Introduction
Cardiac fibrosis is among the common pathological
features of hypertrophic and dilated cardiomyopathy and may result
in ventricular dysfunction, which leads to heart failure (1). Hyperactivity of fibroblasts may
result in the accumulation of extracellular matrix proteins with
adverse effects on cardiac structure and function, including
electrical instability and increased risk of arrhythmogenic cardiac
death (2). Upon activation,
cardiac fibroblasts (CFs) are transformed into myofibroblasts and
thus contribute to the fibrotic response by secreting key
fibrogenic mediators. Among these, growth factors are the most
widely studied mediators implicated in cardiac fibrosis, including
transforming growth factor β (TGFβ) and platelet-derived growth
factor (3). The main
manifestations of cardiac fibrosis include excessive viability of
CFs and the abundant accumulation of extracellular matrix (ECM).
Cardiac fibrosis is also characterized by trans-differentiation of
CFs into activated myofibroblasts and overexpression of α-smooth
muscle actin (α-SMA), as well as ECM components (4). Despite the great efforts that have
been made to slow down the progression of cardiac fibrosis during
the past decades, it remains a threat to public health.
Long noncoding RNAs (lncRNAs) are a category of RNAs
that lack the potential to code proteins and contain >200
nucleotides (5,6). To date, lncRNAs have been
demonstrated to serve significant roles in intracellular and
extracellular activities, such as transcriptional modification,
gene splicing, gene arrangements and tumorigenesis (7–9). It
has been reported that lncRNAs are also involved in the progression
of cardiac fibrosis. The mechanisms by which lncRNAs regulate
cardiac fibrosis remain to be elucidated, although it has been
suggested that the underlying mechanisms may involve the regulation
of pro-fibrogenic factors by lncRNAs, particularly the regulation
of CFs by lncRNAs (10). The roles
of lncRNAs in cardiac fibrosis have recently received increasing
attention. For instance, Zhang et al (11) demonstrated that depletion of
interleukin (IL)-17 alleviated cardiac fibrosis and ameliorated
cardiac function through inhibiting lncRNA AK081284 in diabetic
mice. Furthermore, PFL was identified as a pro-fibrotic lncRNA in
respect to cardiac fibrosis in mice (12).
Homeobox A11 antisense (HOXA11-AS) is a recently
identified lncRNA, and studies have mainly examined its role in
tumorigenesis. For instance, in human non-small cell lung cancer
(NSCLC), it was reported to promote cell viability and invasion
through regulating the expression of microRNA-124 (13). Upregulation of HOXA11-AS was
observed to increase cell viability via the HOXA11-AS-LATS1 axis in
human hepatocellular carcinoma (14). However, to the best of our
knowledge, the role of HOXA11-AS in cardiac fibrosis has not been
identified to date.
The current study aimed to explore the effects of
HOXA11-AS on cell viability and metastasis through upregulation and
knockdown methods. Colony formation, cell viability and Transwell
assays were used to investigate the underlying association of
HOXA11-AS and TGFβ1. The results may help identify novel methods to
diagnose and treat patients with cardiac fibrosis in clinical
practice.
Materials and methods
Mice
The present study was approved by the Ethics
Committee of the Department of Cardiology, Zhejiang Hospital
(Hangzhou, China). A total of 40 neonate mice (C57BL6/J) were
purchased from the Model Animal Research Center of Nanjing
University (Nanjing, China) and housed under standard conditions of
21±2°C, 55% humidity and a 12-h light/dark cycle. The mice were
sacrificed before 1 week of age. Briefly, neonate mice were
anesthetized by placing them in a sealed isoflurane chamber, at an
isoflurane concentration of 2% v/v and a flow rate of 0.6 l/min.
After ~30 sec, mice ceased movement and were placed on wet ice to
induce hypothermic anesthesia as previously reported (15). The ventricular apex was harvested
and subject to isolation of primary cells via trypsinization and
differential centrifugation steps, similar to those reported in a
previous study (16). Following
the isolation of cells, mice were sacrificed by cervical
dislocation (16).
Cell culture and transfection
Mouse CFs were isolated with collagenase type I
(40507ES60; Shanghai Shengsheng Biotechnology Co., Ltd., Shanghai,
China) and cultured in Dulbecco's modified Eagle's medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.)
at 37°C and an atmosphere that contained 5% CO2,
according to a previous study (17). Cell transfections were performed
with Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. At 6 h
after transfection, the culture medium was replaced with fresh
medium. For upregulation, the HOXA11-AS-expressing plasmid was
cloned into a pcDNA3.1 vector with HindIII and XhoI
restricted enzyme sites. For knockdown, specific small interfering
RNAs (siRNAs) against HOXA11-AS (siHOXA11-AS) and TGFβ1 (siTGFβ1)
were used, which were designed and synthesized by GenePharma Co.,
Ltd. (Shanghai, China). The siHOXA11-AS targeting sequence was
5′-CGGAAUAUCGGAAUAAAGUUU-3′, and the siTGFβ1 targeting sequence was
5′-CCAACUAUUGCUUCAGCUC-3′. The siRNAs were dissolved into a
concentration of 20 µM using ddH2O and diluted into 20
nM when used in the cellular experiments. Transfection was
performed 48 h prior to the assessments, unless otherwise stated.
TGFβ1 recombinant protein (cat. no. 10804-H08H) was purchased from
Sino Biological Inc., Wuhan, China.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA from cultured CFs was isolated by TRIzol
reagent (Thermo Fisher Scientific, Inc.) at 1 ml/well in 6-well
plates. RNA concentration was quantified with Nanodrop 2000
spectrophotometer (Thermo Fisher Scientific, Inc.), and a total of
1 µg RNA was reversely transcribed into cDNA with
PrimeScript™ RT reagent kit (Takara Biotechnology Co.,
Ltd., Dalian, China) using the following protocol: 37°C for 15 min
and 85°C for 5 sec. Next, qPCR assays were performed with SYBRGreen
reagent (Takara Biotechnology Co., Ltd.) in a ABI 7900 machine
(Thermo Fisher Scientific, Inc.) according to the following
procedure: 95°C for 2 min, followed by 40 cycles of 95°C for 15 sec
and 60°C for 30 sec. The primers used in qPCR were as follows:
TGFβ1 forward, 5′-CTCCCGTGGCTTCTAGTGC-3′, and reverse,
5′-GCCTTAGTTTGGACAGGATCTG-3′; α-SMA forward,
5′-GTCCCAGACATCAGGGAGTAA-3′, and reverse,
5′-TCGGATACTTCAGCGTCAGGA-3′; type I collagen (COL1) forward,
5′-GCTCCTCTTAGGGGCCACT-3′, and reverse, 5′-CCACGTCTCACCATTGGGG-3′;
fibronectin (FN) forward, 5′-ATGTGGACCCCTCCTGATAGT-3′, and reverse,
5′-GCCCAGTGATTTCAGCAAAGG-3′; GAPDH forward,
5′-AGGTCGGTGTGAACGGATTTG-3′, and reverse,
5′-TGTAGACCATGTAGTTGAGGTCA-3′. The results were quantified with the
2−ΔΔCq method (18).
Each experiment was repeated three times in triplicate.
Luciferase reporter assay
Luciferase reporter assays were performed with
Dual-luciferase Reporter Assay kit (E1910; Promega Corporation,
Madison, WI, USA) according to the manufacturer's protocol.
Briefly, CFs transfected with TGFβ1 luciferase plasmid and
Renilla plasmid (control plasmid) were treated with
siHOXA11-AS or HOXA11-AS-expressing plasmid for 48 h, and then
lysed for 20 min at room temperature using a shaker at 150 rpm.
Subsequently, the lysate was incubated with luciferase assay
reagent II, and the absorbance of all wavelengths was collected
immediately. The Stop & Glo reagent (Thermo Fisher Scientific,
Inc.) was then added, and the absorbance of the plate re-read; the
first reading was the TGFβ1 luciferase and the second one was the
Renilla luciferase (the control luciferase). The luciferase
activity was calculated based on the ratio of two readings.
Colony formation assay
A total of 100 CFs were seeded into 12-well plates,
and transfected with siHOXA11-AS or with HOXA11-AS-expressing
plasmid in the presence or absence of TGFβ1 recombinant protein (10
ng/ml) or siTGFβ1 (20 ng/ml) in triplicate. Following incubation
for 14 days, the colonies were fixed with ice-cold methanol for 10
min at room temperature and stained with crystal violet for 15 min
(1%). Subsequently, the number of colonies was assessed under a
light microscope at a magnification of ×200 (Nikon Corporation,
Tokyo, Japan). Only colonies containing >50 cells were
considered.
Cell viability assay
Cell viability was assessed by MTT assays. Briefly,
a total of 1×103 CFs/well were seeded in 96-well plates
and treated with siRNA against HOXA11-AS (siHOXA11-AS) or negative
control siRNA (siNC) for 48 h. Next, 10 µl MTT (5 µg/ml) was added
into each well, and cells were cultured for an additional 3 h at
37°C in darkness. Next, the formazan crystals were dissolved in 100
µl dimethyl sulfoxide, and the absorbance of the 96-well plate was
read at 570 nM with a plate reader (Thermo Fisher Scientific,
Inc.).
Transwell assay
A total of 5,000 CFs/well were seeded into 12-well
plates and transfected with siHOXA11-AS or HOXA11-AS-expressing
plasmid for 48 h. Next, cells were collected and diluted into a
concentration of 5×105 cells/ml with culture medium
without FBS. For the migration assay, a 150 µl of cells was added
into the upper Transwell chambers (Corning, Inc., Corning, NY, USA)
and 600 µl medium with 10% FBS was added into the lower chamber.
After 8 h of incubation in a 37°C incubator, the chambers were
washed three times with warm PBS, fixed with methanol at room
temperature for 5 min and stained with crystal violet (1%) for 5
min. For the invasion assay, the upper chambers were first coated
with Matrigel (Corning, Inc.) for 6 h at 37°C. Following staining,
images of the cells on the lower surface of the chamber were
obtained with a Nikon light microscope, and the number of cells in
six random fields-of-view was counted. All experiments were
repeated in triplicate.
Western blot analysis
Western blot analysis was performed in CFs
transfected with siHOXA11-AS or HOXA11-AS-expressing plasmid for 48
h. Briefly, total proteins from cultured cells were extracted by
radioimmunoprecipitation assay lysis buffer (Beyotime Institute of
Biotechnology, Nanjing, China). Protein concentration was monitored
with bicinchoninic acid methods (Thermo Fisher Scientific, Inc.).
Next, a total of 30 µg protein was loaded onto a 12% SDS-PAGE gel
and transferred to a polyvinylidene difluoride membrane. Subsequent
to blocking with 5% milk at room temperature for 1 h, the membranes
were incubated with primary antibodies against TGFβ1 (ab64715;
Abcam, Cambridge, MA, USA), α-SMA (ab5694; Abcam), COL1 (ab6308;
Abcam) and FN (ab18265; Abcam), as well as GAPDH (sc-47724; Santa
Cruz Biotechnology, Inc., Dallas, TX, USA) at 4°C overnight. The
dilution was 1:1,000 for all primary antibodies. Samples were then
incubated with horseradish peroxidase-conjugated IgG secondary
antibodies, which were purchased from Santa Cruz Biotechnology,
Inc. for 1 h at room temperature (sc-2004 and sc-2005; dilution,
1:2,000). Immunoreactivities were determined by enhanced
chemiluminescent autoradiography (Thermo Fisher Scientific, Inc.)
with the ImageQuant LAS 4000 device (GE Healthcare Bio-Sciences,
Uppsala, Sweden).
Statistical analysis
All data are presented as the mean ± standard
deviation. A two-tailed Student's t-test was used to compare the
means of two groups, whereas one-way analysis of variance was used
for comparisons among multiple groups, followed by the
least-significant-difference post hoc test. P<0.05 was
considered to denote a statistically significant difference.
Results
LncRNA HOXA11-AS positively regulates
the expression of TGFβ1 in mouse CFs
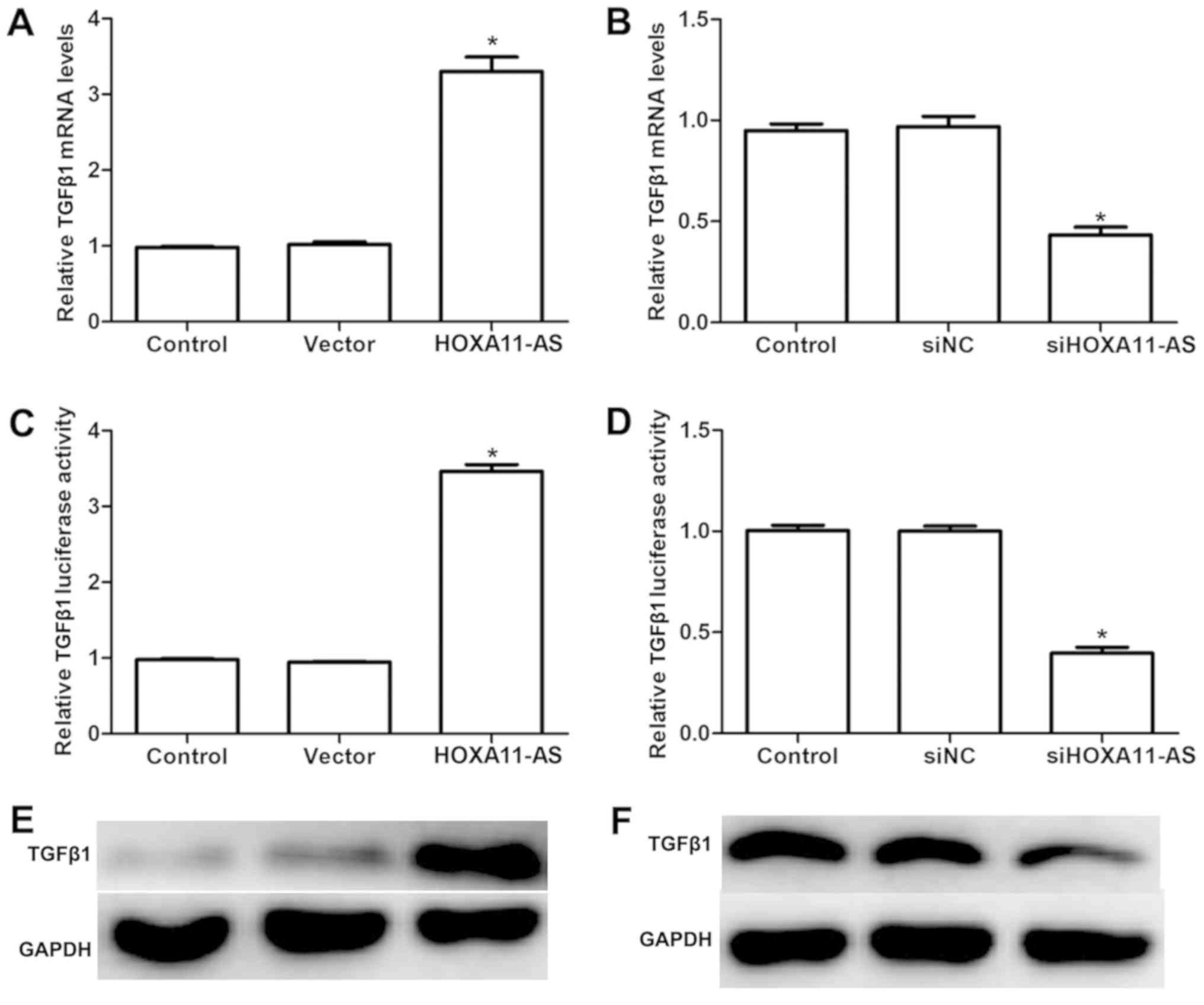
Firstly, the present study examined the effects of
HOXA11-AS on the expression of TGFβ1. As shown in Fig. 1A, when CFs were transfected with
HOXA11-AS-expressing plasmid, the mRNA level of TGFβ1 was
significantly increased, while TGFβ1 mRNA was significantly
decreased in cells transfected with siHOXA11-AS, as compared with
the levels of the control groups (Fig.
1B). Dual-luciferase reporter assays also demonstrated that
overexpression of HOXA11-AS in CFs markedly upregulated the
luciferase activity of TGFβ1, whereas depletion of HOXA11-AS with
specific siRNA knocked down the luciferase signal of TGFβ1
(Fig. 1C and D). Subsequently,
western blot analysis was also performed to determine the
regulatory effect of HOXA11-AS on the pro-inflammatory cytokine
TGFβ1. As shown in Fig. 1E and F,
transfection with HOXA11-AS-expressing plasmid elevated the protein
levels of TGFβ1, while knockdown of HOXA11-AS in CFs suppressed
this protein. All of these results suggested that the lncRNA
HOXA11-AS positively regulated the expression of the key
profibrotic factor TGFβ1.
LncRNA HOXA11-AS regulates the protein
and mRNA levels of TGFβ1 signaling pathway
Based on the results displayed in Fig. 1, the role of HOXA11-AS in the TGFβ1
downstream signaling pathway was further examined. As shown in
Fig. 2A, the protein levels of
α-SMA, COL1 and FN were all upregulated by transfection of
HOXA11-AS-expressing plasmid in CFs, while the inner control GAPDH
level remained stable. Similarly, the expression levels of these
downstream genes of TGFβ1 were inhibited when CFs were treated with
siHOXA11-AS (Fig. 2B). The mRNA
levels were also detected with RT-qPCR analysis, and it was
revealed that the mRNA levels of α-SMA, COL1 and FN were markedly
increased when HOXA11-AS was overexpressed, while these levels
significantly decreased when HOXA11-AS was downregulated in mouse
CFs (Fig. 2C and D). These
results, along with the findings displayed in Fig. 1, suggested that HOXA11-AS regulated
the TGFβ1 signaling pathway in mouse CFs.
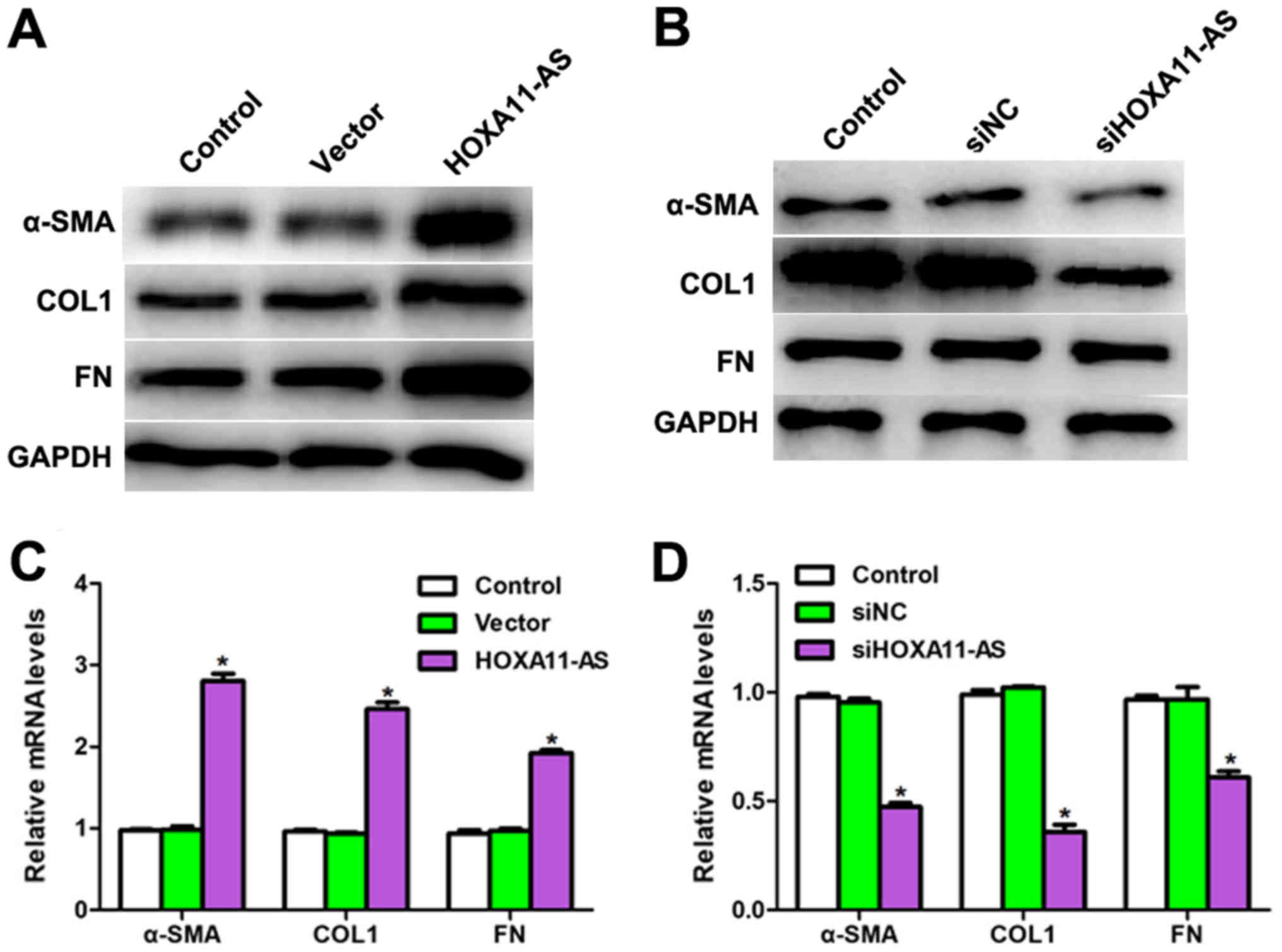 | Figure 2.Long noncoding RNA HOXA11-AS regulated
the levels of proteins and mRNAs associated with the TGFβ1
signaling pathway. Western blot analysis examined the protein
levels of α-SMA, COL1, FN and GAPDH in CFs exhibiting (A) HOXA11-AS
overexpression by plasmid transfection, and (B) HOXA11-AS knockdown
by siRNA transfection. mRNA levels of α-SMA, COL1 and FN were
detected in CFs transfected with (C) HOXA11-AS-expressing plasmid
or (D) siRNA. *P<0.05 vs. control group. CFs, cardiac
fibroblasts; HOXA11-AS, homeobox A11 antisense; TGFβ1, transforming
growth factor β1; siRNA, small interfering RNA; α-SMA, α-smooth
muscle actin; COL1, type I collagen; FN, fibronectin; NC, negative
control. |
Transcript levels of HOXA11-AS are
associated with the viability of mouse CFs
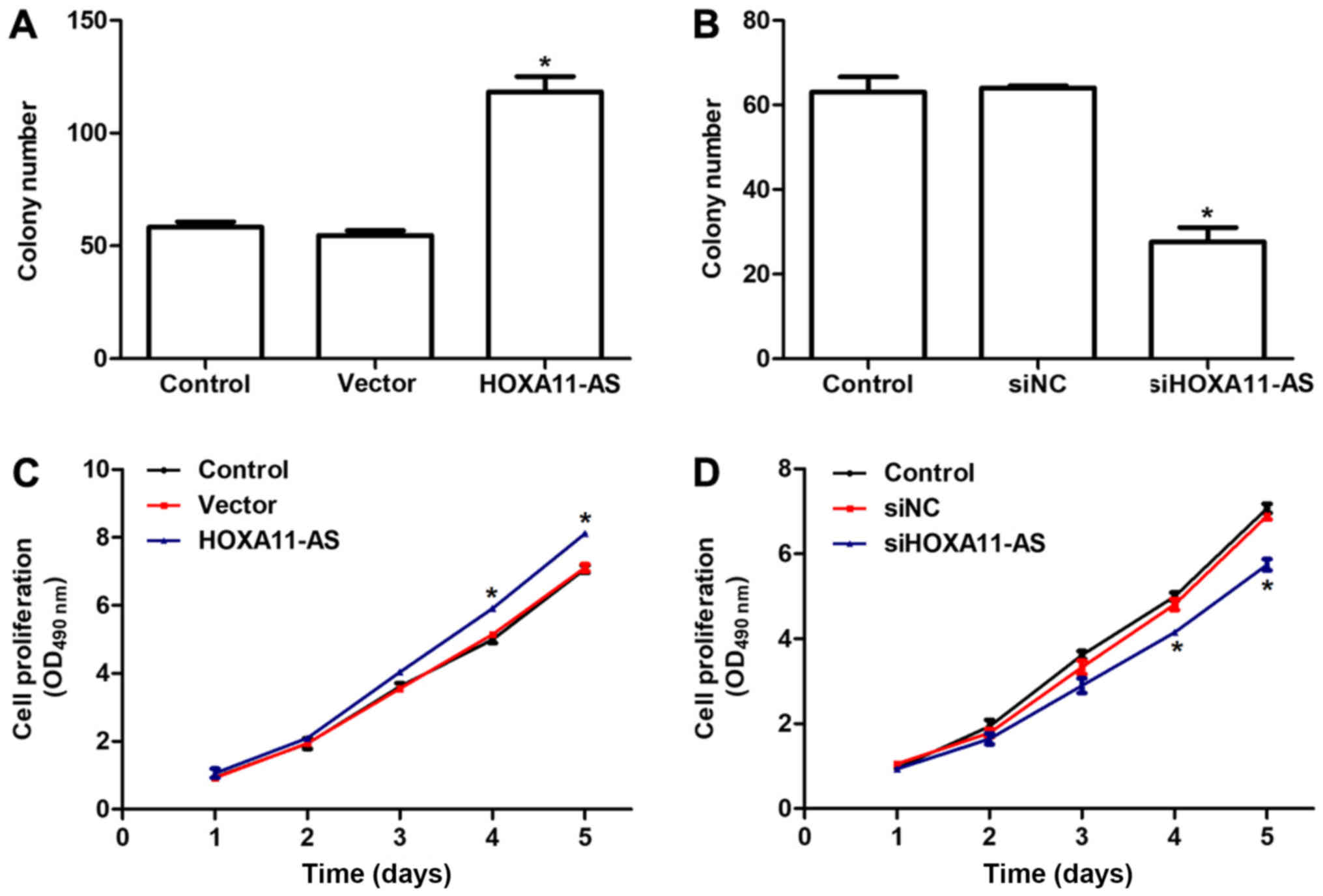
Next, the role of HOXA11-AS on the viability of
mouse CFs was examined by colony formation and MTT assays. It was
observed that overexpression of HOXA11-AS in CFs increased the
colony number by >2-fold (Fig.
3A), whilst knockdown of HOXA11-AS with specific siRNA
suppressed the ability of cells to form colonies, with only ~30
colonies observed in the siHOXA11-AS-treated group (Fig. 3B). Furthermore, there were no
notable differences between three groups in the first 3 days of the
cell viability assays (Fig. 3C and
D). However, overexpression of HOXA11-AS promoted the
proliferative rate by 21% on day 4 and 27% on day 5 in mouse CFs
(Fig. 3C). Similarly, knockdown of
HOXA11-AS in CFs significantly inhibited the cell proliferative
rate on days 4 and 5 (Fig. 3D).
The aforementioned observations indicated that the transcript
levels of HOXA11-AS positively regulated the viability of mouse
CFs.
HOXA11-AS expression is positively
correlated with metastasis in mouse CFs
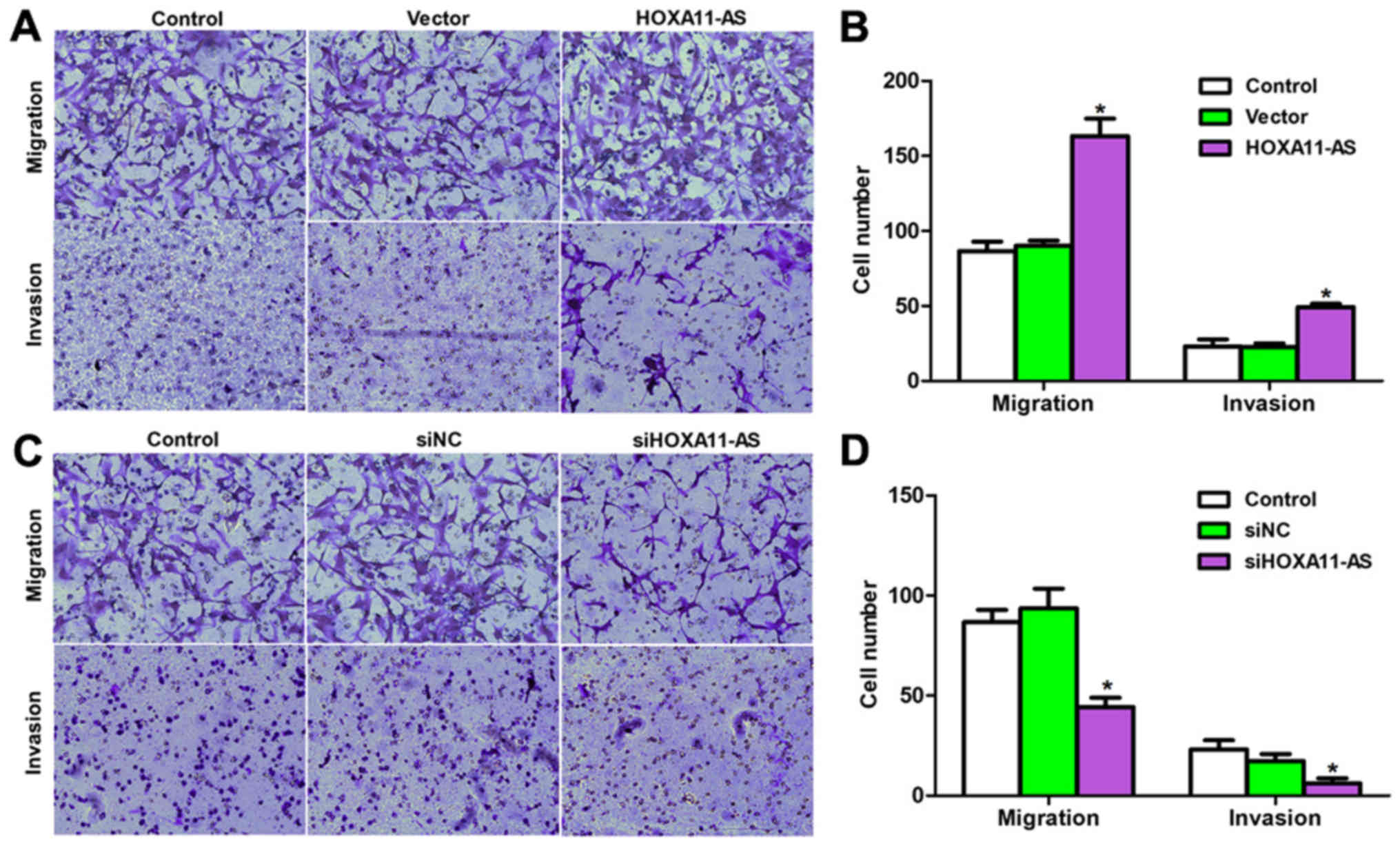
Cell viability and metastasis are two main
manifestations of cardiac injuries. Therefore, the study next
detected the effects of HOXA11-AS on cell metastasis with a
Transwell assay. As shown in Fig. 4A
and B, approximately 85 and 15 migrating and invading cells,
respectively, were observed on the lower surface of the chamber in
the control groups. However, more than 160 and 50 cells that
migrated and invaded, respectively, through the membrane of the
8-µm pores were counted following HOXA11-AS overexpression in CFs.
Similarly, transfection of mouse CFs with specific siRNA against
HOXA11-AS inhibited the cell migration and invasion by approximate
50%, as compared with the control cells (Fig. 4C and D). These data suggested that
the lncRNA HOXA11-AS positively regulated the migration and
invasion of mouse CFs.
HOXA11-AS regulates cell viability and
metastasis through the TGFβ1 signaling pathway in mouse CFs
To explore the detailed mechanisms underlying the
effect of HOXA11-AS in mouse CFs, further investigations were
conducted with TGFβ1 recombinant protein treatment or transfection
with siTGFβ1. As shown in Fig. 5A,
the mRNA levels of HOXA11-AS, TGFβ1 and α-SMA were upregulated when
CFs were transfected with HOXA11-AS-expressing plasmid; however,
when cells were co-transfected with siTGFβ1 and
HOXA11-AS-expressing plasmid, the HOXA11-AS level remained stable,
while TGFβ1 and α-SMA mRNA levels were significantly downregulated.
The opposite results were observed when HOXA11-AS was depleted in
the presence or absence of TGFβ1 recombinant protein treatment
(Fig. 5B). In addition, the colony
formation assay indicated that overexpression of HOXA11-AS promoted
the number of colonies by up to 2-fold compared with the control
group, whereas co-treatment with siTGFβ1 reversed the effects of
HOXA11-AS on colony formation (Fig.
5C). It was also demonstrated that knockdown of HOXA11-AS in
mouse CFs inhibited colony formation, which was consistent with the
earlier observations (Fig. 3);
however, the colony number was markedly increased upon
co-stimulation with TGFβ1 recombinant protein (Fig. 5D). Furthermore, the regulatory
effects of HOXA11-AS on cell migration and invasion were examined
by co-treatment with siTGFβ1 or TGFβ1 recombinant protein (Fig. 5E and F); the results demonstrated
that HOXA11-AS promoted cell metastasis and that the effects were
reversed by TGFβ1 knockdown. Taken together, these data indicated
that HOXA11-AS regulated cell functions through the TGFβ1 signaling
pathway in mouse CFs.
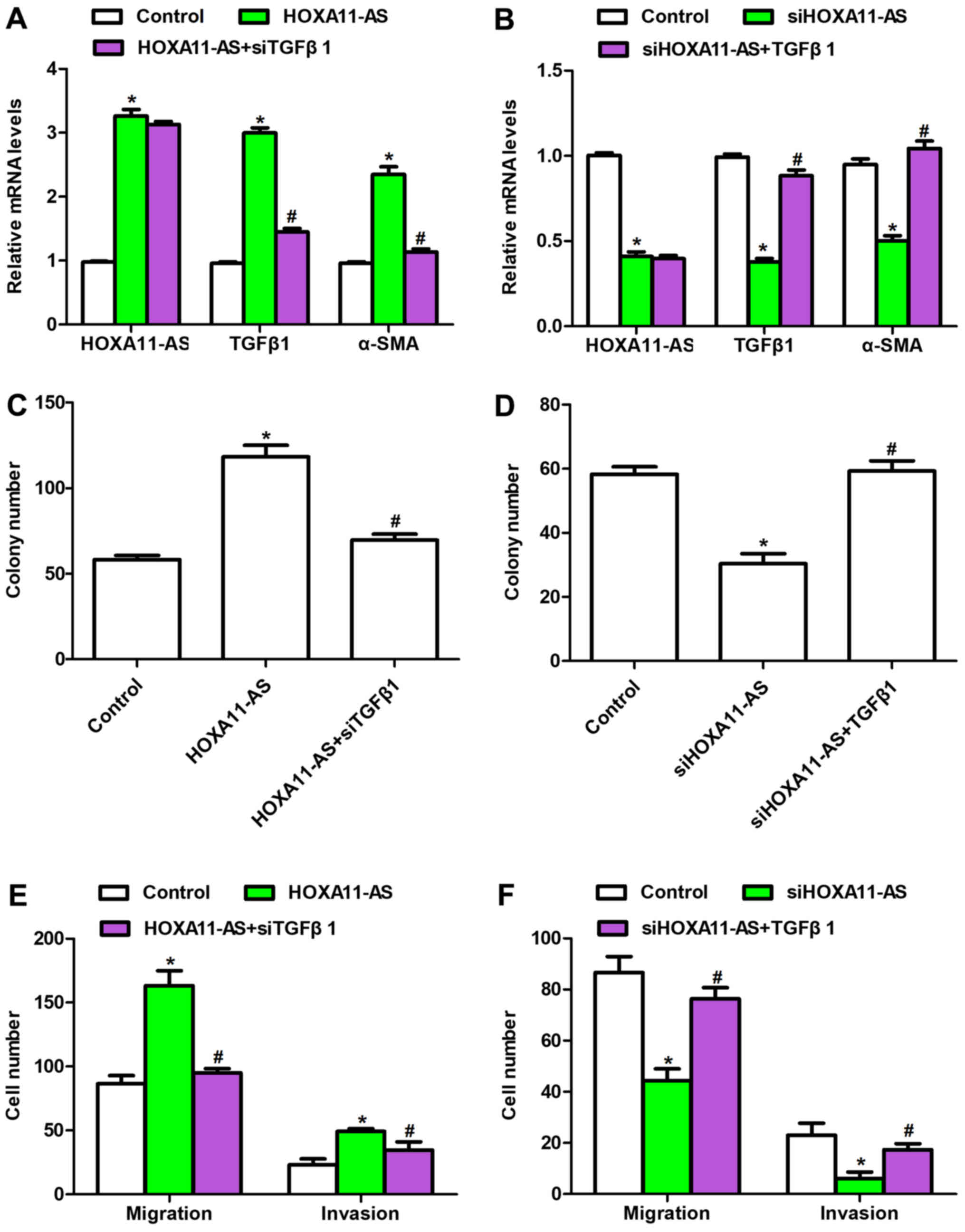 | Figure 5.HOXA11-AS regulated the viability and
metastasis of mouse CFs through the TGFβ1 signaling pathway.
Relative mRNA levels of HOXA11-AS, TGFβ1 and α-SMA were examined by
RT-qPCR in cells with (A) HOXA11-AS overexpression in the presence
or absence of TGFβ1 knockdown, or (B) HOXA11-AS knockdown with or
without TGFβ1 recombinant protein treatment. Colony formation
assays were performed in cells with (C) HOXA11-AS overexpression
with or without siTGFβ1, or (D) HOXA11-AS knockdown with or without
TGFβ1 recombinant protein treatment. Transwell assays were also
performed in CFs with (E) HOXA11-AS overexpression in the presence
or absence siTGFβ1 transfection, or (F) HOXA11-AS depletion in the
presence or absence of TGFβ1 recombinant protein treatment.
*P<0.05 vs. control group; #P<0.05 vs. HOXA11-AS
or siHOXA11-AS group. CFs, cardiac fibroblasts; HOXA11-AS, homeobox
A11 antisense; TGFβ1, transforming growth factor β1; α-SMA,
α-smooth muscle actin; siRNA, small interfering RNA; NC, negative
control; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction. |
Discussion
CFs are considered to be a uniform cell type, widely
distributed in connective tissues, and can be referred to as
mesenchymal original cells that secret multiple ECM components,
such as collagens and FN (19).
CFs are the most common cell type in the heart and serve a key role
in the development of cardiac fibrosis (20). The present study explored the role
of HOXA11-AS in cardiac fibrosis using CFs, and examined the
proliferative rate and metastasis of these cells. The results
indicated that HOXA11-AS may serve as a potential therapeutic
target of cardiac fibrosis.
The TGFβ1 signaling pathway is a classic and
powerful fibrogenic pathway that increases the accumulation of ECM
components, leading to cardiac fibrosis (21,22).
It has also been reported to be involved in various cellular
processes, including cell growth, apoptosis, differentiation and
homeostasis (21,23). TGFβ superfamily ligands bind to the
type II receptor and recruit the phosphorylated type I receptor,
which then phosphorylates receptor-regulated SMADs, finally forming
a complex that accumulates in the nucleus. These ligands then act
as transcription factors and participate in the regulation of the
expression of target genes, including α-SMA, collagen I and FN
(24). In the present study, it
was first observed that overexpression of HOXA11-AS upregulated the
mRNA and protein levels of TGFβ1, whereas knockdown of HOXA11-AS
decreased these levels. The luciferase activity of TGFβ1, which
reflects its transcriptional activity, was also detected. The
luciferase reporter assay results further confirmed that HOXA11-AS
positively regulated the transcription of TGFβ1. Subsequently, the
protein and mRNA expression levels of α-SMA, collagen I and FN were
observed to be increased when CFs were transfected with
HOXA11-AS-expressing plasmid. Thus, the data suggested that
HOXA11-AS directly upregulated the TGFβ1 signaling pathway.
Cell viability and metastasis are two manifestations
of malignancy (25–27), as well as cardiac fibrosis.
Therefore, cell viability and metastasis were examined in cells
transfected with HOXA11-AS-expressing plasmid or siRNA targeting
HOXA11-AS. It was demonstrated that HOXA11-AS promoted the cell
proliferative rate, and increased the potential of cells to migrate
and invade through the Transwell membrane. Taken together with the
former findings that HOXA11-AS upregulated TGFβ1 signaling, it can
be hypothesized that HOXA11-AS served its role through regulating
the TGFβ1 pathway. To this end, TGFβ1 siRNA or recombinant protein
co-treatment with HOXA11-AS-expressing plasmid or siHOXA11-AS,
respectively, in CFs was conducted, and the results revealed that
HOXA11-AS promoted cell viability and metastasis, which was
reversed by knockdown of TGFβ1. These observations directly
suggested that HOXA11-AS promoted cardiac fibrosis progression
through upregulating the TGFβ1 signaling pathway. As for the
detailed mechanism of the regulatory effects of HOXA11-AS on TGFβ1,
further investigation is required in future studies.
In conclusion, the current study demonstrated that
HOXA11-AS promoted cell proliferation and metastasis by increasing
the activity of TGFβ1 in CFs, which further identified the specific
role of HOXA11-AS in cardiac fibrosis. The present study attempted
to explain the detailed mechanism, which may provide novel evidence
for the clinical diagnosis and treatment of cardiac fibrosis in the
near future.
Acknowledgements
Not applicable.
Funding
The present study was funded by major Research and
Development Projects for the Zhejiang Science and Technology Agency
(grant no. 2017C03034) and Scientific and Technological Projects
for Medical and Health of Zhejiang Province (grant no.
2015128660).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
JW and XL performed the experiments with assistance
from QZ, RP, LZ and ZC. LT provided the funding and designed the
project.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Malgija B, Kumar NS and Piramanayagam S:
Collective transcriptomic deregulation of hypertrophic and dilated
cardiomyopathy-importance of fibrotic mechanism in heart failure.
Comput Biol Chem. 73:85–94. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Prinz C, Farr M, Laser KT, Esdorn H, Piper
C, Horstkotte D and Faber L: Determining the role of fibrosis in
hypertrophic cardiomyopathy. Expert Rev Cardiovasc Ther.
11:495–504. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Khalil H, Kanisicak O, Prasad V, Correll
RN, Fu X, Schips T, Vagnozzi RJ, Liu R, Huynh T, Lee SJ, et al:
Fibroblast-specific TGF-β-Smad2/3 signaling underlies cardiac
fibrosis. J Clin Invest. 127:3770–3783. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schafer S, Viswanathan S, Widjaja AA, Lim
WW, Moreno-Moral A, DeLaughter DM, Ng B, Patone G, Chow K, Khin E,
et al: IL-11 is a crucial determinant of cardiovascular fibrosis.
Nature. 552:110–115. 2017.PubMed/NCBI
|
|
5
|
Yu J, Fang Q and Meng S: Knockdown of long
noncoding RNA ENST457720 inhibits proliferation of non-small cell
lung cancer cells in vitro and in vivo. Oncol Res. 1–Mar;2018.(Epub
ahead of print). View Article : Google Scholar
|
|
6
|
Lan T, Chang L, Wu L and Yuan Y:
Downregulation of ZEB2-AS1 decreased tumor growth and metastasis in
hepatocellular carcinoma. Mol Med Rep. 14:4606–4612. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sas-Chen A, Aure MR, Leibovich L, Carvalho
S, Enuka Y, Körner C, Polycarpou-Schwarz M, Lavi S, Nevo N,
Kuznetsov Y, et al: LIMT is a novel metastasis inhibiting lncRNA
suppressed by EGF and downregulated in aggressive breast cancer.
EMBO Mol Med. 8:1052–1064. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhuo C, Jiang R, Lin X and Shao M: LncRNA
H19 inhibits autophagy by epigenetically silencing of DIRAS3 in
diabetic cardiomyopathy. Oncotarget. 8:1429–1437. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Sehgal L, Jain N, Khashab T,
Mathur R and Samaniego F: LncRNA MALAT1 promotes development of
mantle cell lymphoma by associating with EZH2. J Transl Med.
14:3462016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jiang X and Zhang F: Long noncoding RNA: A
new contributor and potential therapeutic target in fibrosis.
Epigenomics. 9:1233–1241. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Zhang YY, Li TT, Wang J, Jiang Y,
Zhao Y, Jin XX, Xue GL, Yang Y, Zhang XF, et al: Ablation of
interleukin-17 alleviated cardiac interstitial fibrosis and
improved cardiac function via inhibiting long non-coding
RNA-AK081284 in diabetic mice. J Mol Cell Cardiol. 115:64–72. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Leisegang MS: LET's sponge: How the lncRNA
PFL promotes cardiac fibrosis. Theranostics. 8:874–877. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yu W, Peng W, Jiang H, Sha H and Li J:
LncRNA HOXA11-AS promotes proliferation and invasion by targeting
miR-124 in human non-small cell lung cancer cells. Tumour Biol.
39:10104283177214402017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu J, Hong JF, Kang J, Liao LH and Li CD:
Promotion of LncRNA HOXA11-AS on the proliferation of
hepatocellular carcinoma by regulating the expression of LATS1. Eur
Rev Med Pharmacol Sci. 21:3402–3411. 2017.PubMed/NCBI
|
|
15
|
Blom JN, Lu X, Arnold P and Feng Q:
Myocardial infarction in neonatal mice, a model of cardiac
regeneration. J Vis Exp. 24–May;2016.(doi: 10.3791/54100).
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Parameswaran S, Santhakumar R, Vidyasekar
P and Verma RS: Enrichment of cardiomyocytes in primary cultures of
murine neonatal hearts. Methods Mol Biol. 1299:17–25. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ackers-Johnson M, Li PY, Holmes AP,
O'Brien SM, Pavlovic D and Foo RS: A simplified, langendorff-free
method for concomitant isolation of viable cardiac myocytes and
nonmyocytes from the adult mouse heart. Circ Res. 119:909–920.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Souders CA, Bowers SL and Baudino TA:
Cardiac fibroblast: The renaissance cell. Circ Res. 105:1164–1176.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Moore-Morris T, Guimarães-Camboa N, Yutzey
KE, Pucéat M and Evans SM: Cardiac fibroblasts: From development to
heart failure. J Mol Med (Berl). 93:823–830. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wrana JL, Attisano L, Cárcamo J, Zentella
A, Doody J, Laiho M, Wang XF and Massagué J: TGF beta signals
through a heteromeric protein kinase receptor complex. Cell.
71:1003–1014. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Massagué J: TGFbeta in cancer. Cell.
134:215–230. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Moustakas A: Smad signalling network. J
Cell Sci. 115:3355–3356. 2002.PubMed/NCBI
|
|
24
|
Chen E, Cen Y, Lu D, Luo W and Jiang H:
IL-22 inactivates hepatic stellate cells via downregulation of the
TGF-β1/Notch signaling pathway. Mol Med Rep. 17:5449–5453.
2018.PubMed/NCBI
|
|
25
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dela Cruz CS, Tanoue LT and Matthay RA:
Lung cancer: Epidemiology, etiology, and prevention. Clin Chest
Med. 32:605–644. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|