplin AE and Lang D: PAX3 and FOXD3 Promote
CXCR4 Expression in Melanoma. J Biol Chem. 290:21901–21914. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Weiss MB, Abel EV, Dadpey N and Aplin AE:
FOXD3 modulates migration through direct transcriptional repression
of TWIST1 in melanoma. Mol Cancer Res. 12:1314–1323. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
Elife. 4:e050052015. View Article : Google Scholar :
|
|
39
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M
and Rajewsky N: Combinatorial microRNA target predictions. Nat
Genet. 37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
40
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li H, Yang T, Shang D and Sun Z: miR-1254
promotes lung cancer cell proliferation by targeting SFRP1. Biomed
Pharmacother. 92:913–918. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen H, Yang Y, Wang J, Shen D, Zhao J and
Yu Q: miR-130b-5p promotes proliferation, migration and invasion of
gastric cancer cells via targeting RASAL1. Oncol Lett.
15:6361–6367. 2018.PubMed/NCBI
|