Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor
of the head and neck with a variable incidence rate according to
race, ethnicity and geographical location; particularly high rates
have been identified in the populations of South China and
Southeast Asia (1). A previous
study demonstrated that Epstein-Barr virus infection, genetic
susceptibility and environmental factors are the principal risk
factors associated with the pathogenesis of NPC (2). NPC is characterized by high rates of
local and distant metastasis in early stages. Although the
combination of radiotherapy and chemotherapy has improved NPC
outcomes, survival rates remain poor (3). Therefore, the characterization of the
molecular mechanisms underlying NPC development has attracted
increasing attention from researchers.
Long non-coding RNAs (lncRNAs) are non-coding RNAs
≥200 nucleotides in length that regulate gene expression via
various mechanisms, including the modulation of RNA stability, the
translation rates of mRNA and the regulation of the epigenetic
machinery (4). Aberrant expression
of lncRNAs is associated with a number of malignancies.
Furthermore, various lncRNAs have been demonstrated to serve as
oncogenes or tumor suppressor genes, regulating cancer cell
proliferation, migration, apoptosis and chemotherapy resistance
(5). Previous studies have
demonstrated that numerous lncRNAs, including small nucleolar RNA
host gene 1 and nuclear paraspeckle assembly transcript 1, are
dysregulated in NPC tissues, serving a role in cancer progression
(6,7).
The lncRNA differentiation antagonizing non-protein
coding RNA (DANCR) was identified to be associated with the
dedifferentiation of epidermal cells (8). Additionally, DANCR has been
demonstrated to be involved in tumorigenesis in various types of
cancer, including colorectal cancer, breast cancer and lung
adenocarcinoma, by regulating protein expression or by sponging
microRNAs (9–11). Wen et al (12) observed that DANCR expression was
increased in metastatic NPC cells, suggesting a role for DANCR in
NPC development. However, the molecular mechanism underlying DANCR
in NPC remains unclear. In the present study, DANCR expression
levels were measured in NPC cells, and the effect of DANCR on the
proliferation, migration and apoptosis of NPC cells was
investigated using loss- and gain-of-function models. Furthermore,
DANCR was identified to regulate the protein expression levels of
AKT serine/threonine kinase (AKT), phosphatase and tensin homolog
(PTEN), and a number of apoptosis-associated factors, including
BCL2 apoptosis regulator (BCL2) and BCL2 associated X apoptosis
regulator (BAX).
Materials and methods
Cell lines and transfection
The human NPC cell lines 5-8F, SUNE-1, C666-1 and
the normal human nasopharyngeal epithelial cell line NP69 (Sun
Yat-sen University, Guangzhou, China) were maintained in our
laboratory following authentication. RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.) was
used to culture NPC cell lines in 5% CO2 at 37°C.
Keratinocyte serum-free medium with epidermal growth factor
(Invitrogen; Thermo Fisher Scientific, Inc.) was used to culture
NP69 cells in 5% CO2 at 37°C.
Small interfering RNA targeting DANCR (si-DANCR) and
scrambled negative control (si-NC) were purchased from Shanghai
GenePharma Co., Ltd. (Shanghai, China). Knockdown was performed
using the following siRNAs: si-DANCR: 5′-GCUGGUAUUUCAAUUGACU-3′;
si-NC: 5′-UUCUCCGAACGUGUCACGU-3′. DANCR overexpression plasmid
(pcDNA-DANCR) was constructed by cloning DANCR into a pcDNA vector
(Invitrogen; Thermo Fisher Scientific, Inc.). To amplify the coding
sequence of DANCR, the cDNA from SUNE-1 cells was used as a
template, and the reaction was performed using the PrimeSTAR HS DNA
polymerase (Takara Biotechnology Co., Ltd., Dalian, China) with the
following primers: Forward (F):
5′-CGGGGTACCGCCCTTGCCCAGAGTCTTCCCGG-3′ and reverse (R):
5′-CCGCTCGAGGTCAGGCCAAGTAAGTTTATTAACCT-3′. The thermocycling
conditions were as follows: Initial denaturation at 95°C for 5 min,
followed by 30 cycles at 95°C for 1 min, 50°C for 2 min, 72°C for 2
min and a final extension at 72°C for 10 min. SUNE-1 cells and 5-8F
cells at 40–70% confluence were transfected with 500 ng plasmid or
with si-DANCR/si-NC at a concentration of 50 nM using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Following incubation for 48 h, cells were
harvested for further experimentation.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from NPC cells, NP69 cells
and tumor tissues using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
protocol. A total of 1 ml TRIzol® was incubated with the
cell suspensions or tissues powder (100 mg) for 5 min at room
temperature, subsequently 0.2 ml chloroform was added and incubated
for 10 min at room temperature. Following centrifugation (12,000 ×
g, 15 min, 4°C), the supernatant was precipitated by alcohol and
centrifuged (7,500 × g, 5 min, 4°C). RT was performed using M-MLV
reverse transcriptase (Takara Biotechnology Co., Ltd.) with 2.5 µM
primers, 5X First-Strand Buffer, dNTP Mix and DTT (Takara
Biotechnology Co., Ltd.). The temperature protocol was as follows:
70°C for 3 min, 42°C for 60 min and 70°C for 15 min. qPCR was
conducted using a SYBR Green kit (Takara Biotechnology Co., Ltd.)
and the 7500 Real-Time PCR System (Applied Biosystems; Thermo
Fisher Scientific, Inc.) (13).
The thermocycling conditions were as follows: Initial denaturation
at 95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and at
60°C for 40 sec. The qPCR primers used were as follows: DANCR,
forward 5′-CGTACTAACTTGTAGCAACC-3′, reverse
5′-TCAGCTGCATTGAGTTAGCG-3′; and GAPDH, forward
5′-ACTCACTCAAGATTGTCAGCA-3′, reverse 5′-GGCCATCACGCCACAGCTTT-3′).
Each experiment was performed three times, and relative gene
expression was determined using the 2−ΔΔCq
quantification method (14).
Western blot analysis
Total protein was extracted from 5-8F cells and
SUNE-1 cells using Pierce cell lysis buffer (Pierce; Thermo Fisher
Scientific, Inc.). Protein concentration was quantified using a
bicinchoninic acid assay kit (Takara Biotechnology Co., Ltd.). A
total of 50 µg protein was loaded per lane. Proteins were separated
by 10% SDS-PAGE and transferred onto polyvinylidene difluoride
membranes (Merck KGaA, Darmstadt, Germany). Following blocking in
5% skim milk for 2 h at 37°C, the membranes were incubated
overnight at 4°C with the following primary antibodies:
Phosphorylated (p)-pan-AKT (1:1,000; cat. no. ab38449; Abcam,
Cambridge, UK), PTEN (1:1,000; cat. no. ab31392; Abcam), BCL2
(1:1,000; cat. no. ab196495; Abcam), BAX (1:1,000; ab53154; Abcam)
and β-actin (1:2,000; cat. no. ab8227; Abcam). Subsequently,
membranes were incubated with the appropriate horseradish
peroxidase-conjugated secondary antibody (1:2,000; cat.no. ab6728;
Abcam) for 2 h at room temperature. Immunoreactive bands were
visualized with an enhanced chemiluminescence detection system
(Merck KGaA). Relative amounts of proteins were quantified by
absorbance analysis using ImageJ software (version 1.8.0; National
Institutes of Health, Bethesda, MD, USA), and the level was
normalized to β-actin.
Cell proliferation assay
A total of 2×103 cells/well of
transfected cells were seeded into 96-well plates and cultured at
37°C for 0, 24, 48 and 72 h. Subsequently, 100 µl Cell Counting
Kit-8 (CCK-8; Dojindo Molecular Technologies, Inc., Kumamoto,
Japan) reagent was added, and cells were incubated with CCK-8
reagent for 4 h. Absorbance at 450 nm was measured using a
microplate reader (Thermo Fisher Scientific, Inc.).
Colony formation assay
Transfected cells (1×103 cells/well)
cultured in complete medium were seeded in 6-well plates. Following
14 days, colonies were fixed in 100% methanol for 15 min at room
temperature. The fixing solution was removed and 10% Giemsa stain
solution was used to stain the colonies for 30 min at room
temperature. The plates were subsequently imaged under a light
microscope (BX51; Olympus Corporation, Tokyo, Japan; magnification,
×100) and colonies were counted.
Wound healing assay
A total of 5×105 cells cultured in
serum-free RPMI-1640 medium, to limit proliferation, were seeded in
6-well plates. Subsequently, a scratch was made through the single
cell layer using a 10 µl pipette tip, and the cells were washed
with PBS. The cells were incubated at 37°C for 48 h and
subsequently imaged using an inverted fluorescence microscope
(IX71; Olympus Corporation; magnification, ×100). Each experiment
was performed in triplicate.
Flow cytometric apoptosis
analysis
SUNE-1 and 5-8F cells were transfected with si-DANCR
and pcDNA-DANCR, respectively. Following transfection and a
subsequent incubation of 48 h, cells were harvested and incubated
with 5 µl Annexin V-fluorescein isothiocyanate (FITC) for 20 min,
and subsequently with 10 µl propidium iodide (PI) for 10 min
(FITC/PI kit; R&D Systems, Inc., Minneapolis, MN, USA).
Apoptotic cells were observed and analyzed using a flow cytometer
(Becton, Dickinson and Company, Franklin Lakes, NJ, USA) and FlowJo
software (version 7; Tree Star, Inc., Ashland, OR, USA). All
experiments were performed in triplicate.
NPC xenograft mouse model
A total of 12 male BALB/cnu/nu mice
(weight, 25–30 g; age, 6-weeks) purchased from Beijing Vital River
Laboratory Animal Technology Co., Ltd. (Beijing, China) were
maintained under a 12-h light/dark cycle at 20–22°C and 40–60%
relative humidity with access to food and water ad libitum.
The lentiviral vectors (LV) containing a short hairpin RNA
targeting DANCR (sh-DANCR) and the LV-control (ctrl) were purchased
from Guangzhou RiboBio Co., Ltd. (Guangzhou, China) and transduced
into SUNE-1 cells with a multiplicity of infection of 50. Following
a 72-h incubation, the cells (2×106) were injected into
the right flanks of nude mice. A total of six mice were used in
each group. Tumor size was measured every 7 days for 5 weeks. Tumor
volumes (mm3) were calculated using the following
formula: Volume = 0.5 × length × width2. All animal
experiments were approved by The Institutional Animal Care and Use
Committee of Shanghai Jiaotong University (Shanghai, China).
Statistical analysis
Data are presented as the mean ± standard deviation
from three replicates. Statistical differences were assessed using
Student's t-test or one-way analysis of variance followed by
Student-Newman-Keuls post hoc test for multiple comparisons.
Statistical analysis was conducted using SPSS software (version 23;
IBM Corp., Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
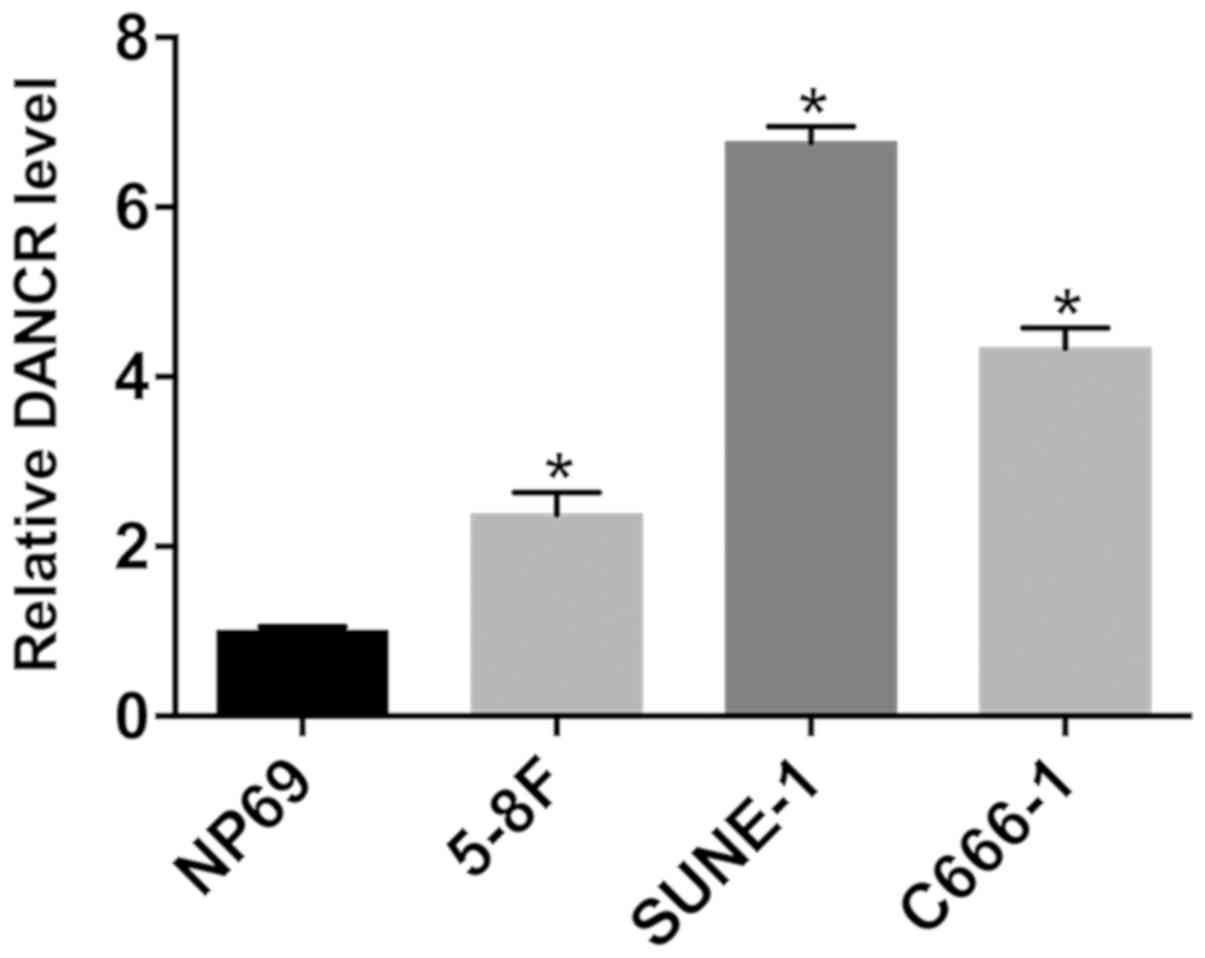
DANCR is upregulated in NPC cells
To investigate DANCR expression in NPC cells,
RT-qPCR was performed in NPC and control NP69 cells. DANCR was
significantly upregulated in NPC cells compared with NP69 cells
(Fig. 1). Among the NPC cell lines
examined, SUNE-1 and 5-8F cells exhibited the highest and lowest
DANCR expression levels, respectively.
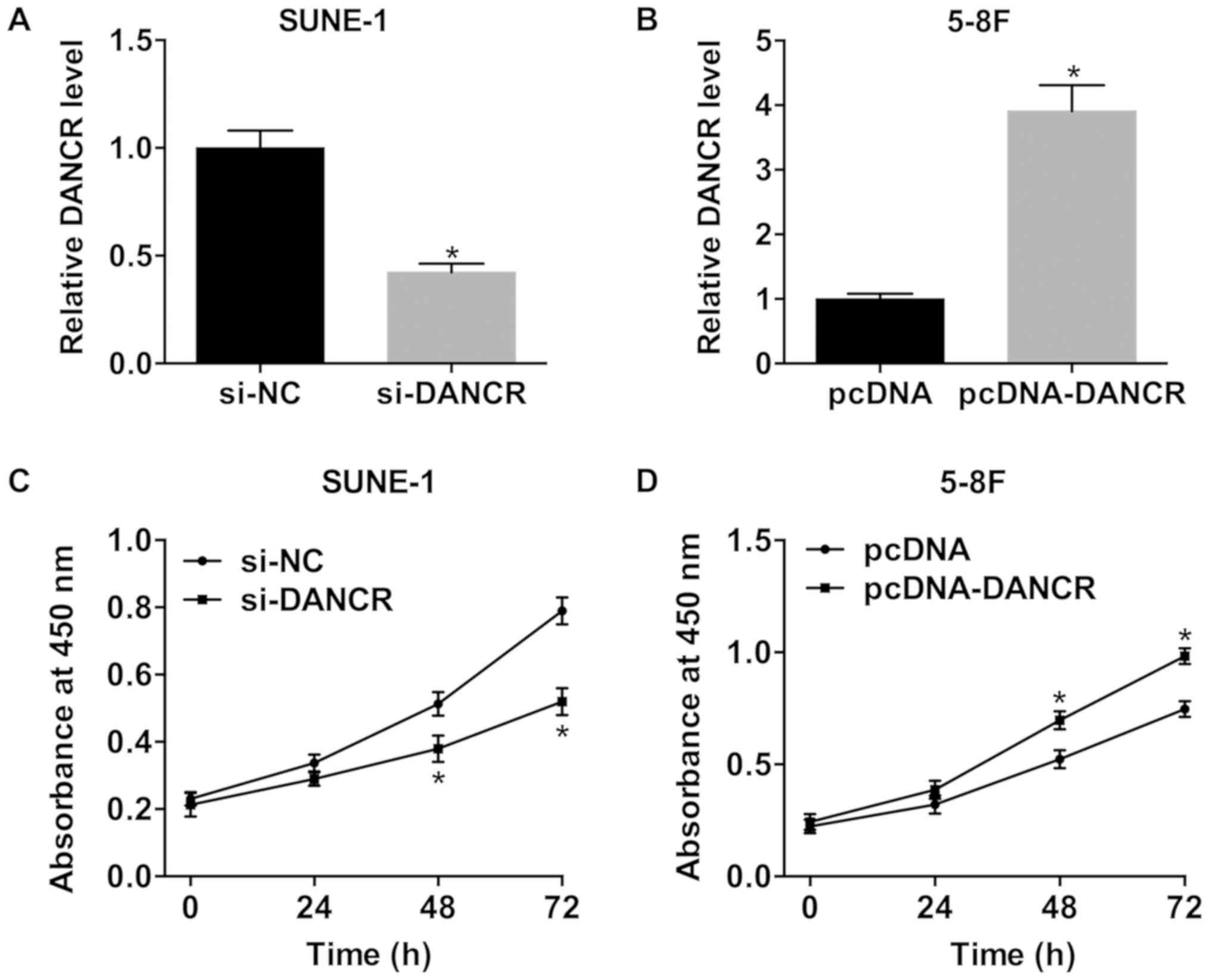
DANCR increases the viability of NPC
cells
SUNE-1 cells were transfected with si-DANCR to
knockdown DANCR and 5-8F cells were transfected with pcDNA-DANCR to
overexpress DANCR. si-DANCR dramatically decreased DANCR expression
in SUNE-1 cells, and DANCR was significantly upregulated in 5-8F
cells transfected with pcDNA-DANCR (Fig. 2A and B). Silencing DANCR
significantly inhibited the viability of SUNE-1 cells (Fig. 2C). Bu contrast, compared with the
negative control, the viability of cells transfected with
pcDNA-DANCR was significantly increased (Fig. 2D).
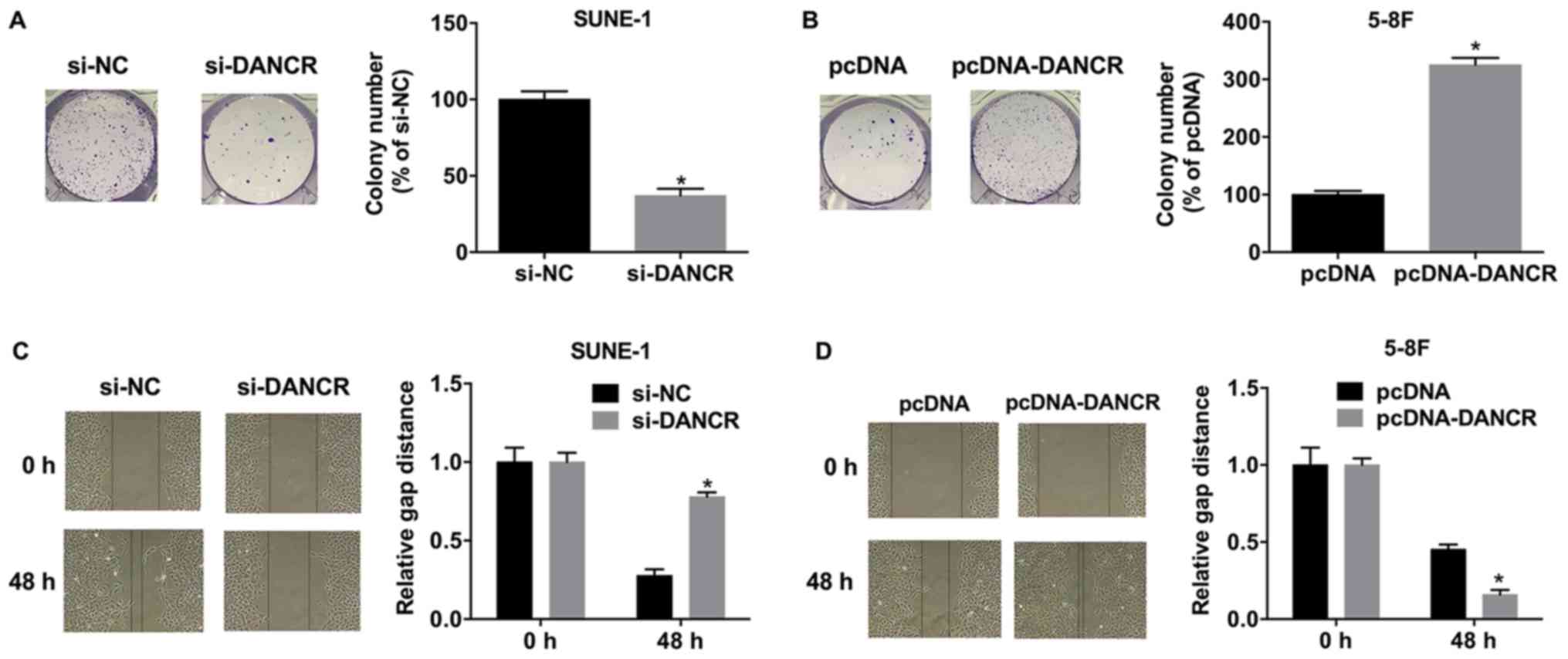
DANCR promotes colony formation and
migration in NPC cells
Silencing DANCR suppressed colony formation in
SUNE-1 cells, whereas, DANCR overexpression significantly increased
colony formation in 5-8F cells (Fig.
3A and B). The migration of SUNE-1 cells transfected with
si-DANCR was significantly decreased compared with si-NC (Fig. 3C), whereas, overexpressing DANCR
significantly increased the migratory ability of 5-8F cells
(Fig. 3D).
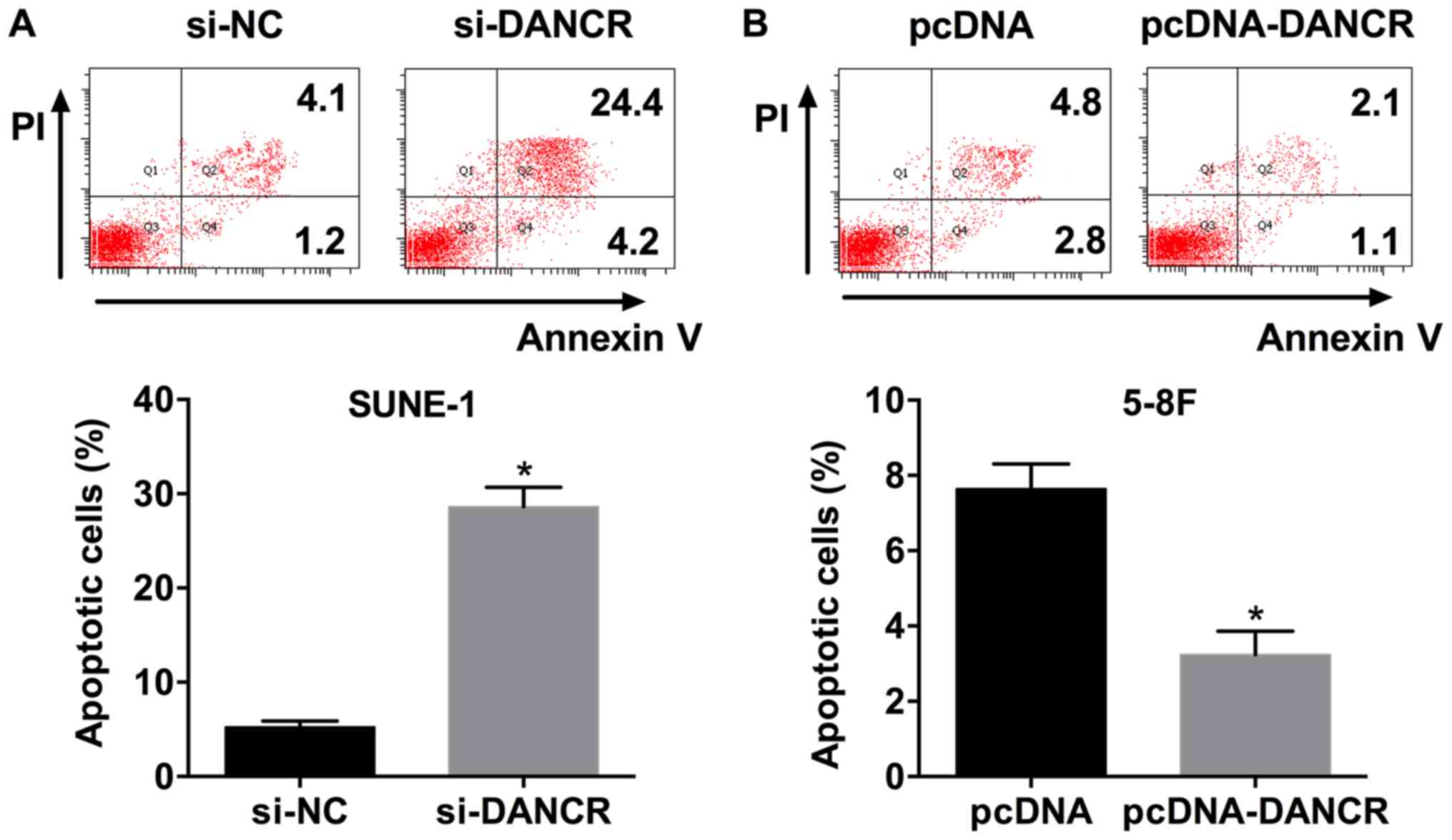
DANCR overexpression suppresses
apoptosis in NPC cells
The apoptosis rate of SUNE-1 cells transfected with
si-DANCR was significantly increased compared with si-NC (Fig. 4A). Additionally, DANCR
overexpression decreased the apoptosis rate of 5-8F cells (Fig. 4B). The present results suggested
that DANCR suppressed apoptosis in NPC cells.
DANCR alters the protein expression
level of AKT, PTEN and apoptosis-associated factors
To investigate the mechanism of DANCR activity in
NPC cell phenotypes, the effect of DANCR on the phosphorylation of
AKT, and protein expression levels of PTEN and the
apoptosis-associated factors BAX and BCL2 was examined. Western
blot analysis suggested that silencing DANCR decreased the
phosphorylation of AKT, but increased total PTEN levels in SUNE-1
cells (Fig. 5A). Overexpression of
DANCR in 5-8F cells resulted in the opposite effects (Fig. 5B). Following DANCR knockdown, BAX
and BCL2 protein expression levels were significantly increased and
decreased, respectively (Fig. 5C).
Conversely, DANCR overexpression produced the opposite results in
5-8F cells (Fig. 5D).
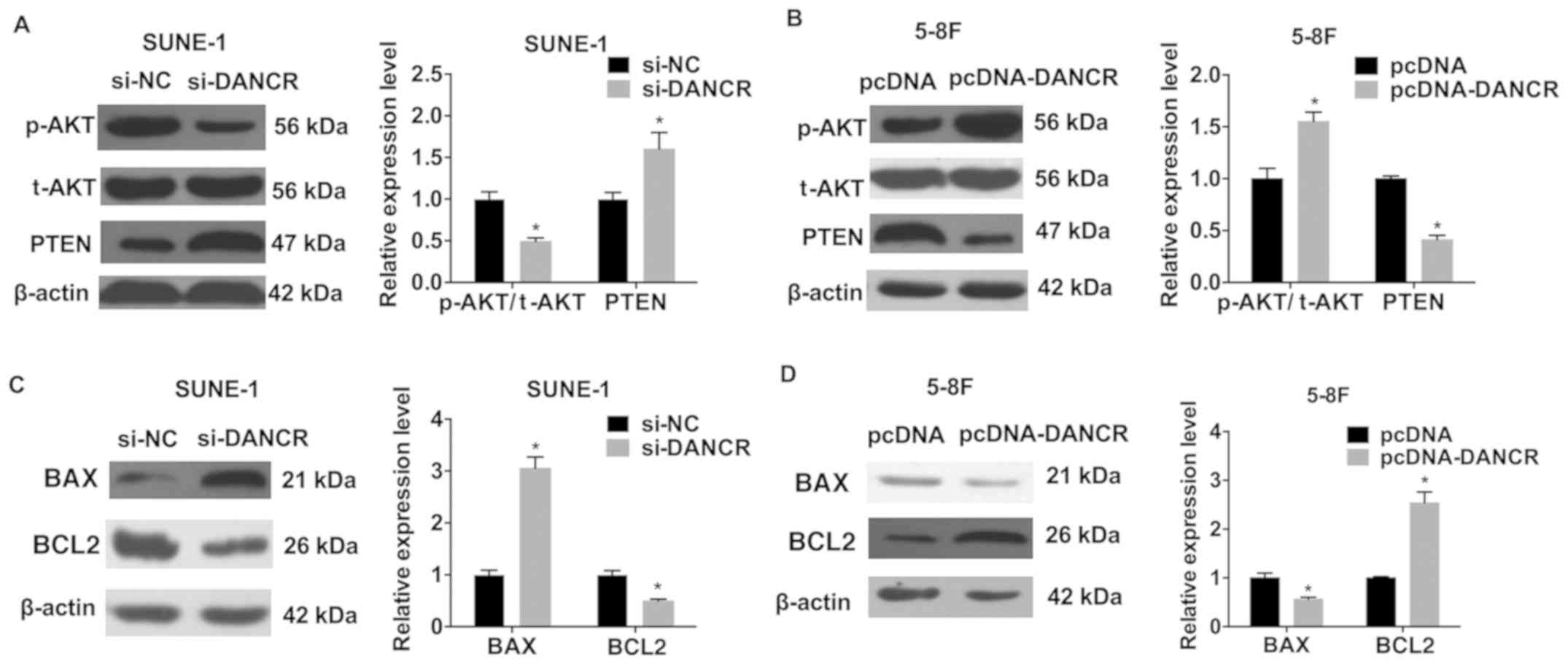 | Figure 5.DANCR regulates the protein expression
levels of AKT, PTEN and apoptosis-associated factors. AKT and PTEN
were detected by western blot analysis in (A) SUNE-1 cells
transfected with si-DANCR and (B) 5-8F cells transfected with
pcDNA-DANCR. BAX and BCL2 protein levels were examined in (C)
SUNE-1 cells transfected with si-DANCR and (D) 5-8F cells
transfected with pcDNA-DANCR. *P<0.05 vs. respective control.
si-, small interfering RNA; NC, negative control; DANCR,
differentiation antagonizing non-protein coding RNA; p-,
phosphorylated; AKT, AKT serine/threonine kinase; t-, total; PTEN,
phosphatase and tensin homolog; BCL2, BCL2, apoptosis regulator;
BAX, BCL2 associated X, apoptosis regulator. |
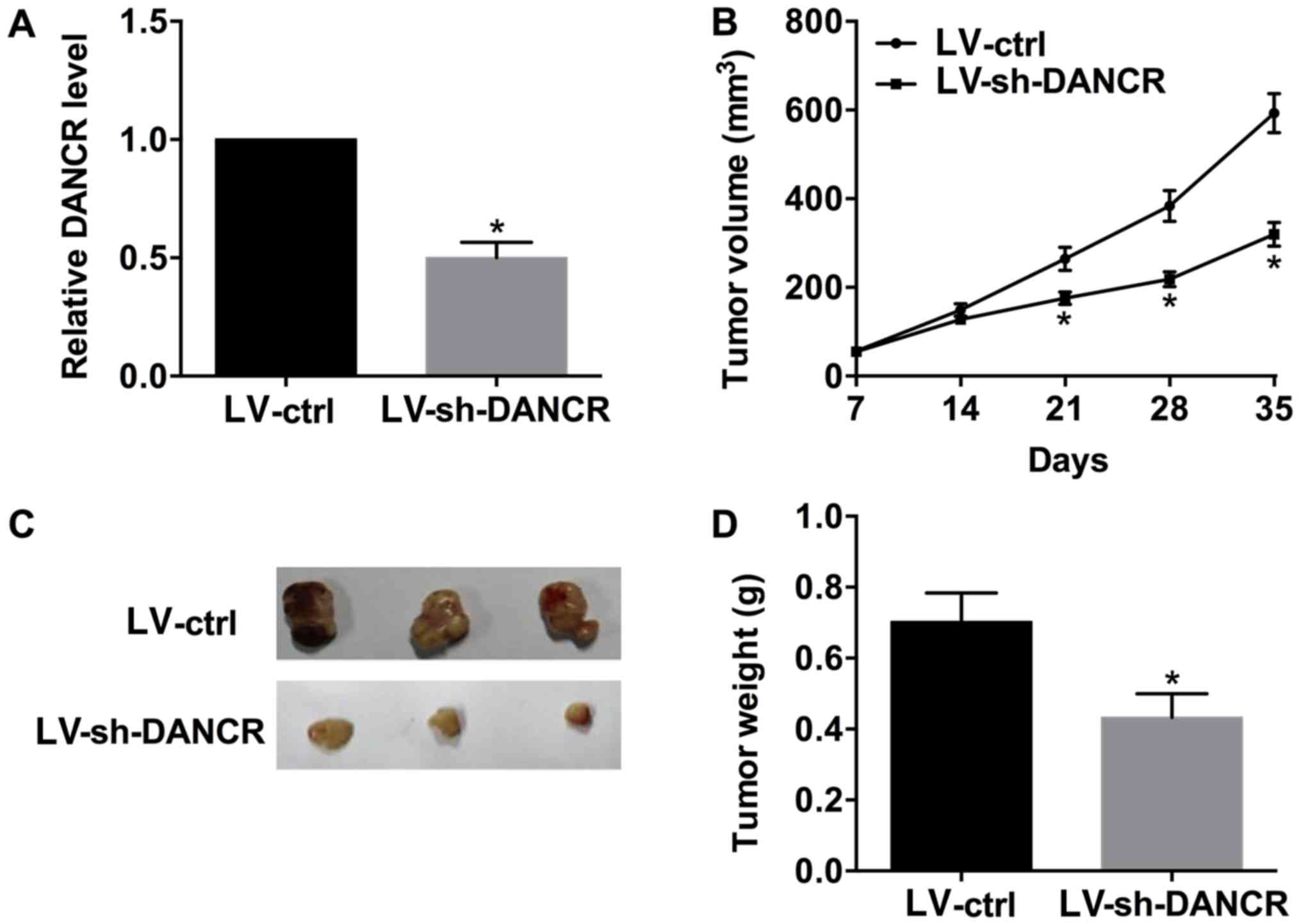
DANCR promotes tumor growth in
vivo
Murine xenograft models were established by
injecting SUNE-1 cells transduced with LV-ctrl or LV-sh-DANCR into
nude mice. Subsequently, the tumor size was measured every 7 days
for 5 weeks. DANCR was significantly downregulated in the tumor
tissues derived from SUNE-1 cells transduced with LV-sh-DANCR
compared with LV-ctrl (Fig. 6A).
The xenografts results suggested that LV-sh-DANCR was able to
inhibit tumor growth in vivo (Fig. 6B-D).
Discussion
lncRNAs serve critical roles in NPC progression and
are associated with cancer prognosis. Ma et al (15) identified that the lncRNA HOX
transcript antisense RNA serves as an oncogene in NPC progression
and recurrence by upregulating fatty acid synthase. NPTN intronic
transcript 1 was identified to be repressed by enhancer of zeste 2
polycomb repressive complex 2 subunit and to serve tumor
suppressive roles in NPC (16).
Additionally, oncogenic functions of HNF1A antisense RNA 1 have
been identified in NPC, and this lncRNA was demonstrated to be
involved in epithelial-mesenchymal transition (17). Although numerous lncRNAs have been
reported to be involved in NPC progression, a complete
understanding of the lncRNAs involved in this type of malignancy is
required to develop effective therapeutic strategies to treat
NPC.
In the present study, it was identified that DANCR
expression was significantly increased in NPC cells.
Gain-of-function experiments suggested that DANCR overexpression
promoted cell proliferation and migration, and inhibited apoptosis
in 5-8F cells. By contrast, silencing DANCR in SUNE-1 cells
suppressed their proliferation and migration, and increased
apoptosis. Furthermore, knockdown of DANCR decreased tumor
xenograft growth in vivo.
DANCR was identified to suppress progenitor
differentiation in epidermal cells (8). Additionally, DANCR may serve as an
oncogene in various types of cancer, including osteosarcoma,
gastric cancer and colorectal cancer (9,18,19).
Notably, DANCR was identified to serve as a tumor suppressor in
others malignancies, including breast cancer and renal cell
carcinoma (11,20). DANCR is involved in the development
of numerous diseases via various mechanisms. DANCR serves as a
competing endogenous RNA for microRNA (miR)-577 in colorectal
cancer and for miR-33a-5p in osteosarcoma (9,19).
In prostate cancer, DANCR promotes cell invasion by epigenetically
suppressing tissue inhibitor of metalloproteinase 2 and 3 (21).
To examine the function of DANCR in NPC, the
mechanism underlying the regulation of AKT phosphorylation and the
protein expression levels of PTEN by DANCR was investigated. The
present results suggested that silencing DANCR decreased the
phosphorylation AKT and increased the protein expression level of
PTEN in SUNE-1 cells; whereas, overexpression of DANCR led to the
opposite effects in 5-8F cells. PTEN is a tumor suppressor in
numerous types of cancer (22). A
previous study suggested that the expression level of PTEN was
decreased in NPC, indicating a tumor suppressive role for this
lncRNA (23). PTEN negatively
regulates the phosphatidylinositol 3-kinase (PI3K)/AKT pathway,
which is associated with proliferation, migration, invasion and
inhibition of apoptosis during cancer development (24). Downregulation of PTEN was
demonstrated to activate the PI3K/AKT pathway in NPC (25). In the present study, the effect of
DANCR on the protein expression levels of the apoptosis-associated
factors BAX and BCL2 was additionally investigated. Following DANCR
knockdown, BAX and BCL2 protein expression levels were increased
and decreased, respectively. By contrast, DANCR overexpression
downregulated BAX expression and upregulated BCL2 expression in
5-8F cells. BAX and BCL2 are proteins associated with apoptosis.
BAX is a pro-apoptotic protein, whereas, BCL2 is an anti-apoptotic
factor (26). DANCR may be
involved in cancer progression via various processes, including the
regulation of mRNA stability and the alteration of epigenetic
marks, and may serve as a competing endogenous RNA for multiple
miRNAs. Although in the present study it was identified that DANCR
regulated the phosphorylation of AKT, and the expression of PTEN
and apoptosis-associated factors, DANCR may regulate the expression
of multiple genes in NPC, and further studies are required to
investigate this possibility.
The present study identified that the lncRNA DANCR
was upregulated in NPC cells. DANCR promoted cell proliferation and
migration, and inhibited apoptosis in NPC cells. Mechanistically,
DANCR regulated the levels of p-AKT, PTEN, BAX and BCL2 in NPC
cells. The present results suggested that DANCR may represent a
promising therapeutic target for the treatment of NPC.
Acknowledgements
Not applicable.
Funding
The present study was supported by The Science
Identification of Shanghai (grant no. 2016034A; China).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YH, HZ and XJ designed the study. YH performed the
experiments and wrote the paper. HZ performed the statistical
analyses. PH, JZ, QD, WS and MZ contributed in performing the cell
and animal experiments. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by The Clinical
Research Ethics Committee of Shanghai Jiaotong University
(Shanghai, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Katano A, Takahashi W, Yamashita H,
Yamamoto K, Ando M, Yoshida M, Saito Y, Abe O and Nakagawa K:
Radiotherapy alone and with concurrent chemotherapy for
nasopharyngeal carcinoma: A retrospective study. Medicine
(Baltimore). 97:e05022018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zhang Y, Wang H, Liu Y, Wang C, Wang J,
Long C, Guo W and Sun X: Baicalein inhibits growth of Epstein-Barr
virus-positive nasopharyngeal carcinoma by repressing the activity
of EBNA1 Q-promoter. Biomed Pharmacother. 102:1003–1014. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lee AW, Ma BB, Ng WT and Chan AT:
Management of nasopharyngeal carcinoma: Current practice and future
perspective. J Clin Oncol. 33:3356–3364. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hu G, Niu F, Humburg BA, Liao K, Bendi S,
Callen S, Fox HS and Buch S: Molecular mechanisms of long noncoding
RNAs and their role in disease pathogenesis. Oncotarget.
9:18648–18663. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huang G, Jiang H, Lin Y, Wu Y, Cai W, Shi
B, Luo Y, Jian Z and Zhou X: lncAKHE enhances cell growth and
migration in hepatocellular carcinoma via activation of NOTCH2
signaling. Cell Death Dis. 9:4872018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu F, Tai Y and Ma J: lncRNA
NEAT1/let-7a-5p axis regulates the cisplatin resistance in
nasopharyngeal carcinoma by targeting Rsf-1 and modulating the
Ras-MAPK pathway. Cancer Biol Ther. 19:534–542. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lan X and Liu X: lncRNA SNHG1 functions as
a ceRNA to antagonize the effect of miR-145a-5p on the
down-regulation of NUAK1 in nasopharyngeal carcinoma cell. J Cell
Mol Med. Mar 25–2018.(Epub ahead of print). doi:
10.1111/jcmm.13497. View Article : Google Scholar
|
|
8
|
Kretz M, Webster DE, Flockhart RJ, Lee CS,
Zehnder A, Lopez-Pajares V, Qu K, Zheng GX, Chow J, Kim GE, et al:
Suppression of progenitor differentiation requires the long
noncoding RNA ANCR. Genes Dev. 26:338–343. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang Y, Lu Z, Wang N, Feng J, Zhang J,
Luan L, Zhao W and Zeng X: Long noncoding RNA DANCR promotes
colorectal cancer proliferation and metastasis via miR-577
sponging. Exp Mol Med. 50:572018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhen Q, Gao LN, Wang RF, Chu WW, Zhang YX,
Zhao XJ, Lv BL and Liu JB: lncRNA DANCR promotes lung cancer by
sequestering miR-216a. Cancer Control. 25:10732748187698492018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sha S, Yuan D, Liu Y, Han B and Zhong N:
Targeting long non-coding RNA DANCR inhibits triple negative breast
cancer progression. Biol Open. 6:1310–1316. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wen X, Tang X, Li Y, Ren X, He Q, Yang X,
Zhang J, Wang Y, Ma J and Liu N: Microarray expression profiling of
long non-coding RNAs involved in nasopharyngeal carcinoma
metastasis. Int J Mol Sci. 17:E19562016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu SY, Huang X and Cheong KL: Recent
advances in marine algae polysaccharides: Isolation, structure, and
activities. Mar Drugs. 15:E3882017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ma DD, Yuan LL and Lin LQ: lncRNA HOTAIR
contributes to the tumorigenesis of nasopharyngeal carcinoma via
up-regulating FASN. Eur Rev Med Pharmacol Sci. 21:5143–5152.
2017.PubMed/NCBI
|
|
16
|
Sun Q, Liu H, Li L, Zhang S, Liu K, Liu Y
and Yang C: Long noncoding RNA-LET, which is repressed by EZH2,
inhibits cell proliferation and induces apoptosis of nasopharyngeal
carcinoma cell. Med Oncol. 32:2262015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhuang K, Wu Q, Jin CS, Yuan HJ and Cheng
JZ: Long non-coding RNA HNF1A-AS is upregulated and promotes cell
proliferation and metastasis in nasopharyngeal carcinoma. Cancer
Biomark. 16:291–300. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Pan L, Liang W, Gu J, Zang X, Huang Z, Shi
H, Chen J, Fu M, Zhang P, Xiao X, et al: Long noncoding RNA DANCR
is activated by SALL4 and promotes the proliferation and invasion
of gastric cancer cells. Oncotarget. 9:1915–1930. 2017.PubMed/NCBI
|
|
19
|
Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu
J, Miao N, Shen J and Peng T: lncRNA DANCR promotes tumor
progression and cancer stemness features in osteosarcoma by
upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 405:46–55.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin L, Fu H, Quan J, Pan X, He T, Hu J, Li
Y, Li H, Yang Y, Ye J, et al: Overexpression of long non-coding RNA
differentiation antagonizing non-protein coding RNA inhibits the
proliferation, migration and invasion and promotes apoptosis of
renal cell carcinoma. Mol Med Rep. 16:4463–4468. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jia J, Li F, Tang XS, Xu S, Gao Y, Shi Q,
Guo W, Wang X, He D and Guo P: Long noncoding RNA DANCR promotes
invasion of prostate cancer through epigenetically silencing
expression of TIMP2/3. Oncotarget. 7:37868–37881. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Salmena L: PTEN: History of a tumor
suppressor. Methods Mol Biol. 1388:3–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gao Q, Tang L, Wu L, Li K, Wang H, Li W,
Wu J, Li M, Wang S and Zhao L: LASP1 promotes nasopharyngeal
carcinoma progression through negatively regulation of the tumor
suppressor PTEN. Cell Death Dis. 9:3932018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carnero A, Blanco-Aparicio C, Renner O,
Link W and Leal JF: The PTEN/PI3K/AKT signalling pathway in cancer,
therapeutic implications. Curr Cancer Drug Targets. 8:187–198.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Song LB, Li J, Liao WT, Feng Y, Yu CP, Hu
LJ, Kong QL, Xu LH, Zhang X, Liu WL, et al: The polycomb group
protein Bmi-1 represses the tumor suppressor PTEN and induces
epithelial-mesenchymal transition in human nasopharyngeal
epithelial cells. J Clin Invest. 119:3626–3636. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Edlich F: BCL-2 proteins and apoptosis:
Recent insights and unknowns. Biochem Biophys Res Commun.
500:26–34. 2018. View Article : Google Scholar : PubMed/NCBI
|