Introduction
The rhesus (Rh) blood group system is crucial for
the safety of transfusion medicine for RhD− recipients
(1). The Rh system is the largest
and most polymorphic blood group system containing 55 antigens, of
which the D antigen is the most clinically relevant (2,3). The
RhD antigen has greater immunogenicity than virtually all other red
blood cell (RBC) antigens (4). A
previous study reported that 30 µl of RhD+ RBCs can
induce potent humoral responses in a RhD− recipient
(5). To ensure transfusion safety,
RhD− individuals should receive RhD− donor
blood. However, there is a clear shortage of RhD− blood,
and RhD− individuals have a frequency of only 0.1–0.4%
in China (6–8). This, together with an increase in the
demand for clinical transfusion, increases the risk for
RhD− individuals, particularly in emergency conditions
(9–11). Transfusion with RhD+
blood to RhD− individuals can be life-saving in
emergencies, but may cause hemolytic anemia induced by endogenous
RhD-specific alloantibodies (12,13).
Therefore, the development of new therapies to prevent RhD-related
hemolytic anemia will be important in the management of blood
transfusion for RhD− individuals.
Cell-based systematic evolution of ligands by
exponential enrichment (cell-SELEX) has been demonstrated to be an
effective strategy in generating universal RBCs with shielded
antigens (14,15). Aptamers are short single-stranded
DNA (ssDNA) or RNA oligonucleotides that can bind to target
molecules with high affinity and specificity (15). Aptamers can be generally screened
from a large nucleic acid library containing random sequences
through the iterative in vitro selection and amplification
process of SELEX (16). Aptamers
are also called ‘chemical antibodies’ and have been successfully
used as a targeted therapy in clinical treatment (17,18).
In theory, specific aptamers targeting RhD antigen epitopes can be
obtained by using RhD+ RBCs as target cells to screen a
nucleic acid library through appropriate cell-SELEX procedures.
However, there is no available aptamer to mask RhD antigen epitopes
for transfusion medicine in the clinic.
In the present study, the hypothesis that ssDNA
aptamers could be screened using RhD+ RBC-SELEX
procedures was tested. It was also assessed whether these
RhD-specific ssDNA aptamers could mask RhD antigen epitopes on RBCs
to prevent the binding of RhD-specific alloantibodies and eliminate
the immunoreactivity of RhD+ RBCs.
Materials and methods
RBCs
O-type RhD+ RBC samples were obtained
from 10 non-parental healthy donors (four females and six males,
aged 22–41 years old) from Shenzhen Blood Center in August 2016.
RBCs were washed three times with saline and then made into 3%
saline suspension of RBCs by adding 50 µl 3% RBC saline suspension
and 50 µl IgM anti-RhD (cat. no. 20163402337; Shanghai
Hemo-Pharmaceutical & Biological) reagent into a clean tube,
and the agglutination was observed after centrifugation at 800 × g
for 15 sec. The RBCs showed strong agglutination in the presence of
anti-RhD IgM. After being washed, the RBCs were adjusted at
4×106 RBCs/ml in citrate-phosphate-dextrose-adenine
solution and used as the target cells for cell-SELEX. Similarly,
AB-type RhD−
(RHD−/RHD−) RBC samples
obtained from 10 non-parental healthy donors (five females and five
males, aged 22–35 years old) were prepared in the same manner as
the control cells. All donors provided written consent for this
study.
RhD antibodies
The IgG monoclonal anti-RhD antibody was purchased
from Shanghai Hemo-Pharmaceutical & Biological (cat. no.
20160725). Human RhD alloantibodies were obtained from the serum of
three female donors (named 1–3, aged 27–34 years old), who had
fetuses and/or newborns with RhD-associated hemolytic disease at
the Shaanxi Institute of Transfusion Medicine in July 2016. The
titers of all antibodies were determined by an antibody titration
procedure, conducted as previously described, to reduce
inter-laboratory variation (19).
The monoclonal anti-RhD antibody at 1:128 dilution had a strong
agglutination activity with a titer of 512 [1,024 (titer of the
highest dilution of serum that gives a reaction), score 109 (sum of
scores for all tubes in the titration study)], and RhD alloantibody
1 at 1:8 dilution displayed a strong agglutination activity with a
titer of 64 (128, score 80). RhD alloantibody 2 at 1:2 dilution had
a titer of 64 (256, score 70), and RhD alloantibody 3 at 1:1
dilution had a titer of 32 (64, score 57).
SELEX ssDNA library and PCR
primers
The ssDNA library and primers for PCR were
synthesized by Invitrogen; Thermo Fisher Scientific, Inc., and
purified by high performance liquid chromatography. Individual
ssDNAs in the library contained a central randomized sequence of 40
nucleotides (nt) flanked by 21-nt primer sequences
(5′-AGAGACGGACACAGGATGAGC-40nt-CCTTCCCCAAGACAGCATCCA-3′). The
selected ssDNA pool was amplified by symmetric PCR using the
sequences of biotinylated primers (forward
5′-biotin-AGAGACGGACACAGGATGAGC-3′, reverse
5′-biotin-TGGATGCTGTCTTGGGGAAGG-3′). The non-biotinylated excessive
forward primer (5′-AGAGACGGACACAGGATGAGC-3′) and partially
biotinylated reverse primer (5′-biotin-TGGATGCTGTCTTGGGGAAGG-3′)
were used in asymmetric PCR at a ratio of 20:1 (20).
Generation and purification of ssDNA
sub-libraries
The ssDNA sub-libraries were generated by an
indirect purification method, as described previously (20,21).
Briefly, the ssDNA library was first selected by the symmetric PCR
in a 50-µl reaction volume containing 10 µl template, 1X PCR buffer
(10 mM Tris-HCl, 50 mM KCl, pH 9.0), 1.5 mM MgCl2, 400
µM each dNTP, 2.5 U Taq DNA polymerase (Promega Corporation), and
50 pmol each biotinylated primer. The PCR was performed at 94°C for
5 min, followed by 11 cycles of 94°C for 30 sec, 61°C for 30 sec
and 72°C for 30 sec, followed by 72°C for 5 min. Subsequently, the
PCR products were selected by asymmetric PCR in a 20-µl reaction
volume containing 1 µl symmetric PCR product as the template, 2 µl
forward primer (10 pmol/µl), 2 µl biotinylated reverse primer (0.5
pmol/µl; primers at ratio of 20:1), 10 µl of AmpliTaq Gold Fast PCR
master mix (Thermo Fisher Scientific, Inc.). The asymmetric PCR
reactions were performed at 96°C for 10 min, then 30 cycles of 96°C
for 3 sec, 59°C for 3 sec and 68°C for 3 sec, followed by 72°C for
10 sec. The asymmetric PCR products were selected by
streptavidin-coated magnetic beads (Z5482, Promega Corporation) to
eliminate biotinylated double-stranded DNA and by-products.
Briefly, streptavidin-coated magnetic beads (0.48 mg) were washed
three times with PBS-Tween 20 (pH 7.4, with 0.02% Tween-20) and
were captured using a magnetic stand after each wash. Asymmetric
PCR products (320 µl) were added to washed beads and incubated for
20 min at 25°C. After magnetic separation, the supernatants were
collected. The unbound ssDNAs were precipitated with sodium acetate
(3 M, pH 5.5) and 70% ethanol. Finally, the ssDNAs were dissolved
in normal saline as ssDNA sub-libraries for the next round of
screening (Table I).
 | Table I.Selection conditions for the
cell-systematic evolution of ligands by exponential enrichment
process. |
Table I.
Selection conditions for the
cell-systematic evolution of ligands by exponential enrichment
process.
| Round | ssDNA (pmol) | RhD−
cells (×106) | Counter selection
time (min) | RhD+
cells (×106) | Positive selection
time (min) |
|---|
| 1 | 2,750 | 2 | 30 | 2 | 30 |
| 2 | 1,000 | 4 | 30 | 4 | 30 |
| 3–5 | 500 | 4 | 30 | 4 | 30 |
| 6–8 | 100 | 2 | 30 | 4 | 30 |
| 9–11 | 50 | 2 | 30 | 2 | 30 |
| 12–13 | 50 | 1 | 30 | 2 | 10 |
| 14 | 50 | 0 | 0 | 2 | 5 |
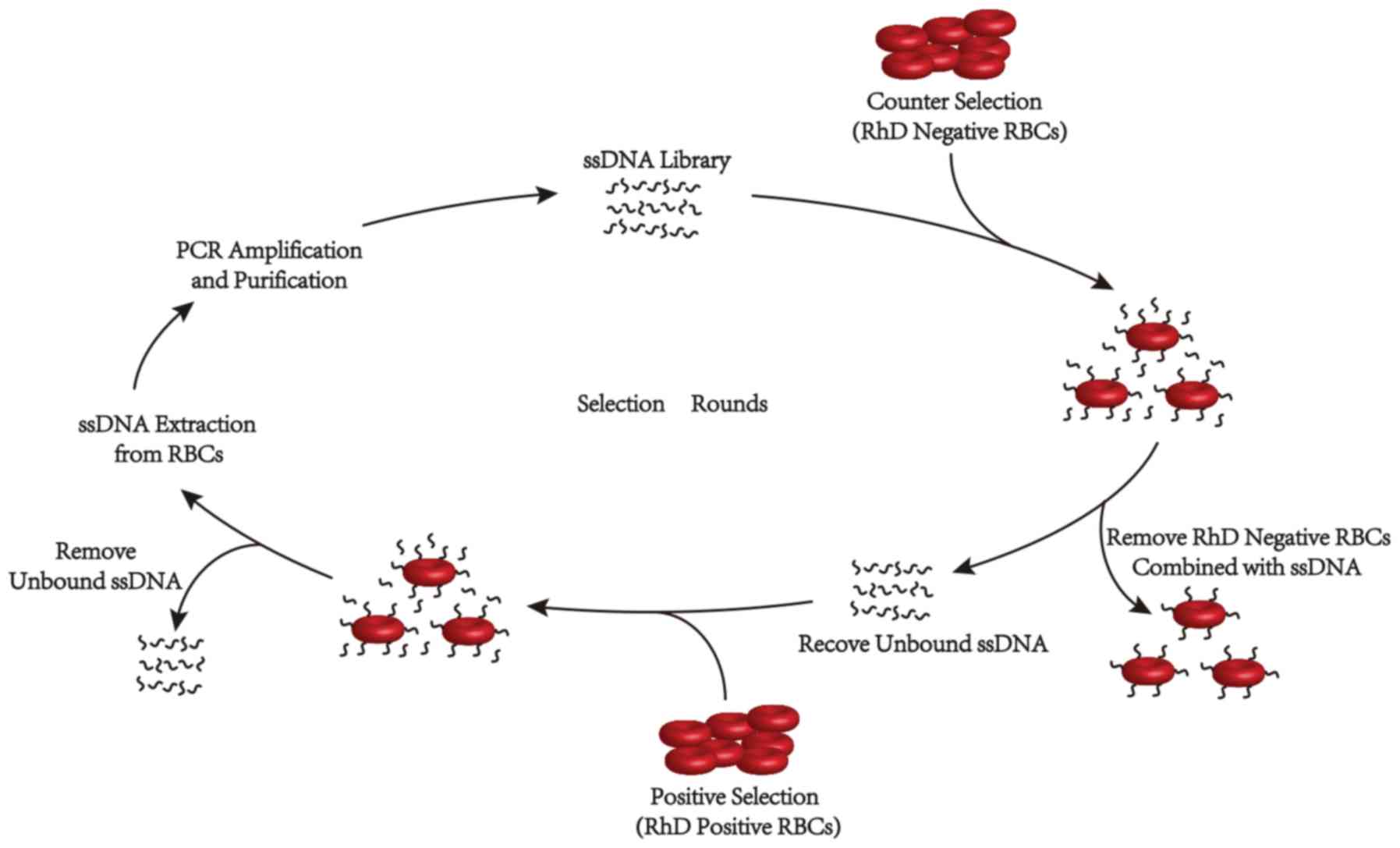
In vitro cell-SELEX
The ssDNA sub-libraries in normal saline were
further screened by the cell-SELEX strategy described previously,
with some modifications (Fig. 1)
(22,23). Briefly, the ssDNA sub-libraries
were heat-denatured at 95°C for 5 min, snap-cooled and reacted with
RhD− RBCs in the presence of 0.1 mg/ml salmon sperm DNA
(Thermo Fisher Scientific, Inc.) for 30 min at 25°C. After
centrifugation at 1,000 × g for 1 min at 25°C, the supernatants
were recovered and incubated with RhD+ RBCs for 30 min
at 25°C. The RBCs were magnetized for 2 min using 200 µl MagneLys
solution (Diagast) following which they were washed with normal
saline five times, and then the ssDNAs bound on the magnetized RBCs
were extracted. These were used as templates for PCR amplification
in order to generate ssDNA sub-libraries using the Dynabeads SILANE
viral NA kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. To increase the stringency of selection,
the concentrations of ssDNAs and positive selection time were
gradually reduced throughout the selection process (Table I).
Enrichment analysis
The enrichment efficiency of ssDNAs was determined
by quantitative PCR (qPCR) using a LightCycler 480II (Roche
Diagnostics). The reaction mixtures consisted of 1 µl magnetic
ssDNA, 10 µl SYBR Green I master mix (Roche Diagnostics GmbH), 1 µl
mixed primers (forward, 5′-AGAGACGGACACAGGATGAGC-3′ and reverse,
5′-TGGATGCTGTCTTGGGGAAGG-3′; each primer 5 pmol/µl) and 8 µl
nuclease-free water. The reactions were performed at 95°C for 5 min
and subjected to 45 cycles of 95°C for 10 sec, 61°C for 10 sec, and
72°C for 10 sec. Nuclease-free water served as a negative control.
Each test was repeated three times to verify the data repeatability
and the mean value was analyzed using LightCycler 480 software
V1.5.1.62 (Roche Diagnostics), and the copy number of ssDNA was
determined by comparing Ct values with those from
the standard curves.
Sequencing of ssDNA aptamers
After 14 rounds of selections, the purified ssDNAs
were sequenced, as described previously (21). In brief, the ssDNA was amplified by
PCR, as described above, in the presence of the adapter sequences
(5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′,
5′-CCTCTCTATGGGCAGTCGGTGAT-3′, 5′-CCATCTCATCCCTGCGTGTCTCCGACTCAG-3′
and 5′-CCTCTCTATGGGCAGTCGGTGAT-3′) and sequenced by the Personal
Genome Machine (PGM) System. Individual sequence reads were
filtered by PGM (Torrent suite software v3.0) to remove low-quality
(<10 copies) and polyclonal sequences and analyzed by Ion
reporter server system software 5.0 (Thermo Fisher Scientific,
Inc.). The remaining reads were clustered, and the most abundant
sequences were chemically synthesized for further characterization.
All sequencing was performed and analyzed by Thermo Fisher
Scientific, Inc.
Immunofluorescence
The masking efficacy and affinity of the aptamer
candidates binding to RhD antigen epitopes were determined by
direct and indirect immunofluorescence. Briefly, RhD+
RBCs (4×106 cells/well) were blocked in triplicate with
ssDNAs (500 pmol, 100 µl saline) in 96-well plates at 25°C for 60
min and incubated with 50 µl standardized monoclonal anti-RhD
antibody (1:128) for 30 min at 25°C. After being washed, the
antibody bound to RBCs was detected with FITC-conjugated goat
anti-mouse IgG F(ab′)2 fragment (1:2,000; cat. no.
AB616780; Bio-Rad Laboratories, Inc.) for 30 min at 25°C.
Subsequently, the fluorescent signals in RBCs were detected
analyzed by Attune Nxt acoustic focusing cytometry and Attune
software v1.1.0 (Thermo Fisher Scientific, Inc.). RhD+
RBCs incubated with FITC-conjugated goat anti-mouse IgG were used
as the blank controls, and normal saline replacing ssDNA aptamer
candidates served as the positive control. Data were expressed as
fluorescence intensity (FI) using the following formula: FI=FI
(test group RBCs)-FI (blank control). Individual ssDNA aptamers,
which led to reduced detection of FI compared with the positive
control, were considered to be bioactive candidates.
The affinity of each ssDNA candidate binding to RhD
antigens was determined by direct immunofluorescence. Individual
ssDNA candidates were labeled with Alexa Fluor® 488,
these were synthesized by Invitrogen; Thermo Fisher Scientific,
Inc. RhD+ RBCs (2% suspension, 100 µl/well) were stained
with different concentrations (200–1,200 nM) of Alexa Fluor
488-labeled ssDNA at 37°C for 60 min. After being washed, the cells
were analyzed by Attune Nxt acoustic focusing cytometry and Attune
software v1.1.0 (Thermo Fisher Scientific, Inc.). The dissociation
constant (Kd) was analyzed by plotting
fluorescence intensity (y-axis) against ssDNA aptamer
concentrations (x-axis) using GraphPad Prism V6 software (GraphPad
Software, Inc.). The equation for the calculation was Y =
Bmax × X / Kd + X
(Bmax is the degree of saturation with maximum
binding in the same units as Y).
Shielding RhD antigens
The ability of ssDNA aptamers to mask RhD antigens
was determined by indirect immunofluorescence and the indirect
agglutination test (IAT). The procedure of indirect
immunofluorescence was performed, similar to that described above,
except for using varying concentrations (0, 200, 300, 400 or 500
pmol) of each ssDNA aptamer.
An IAT was performed, as described previously
(24). Briefly, RhD+
RBCs (4×106 cells/well) were incubated with different
concentrations of ssDNA aptamers in 100 µl saline for 60 min and
incubated with 100 µl standardized human anti-RhD alloantibodies at
37°C for 30 min. After being washed, 100 µl anti-globulin (cat. no.
20153401143; Shanghai Hemo-Pharmaceutical & Biological) was
added to the dry RBCs and centrifuged immediately at 1,000 × g for
15 sec at 25°C. The formed RBC clusters were re-suspended by gentle
shaking and the degree of agglutination was evaluated with a
Axioscope A1 microscope (Zeiss AG), using magnification, ×10). The
reactive system without ssDNA aptamer or RBC served as positive and
negative controls, respectively.
Monocyte monolayer assay (MMA)
A MMA was performed, as previously described
(25). In brief, peripheral blood
mononuclear cells (PBMCs) from 6 healthy donors (four females and
two males, aged 19–35 years old) were isolated by Ficoll-Hypaque
density gradients (500 × g for 25 min at 25°C) and used as the
effectors. After being washed, the PBMCs from the 6 donors were
mixed and adjusted to a concentration of 1×106 cells/ml.
RhD+ RBCs were used as the targets. Simultaneously,
RhD+ RBCs were incubated with 2-fold volumes of saline
or standardized human anti-RhD antibodies at 37°C for 30 min as the
un-sensitized and sensitized RBCs, respectively. Some
RhD+ RBCs were pre-incubated with different
concentrations (200–500 pmol) of ssDNA aptamers for 30 min at 25°C
and sensitized with standardized human anti-RhD antibodies as the
experimental RBCs. After being washed, the RBCs were adjusted to a
concentration of 1×107 cells/ml. Subsequently, the PBMC
suspension (50 µl) was cultured in RPMI 1640 (Corning Inc.) with
10% FBS (Corning Inc.) on 8-well Lab-Tek™ II chamber slides (Thermo
Fisher Scientific, Inc.) for 60 min at 37°C in 5% CO2,
washed, and co-cultured in triplicate with 100 µl of sensitized,
control un-sensitized or experimental RhD+ RBCs for 90
min in 5% CO2 at 37°C. The slides were washed,
air-dried, stained with Wright-Giemsa for 5 min at room
temperature, and images were captured using a light microscope
(magnification, ×40), at least 500 monocytes in each sample were
examined under multiple fields. The number of RBC-bound monocytes
in individual samples was counted, and at least 500 monocytes in
each sample were examined in a blinded manner. Based on reactivity
of un-sensitized RBCs, 3% of monocytes with RBC binding was
considered a positive MMA result (26).
Statistical analysis
Statistical analyses were performed using IBM SPSS
Statistics V22 software (IBM Corp.). Each experiment was repeated
three times and data are expressed as mean ± SD. The difference
among groups was analyzed by ANOVA with 95% confidence and post hoc
Tukey's test. Statistical significance was defined as
P<0.05.
Results
Enrichment and sequencing of ssDNA
aptamer candidates
To examine the enrichment efficacy of in
vitro ssDNA aptamer candidates targeting RhD antigens in each
cell-SELEX round, the ssDNA copies were quantified by qPCR. The
qPCR results showed that the copy numbers of ssDNA aptamer
candidates significantly increased near 1,000 folds from
5.43±0.34×105 copies/µl in the first round to
5.01±0.22×108 copies/µl in the last round (P<0.001;
data not shown). Sequencing analysis indicated 12 dominant ssDNA
aptamer sequences (copies >1,000, random sequence length at 40
bp) after 14 rounds of the ssDNA motif library. The sequences of
the most abundant ssDNA aptamers were selected and synthesized.
ssDNA aptamer candidates have the
ability to shield RhD antigens
The ability of ssDNA aptamer candidates to shield
anti-RhD RhD antigen-specific immune recognition was evaluated by
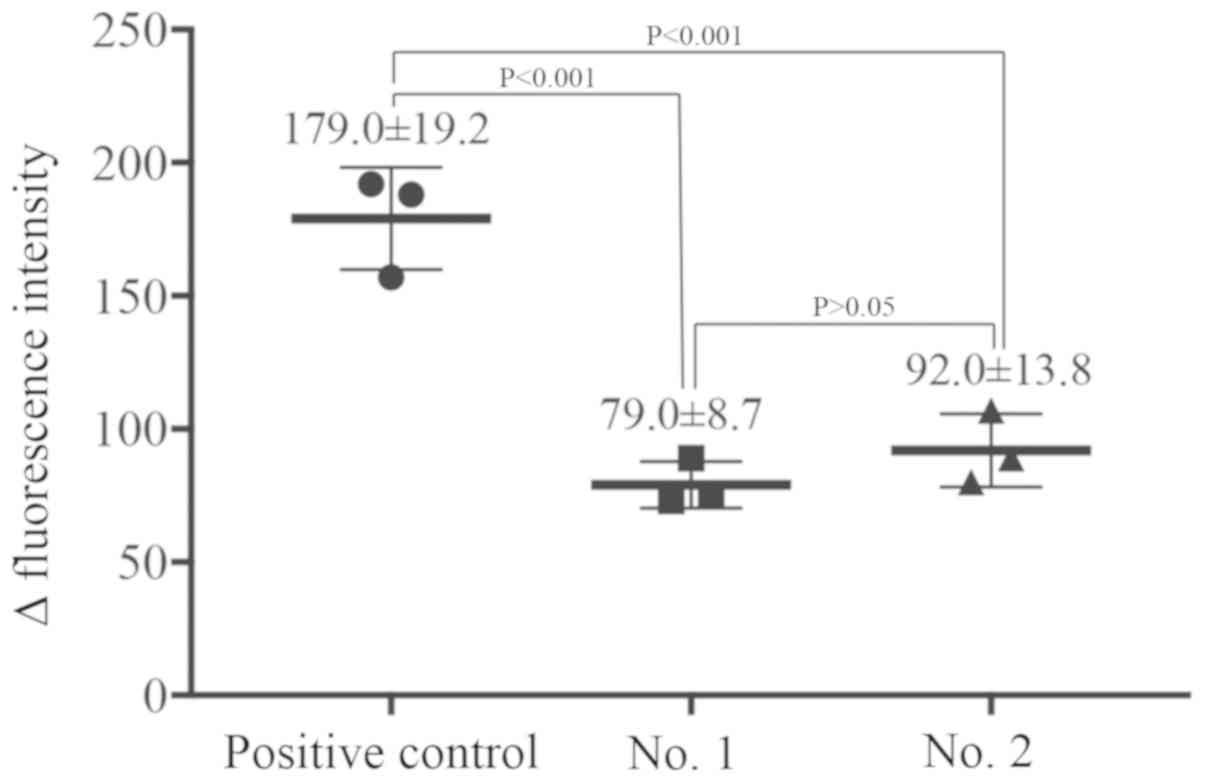
indirect immunofluorescence. The results exhibited that 2 (termed
No. 1 and 2) out of 12 advanced ssDNA aptamer candidates
significantly decreased the fluorescence intensity of monoclonal
anti-RhD binding to RhD antigens (P<0.001; Fig. 2). The central sequences of 40 nt
between the upstream and downstream primers were
5′-GGCCTGGTCTGTTAGCCGGGTAGCAGCCCCGGCACCTATT-3′ and
5′-GGGTAGCAGCCCCGCGGAGGGTCGGCTATAAGAACCAAGA-3′, respectively
(Table II). The aim of further
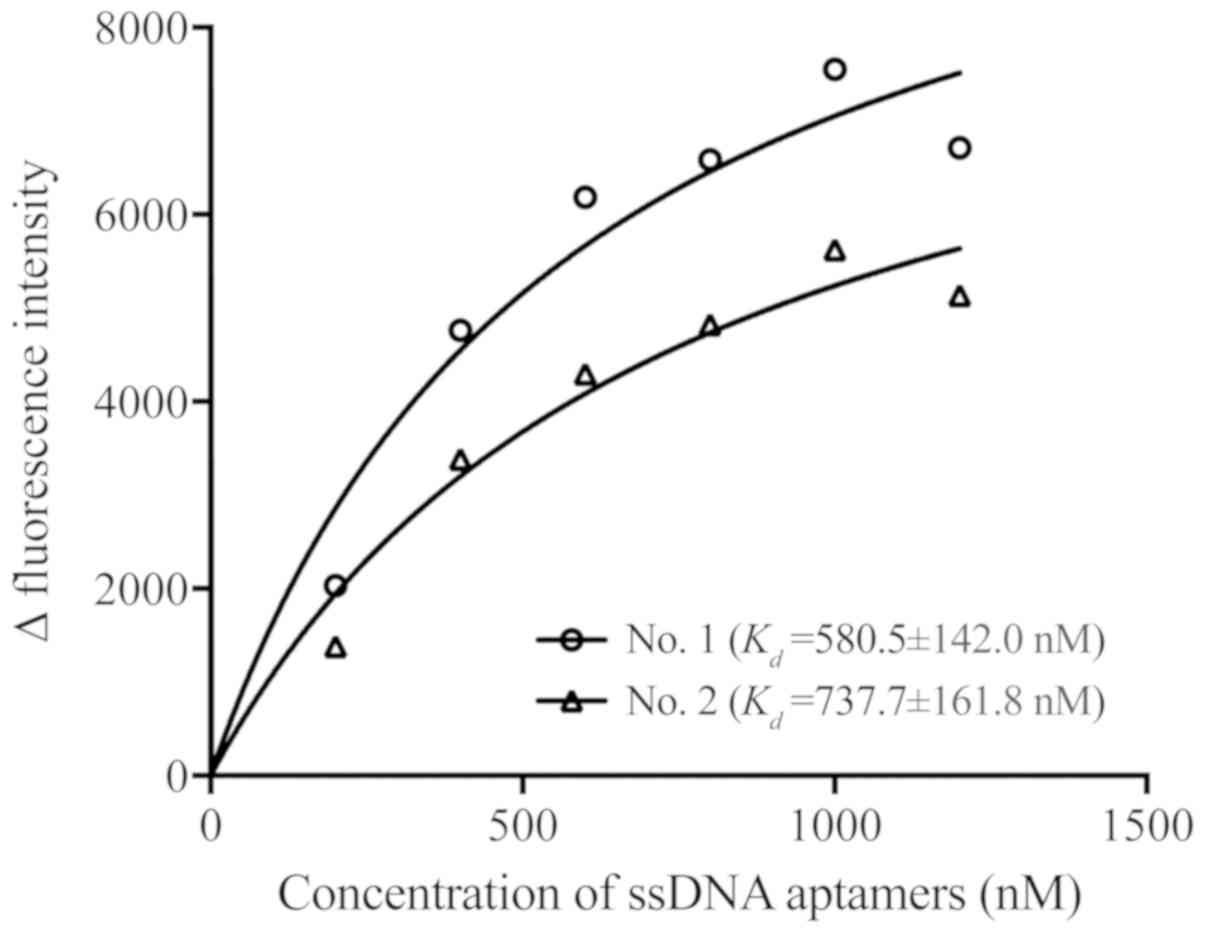
direct immunofluorescence was to evaluate the affinity of the two
selected specific ssDNA aptamers, it was revealed that treatment
with different concentrations of ssDNA aptamers increased the
fluorescence intensity in a dose-dependent manner, and the No. 1
and 2 ssDNA aptamers binding to RhD antigens had
Kd values 580.5±142.0 and 737.7±161.8 nM,
respectively (Fig. 3).
 | Table II.Sequences of variable regions in the
advanced ssDNA aptamer candidates. |
Table II.
Sequences of variable regions in the
advanced ssDNA aptamer candidates.
| No. | Variable region
sequences (5′→3′) | nt |
|---|
| 1a |
GGCCTGGTCTGTTAGCCGGGTAGCAGCCCCGGCACCTATT | 40 |
| 2a |
GGGTAGCAGCCCCGCGGAGGGTCGGCTATAAGAACCAAGA | 40 |
| 3 |
CTATTCCCCACGTCACTTTTCCCGTAGGTTGGACTCGACC | 40 |
| 4 |
GGTGCAGGGGGGTCGGAGAAGAGGTTGAGGGGAGAGGGGT | 40 |
| 5 |
ACGGCCTCTGTATAATGCTGGCCTTGACGCTTGTCCCTTG | 40 |
| 6 |
ACGGCCTCTGTACAATGCTGGCCTTGACGCTTGTCCCTTG | 40 |
| 7 |
TACACCAATCTCCCCCCTACATTCTCCCACCAGCACTCCA | 40 |
| 8 |
GGGTAGCAGCCCCGCGGAGGGTCGGCTATAAGAACTAGGA | 40 |
| 9 |
GGGTAGCAGCCCCGCGGAGGGTCGGCTATAAGAACCAGGA | 40 |
| 10 |
GGGTAGCAGCCCCGCGGAGGGTCAGCTATAAGAACCAGGA | 40 |
| 11 |
GGGTAGCAGCCCCGCGGAGGGTCGGCTATAGGAACCAGGA | 40 |
| 12 |
CTATTCCCCACGTCACTTTTCCCGTAGGCTGGACTCGACC | 40 |
ssDNA aptamers mask RhD antigens for
their immunoreactivity
Due to relatively small molecule of ssDNA,
combination of ≥2 ssDNA aptamers often has superior efficacy in
antigen shielding (21,27). It was observed that treatment with
both No. 1 and 2 ssDNA aptamers (200–500 pmol each) significantly
decreased the fluorescent signals of anti-RhD binding to
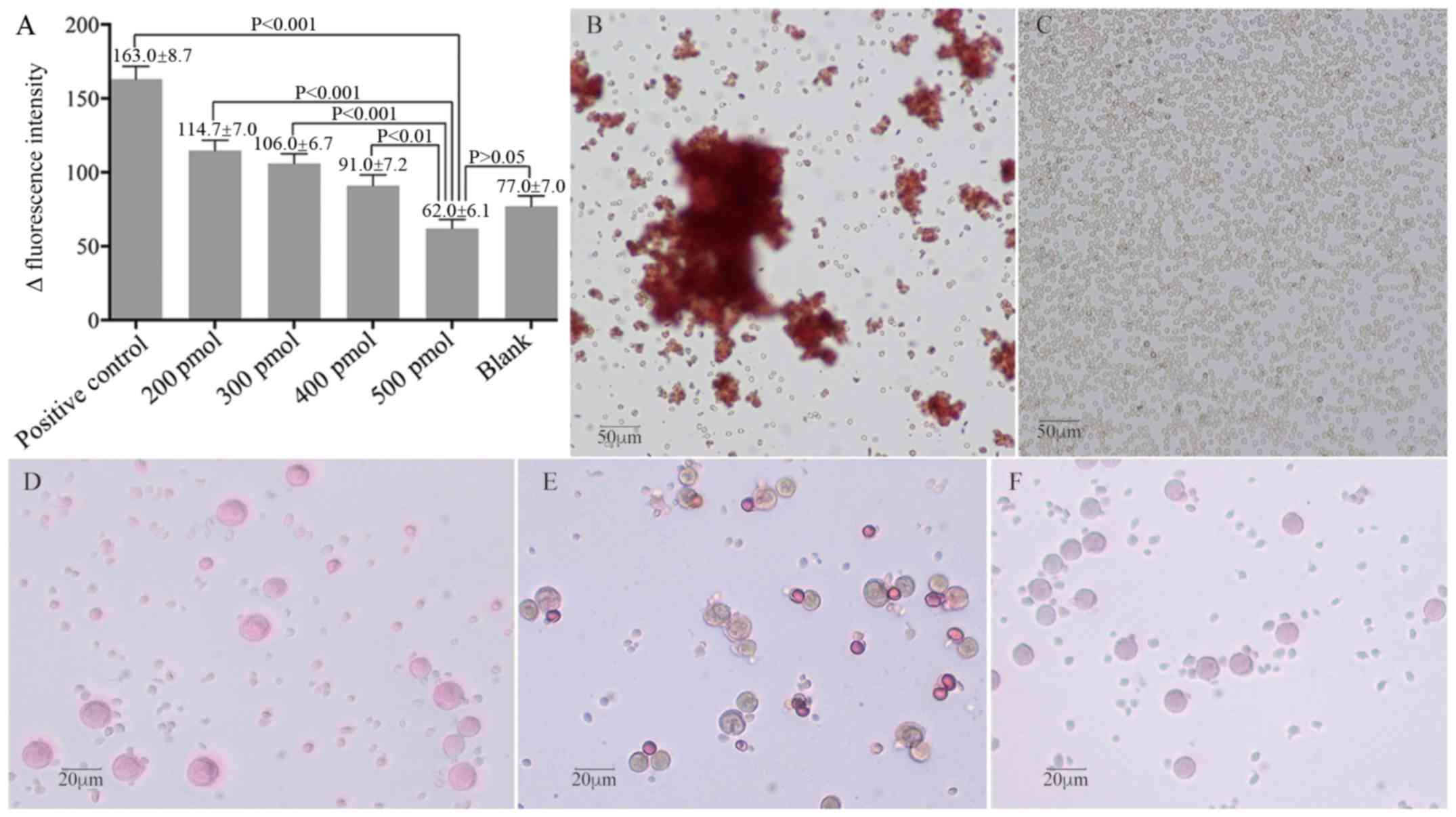
RhD+ RBCs in a dose-dependent manner (Fig. 4A). Treatment with both ssDNA
aptamers at 500 pmol blocked the fluorescence intensity of anti-RhD
(P<0.001 vs. positive control, P>0.05 vs. blank). A similar
pattern of antigen shielding ability of ssDNA aptamers was observed
by IAT. While positive control RBCs (without aptamer treatment)
display varying sizes of agglutinative clusters (Fig. 4B), treatment with 400 pmol ssDNA
aptamers resulted in few RBC clusters (data not shown), and
treatment with 500 pmol completely prevented the formation of
anti-RhD-mediated RBC agglutination in vitro (Fig. 4C). Further MMA analysis revealed
that treatment of monocytes with sensitized RhD+ RBCs
resulted in 19.5% monocyte reactivity, while treatment of monocytes
with un-sensitized (2.2%) or experimental (2.5%) RhD+
RBCs failed to achieve a positive monocyte reactivity (>3.0%;
Fig. 4D-F). Such data indicated
that the ssDNA aptamers blocked the sensitization of anti-RhD
antibodies, leading to a failure of monocytes to bind and
phagocytize RhD+ RBCs. Collectively, these data
demonstrated that the ssDNA aptamers at an optimal dose effectively
masked RhD antigens on RhD+ RBCs to prevent their
immunoreactivity with anti-RhD antibodies.
Discussion
In the present study, two ssDNA aptamers containing
a randomized central sequence of 40 nt flanked by 21-nt primer
sequences that have been synthesized and screened by human
RhD+ RBC-SELEX are reported. These ssDNA aptamers
effectively masked human RhD antigen epitopes on RhD+
RBCs to prevent the binding of an anti-RhD antibody and human
RhD-specific alloantibodies to RhD+ RBCs. Conceivably,
RhD+ RBCs may be modified by these ssDNA aptamers and
survive in RhD− recipients even if they have
RhD-specific alloantibodies. To the best of our knowledge, this was
the first study reporting the generation and biological
characterization of RhD-specific ssDNA aptamers. These novel
bioactive ssDNA aptamers may be valuable in the modification of
RhD+ RBCs for transfusion into RhD−
recipients, thereby reducing the problem of RhD− blood
shortages in emergency situations.
First, two ssDNA aptamers were successfully obtained
using the cell-SELEX procedure. Each aptamer achieved a 1,000-fold
enrichment, supporting the notion that the cell-SELEX procedure is
a powerful tool in screening an ssDNA aptamer library. These two
novel ssDNA aptamers had a high binding affinity to RhD+
RBCs at a nanomolar concentration (Kd 580.5±142.0
and 737.7±161.8 nM for ssDNA aptamers No. 1 and 2, respectively).
The affinity to these concentrations indicated that the aptamers
have a strong binding force, which is consistent with previous
findings (28). However, other
advanced ssDNA aptamers did not effectively block the binding of
the monoclonal anti-RhD antibody to RhD+ RBCs. This may
stem from the lack of D-antigen specificity, low affinity or high
Kd binding of these ssDNA aptamers.
The IAT can analyze in vitro antibody-antigen
interactions, and has been used to detect antibody-mediated RBC
agglutination that may cause hemolytic reactions following
transfusion (29,30). In the current study, it was found
that treatment with ssDNA aptamers resulted in a negative IAT. It
is possible that ssDNA aptamers may bind to RhD antigen epitopes to
prevent the binding of anti-RhD and RhD-specific alloantibodies,
thus leading to a failure of these antibodies to interact with
their targets on RhD+ RBCs. However, a combination of
aptamers No. 1 and 2 at 500 pmol each was less effective in masking
RhD antigen epitopes than each type of aptamer on the binding of
anti-RhD monoclonal antibody to prevent RBC agglutination. This may
be due to different specificity characteristics of polyclonal
alloantibodies and monoclonal anti-RhD antibody. It may be required
for more aptamers to mask D antigen epitopes recognized by
polyclonal alloanti-RhD. Furthermore, it was observed that
treatment with ssDNA aptamers (No. 1 and 2 together at 500 pmol
each) resulted in a trend towards the immunofluorescence intensity
being lower than that of RhD background level. This suggests that
the non-specific binding of fluorescent antibodies to RBCs may be
reduced when ssDNA is bound to RBCs, although this finding was not
significant.
The MMA measures Fcγ receptor-mediated phagocytosis
of alloantibody-sensitized RBCs and has been successfully used to
predict blood transfusion outcomes and avoid immune destruction of
antibody-sensitized RBCs (31–33).
In the present study, it was found that while co-culture of
monocytes with sensitized RhD+ RBCs resulted in a rate
of 19.5% monocyte reactivity, co-culture of monocytes with
RhD+ RBCs that had been pre-treated with ssDNA aptamers
greatly reduced the monocyte reactivity rate to 2.5%, which was
lower than the negative control. This indicated that treatment with
both No. 1 and 2 ssDNA aptamers at 500 pmol each completely masked
the RhD antigen epitopes recognized by RhD-specific alloantibodies
to prevent the RhD-specific alloantibody-mediated phagocytosis by
monocytes. It would be interesting to further investigate the
safety and therapeutic effect of these ssDNA aptamers, and their
stability and immunogenicity in vivo. If successful, the
RhD-specific ssDNA aptamers may be applicable in vivo to
prevent, or at least ameliorate, hemolytic reactions in
RhD− patients, who have fatal blood loss and require
RhD+ blood due to the shortage of RhD−
blood.
In the present study, RhD-specific ssDNA aptamers
were successfully screened using the RhD+ RBC-SELEX
procedure, and these aptamers effectively masked the RhD antigen
epitopes on RhD+ RBCs to prevent the binding of anti-RhD
and human RhD-specific alloantibodies to RhD+ RBCs.
Potentially, the RhD-specific ssDNA aptamers may be valuable in the
modification of RhD+ RBCs for transfusion medicine to
prevent or ameliorate hemolytic reactions in RhD−
patients, who have fatal blood loss and require RhD+
blood due to the unavailability of RhD− blood. The
approach reported in the present study may provide a potential way
of overcoming the problems presented by RhD− RBCs
shortages in an emergency.
Acknowledgements
Not applicable.
Funding
This work was supported by a grant from the Shenzhen
Science and Technology Bureau (grant no.
JCYJ20140403092619633).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon a reasonable
request.
Authors' contributions
YZ conceptualized the study. HX designed the study.
XW, RL and LL performed the experiments and analyzed the data. LW
and HZ analyzed the data. YZ and HZ wrote the manuscript. HX, XW,
LW, RL and LL revised the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
This study was approval by the Ethics Committee of
Shenzhen University General Hospital (Shenzhen, China). All
participates signed written informed consent for the publication of
their data.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Rh
|
rhesus
|
|
ssDNA
|
single-stranded DNA
|
|
PGM
|
personal genome machine
|
|
RBC
|
red blood cell
|
|
qPCR
|
quantitative PCR
|
|
FI
|
fluorescence intensity
|
|
IAT
|
indirect agglutination test
|
|
MMA
|
monocyte monolayer assay
|
|
PBMCs
|
peripheral blood mononuclear cells
|
References
|
1
|
Flegel WA: Molecular genetics and clinical
applications for RH. Transfus Apher Sci. 44:81–91. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kumar M, Chapman A, Javed S, Alam U, Malik
RA and Azmi S: The investigation and treatment of diabetic
gastroparesis. Clin Ther. 40:850–861. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hughes-Jones NC: Quantitation and the Rh
blood group system. Transfus Med. 1:69–76. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gonsalkorale K, Vanhecke C and Badami KG:
Red blood cell phenotyping after transfusion: An in vitro model.
Immunohematology. 29:93–96. 2013.PubMed/NCBI
|
|
5
|
Urbaniak SJ and Robertson AE: A successful
program of immunizing Rh-negative male volunteers for anti-D
production using frozen/thawed blood. Transfusion. 21:64–69. 1981.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lan JC, Chen Q, Wu DL, Ding H, Pong DB and
Zhao T: Genetic polymorphism of RhD-negative associated haplotypes
in the Chinese. J Hum Genet. 45:224–227. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shao CP: Transfusion of RhD-positive blood
in ‘Asia type’ DEL recipients. N Engl J Med. 362:472–473. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang D, Toyofuku WM and Scott MD: The
potential utility of methoxypoly(ethylene glycol)-mediated
prevention of rhesus blood group antigen RhD recognition in
transfusion medicine. Biomaterials. 33:3002–3012. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sun T, Lu SF and Jin GZ: Solving shortage
in a priceless market: Insights from blood donation. J Health Econ.
48:149–165. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Stanger SH, Yates N, Wilding R and Cotton
S: Blood inventory management: Hospital best practice. Transfus Med
Rev. 26:153–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tongmao Z: Attention to emergency blood
transfusion in RhD negative patients. Chin J Blood Transfusion.
23:654–655. 2010.
|
|
12
|
Wang QP, Dong GT, Wang XD, Gu J, Li Z, Sun
AY, Shao CP, Pan ZL, Huang LH, Xie WX, et al: An investigation of
secondary anti-D immunisation among phenotypically RhD-negative
individuals in the Chinese population. Blood Transfus. 12:238–243.
2014.PubMed/NCBI
|
|
13
|
Selleng K, Jenichen G, Denker K, Selleng
S, Mullejans B and Greinacher A: Emergency transfusion of patients
with unknown blood type with blood group O Rhesus D positive red
blood cell concentrates: A prospective, single-centre,
observational study. Lancet Haematol. 4:e218–e224. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Catuogno S and Esposito CL: Aptamer
cell-based selection: Overview and advances. Biomedicines.
5:E492017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Quang NN, Miodek A, Cibiel A and Ducongé
F: Selection of aptamers against whole living cells: From
cell-SELEX to identification of biomarkers. Methods Mol Biol.
1575:253–272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stoltenburg R, Reinemann C and Strehlitz
B: SELEX-a (r)evolutionary method to generate high-affinity nucleic
acid ligands. Biomol Eng. 24:381–403. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ng EW, Shima DT, Calias P, Cunningham ET,
Jr Guyer DR and Adamis AP: Pegaptanib, a targeted anti-VEGF aptamer
for ocular vascular disease. Nat Rev Drug Discov. 5:123–132. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Morita Y, Leslie M, Kameyama H, Volk DE
and Tanaka T: Aptamer therapeutics in cancer: Current and future.
Cancers (Basel). 10:E802018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
American Association of Blood Banks.
Technical manual. Fung MK, Grossman BJ, Hillyer CD and Westhoff CM:
18th. AABB; Bethesda, MD: 2014
|
|
20
|
Zhang Y, Xu H, Zhou H, Wu F, Su Y, Liang Y
and Zhou D: Indirect purification method provides high yield and
quality ssDNA sublibrary for potential aptamer selection. Anal
Biochem. 476:84–90. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Wu F, Wang M, Zhuang N, Zhou H
and Xu H: Single-stranded DNA aptamer targeting and neutralization
of anti-D alloantibody: A potential therapeutic strategy for
haemolytic diseases caused by Rhesus alloantibody. Blood Transfus.
16:184–192. 2018.PubMed/NCBI
|
|
22
|
Sefah K, Shangguan D, Xiong X, O'Donoghue
MB and Tan W: Development of DNA aptamers using cell-SELEX. Nat
Protoc. 5:1169–1185. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tawiah KD, Porciani D and Burke DH: Toward
the selection of cell targeting aptamers with extended biological
functionalities to facilitate endosomal escape of cargoes.
Biomedicines. 5:E512017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Coombs RR, Mourant AE and Race RR: A new
test for the detection of weak and incomplete Rh agglutinins. Br J
Exp Pathol. 26:255–266. 1945.PubMed/NCBI
|
|
25
|
Garner SF, Gorick BD, Lai WY, Brown D,
Taverner J, Hughes-Jones NC, Contreras M and Lubenko A: Prediction
of the severity of haemolytic disease of the newborn. Quantitative
IgG anti-D subclass determinations explain the correlation with
functional assay results. Vox Sang. 68:169–176. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Arndt PA and Garratty G: A retrospective
analysis of the value of monocyte monolayer assay results for
predicting the clinical significance of blood group alloantibodies.
Transfusion. 44:1273–1281. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Osborne SE, Matsumura I and Ellington AD:
Aptamers as therapeutic and diagnostic reagents: Problems and
prospects. Curr Opin Chem Biol. 1:5–9. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Eissa S and Zourob M: Aptamer-based
label-free electrochemical biosensor array for the detection of
total and glycated hemoglobin in human whole blood. Sci Rep.
7:10162017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lin CK, Wong KF, Mak KH, Yuen CM and Lee
AW: Hemolytic transfusion reaction due to Rh antibodies detectable
only by manual polybrene and polyethylene glycol technique. Am J
Clin Pathol. 104:660–662. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hanci H, Igan H and Uyanik MH: Evaluation
of a new and rapid serologic test for detecting brucellosis:
Brucella coombs gel test. Pak J Biol Sci. 20:108–112. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Noumsi GT, Billingsley KL and Moulds JM:
Successful transfusion of antigen positive blood to alloimmunised
patients using a monocyte monolayer assay. Transfus Med. 25:92–100.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tong TN, Cserti-Gazdewich CM and Branch
DR: Value of MMA crossmatch? Transfus Med. 26:301–302. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tong TN and Branch DR: Use of a monocyte
monolayer assay to evaluate Fcγ receptor-mediated phagocytosis. J
Vis Exp. 11:9–15. 2017.
|