Introduction
Tuberculosis (TB) remains a major global public
health problem. According to the 2019 Global Tuberculosis Report,
TB is one of the top 10 causes of death worldwide, followed by AIDS
(1). In 2018, there were an
estimated 1.2 million (range, 1.1–1.3 million) deaths from TB among
HIV-negative people and an additional 251,000 (range,
223,000-281,000) deaths from TB among HIV-positive people.
Additionally, an estimated 10.0 million (range, 9.0–11.1 million)
people fell ill with TB, equivalent to 132 cases (range, 118–146)
per 100,000 population (1). In
order to improve TB management and reduce the transmission of
Mycobacterium tuberculosis (Mtb), early diagnosis is
essential in patients with active pulmonary TB (APTB) (2). However, traditional methods based on
directly detecting Mtb do not meet clinical requirements due
to their low sensitivity and specificity.
Circular RNAs (circRNAs), a novel type of non-coding
RNA lacking 5′ caps and 3′-poly(A) tails, have been recently
reported to participate in the pathological processes of various
human diseases (3–5). circRNAs can be divided into three
categories according to their origin in genomic regions, namely
exon circRNAs, intron circRNAs and exon-intron circRNAs (6–8).
circRNAs are more stable than linear RNA and resist exonuclease
RNase R digestion due to their closed loop structure (9). Their expression is often
tissue-specific and developmental-stage specific (7,10).
These properties suggest that circRNAs are an ideal biomarker for
disease diagnosis.
Recent studies have reported that circRNAs may serve
as potential biomarkers for a number of human diseases. For
example, Bao et al (11)
indicated that hsa_circ_0037911 in whole blood samples could be a
stable biomarker for early diagnosis of essential hypertension.
Hang et al (12) suggested
that a novel plasma circRNA, circFARSA, may be a potential
biomarker for non-small cell lung cancer. Zhao et al
(13) found that hsa_circ_0001275
in peripheral blood mononuclear cells (PBMCs) could serve as a
potential novel diagnostic biomarker for postmenopausal
osteoporosis. Nevertheless, whether circRNAs could act as novel
biomarkers for APTB diagnosis and therapeutic prediction remains
unknown.
In our previous study, circRNA-sequencing (seq) in
PBMCs revealed differential expression of circRNAs in patients with
APTB (14). In the present study,
only hsa_circ_0001380 was evaluated. The results revealed that
hsa_circ_0001380 was significantly downregulated in the PBMCs of
patients with APTB compared with healthy volunteers (HVs).
hsa_circ_0001380 is located at chr3 196,118,683-196,129,890 with a
spliced length of 247 nt, and is derived from exons 3–5 of the
UBXN7 gene. In addition, the potential functions of
hsa_circ_0001380 were explored in silico. A receiver
operating characteristic (ROC) curve was also constructed to
evaluate the diagnostic value of hsa_circ_0001380 in APTB. These
results indicated that hsa_circ_0001380 may be a potential
diagnostic biomarker for APTB.
Materials and methods
Patients and clinical samples
In the present study, a total of 32 patients with
APTB (22 male, 10 female), aged 17–60 years, were recruited from
Guangdong Medical University and Dongguan Sixth Hospital from
February 2018 to August 2018, as previously described (14). APTB was detected in all subjects by
Ziehl-Neelsen acid fast staining of the sputum smears and
Lowenstein-Jensen slant culture according to the ‘Health Standards
for the People's Republic of China-Diagnosis of Tuberculosis’ (WS
288-2017) (15). The diagnosis was
based on sputum smear examination and clinical symptoms. A total of
31 HVs (20 male, 11 female) aged 20–65 years, with no
bacteriological or clinical evidence of APTB, were recruited as
controls from February 2018 to August 2018. A volume of ~5 ml
peripheral blood was collected at the different time points
(patients with TB within 24 h of admission to the hospital and HV
at 6:00-8:00 am.) from each subject. Ethical approval was obtained
from the Ethical Committee of Dongguan Sixth Hospital, Guangdong
Medical University. All subjects provided written informed consent
prior to the study.
Isolation of PBMCs
PBMCs were isolated as previously reported (16). Briefly, PBMCs were freshly isolated
from peripheral blood by density gradient centrifugation on
Ficoll-Paque (TBD Science) at 450 × g for 20 min at 20°C, according
to the protocol of the manufacturer. Cell viability was tested
using the Trypan blue dye exclusion method (>95% in all
experiments). Next, the PBMCs were lysed with TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and stored at
−80°C for further experiments.
RNA sample preparation and quality
control
Total RNA was extracted from the PBMCs using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the instructions of the manufacturer. The
quantity of total RNA was determined at 260/280 nm using NanoDrop
ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.). The RNA
integrity was evaluated by 2% agarose gel electrophoresis, and the
bands were visualized using nucleic acid dye ethidium bromide (data
not shown).
RNase R digestion
To assess the stability of circRNAs, exonuclease
RNase R (Epicentre; Illumina, Inc.) was used to treat the total
RNA, according to the protocol of the manufacturer. Briefly, 2 µg
total RNA was digested with or without 2 U RNase R for 10 min at
37°C in 1X RNase R reaction buffer (Epicentre; Illumina, Inc.), and
reverse transcription-quantitative (RT-q)PCR was subsequently
performed.
RT-qPCR
A total of 500 ng purified total RNA was used for
cDNA synthesis. Total RNA was reverse transcribed into cDNA at 37°C
for 15 min and 85°C for 5 sec using a Primescript RT reagent kit
with random primers according to the protocol of the manufacturer
(Takara Bio, Inc.). Next, qPCR was performed using the Applied
Biosystems QuantStudio 6 Flex System (Thermo Fisher Scientific,
Inc.) with TransStart Tip Green qPCR SuperMix (Beijing Transgen
Biotech Co., Ltd.) at 95°C for 2 min, followed by 40 cycles of 95°C
for 5 sec, 60°C for 30 sec, 72°C for 30 sec. Also, the melting
curve of RT-qPCR was used to evaluate the specificity of the
amplification product using the software of QuantStudio 6 Flex
Real-time PCR System. In order to detect its specific expression,
divergent primers crossing the back-splice junction were designed.
The primer sequences used in the present study are listed in
Table I. The primers were
synthesized by Sangon Biotech Co., Ltd. GAPDH was used as an
internal control. All RT-qPCR reactions were performed in
triplicate. The data were analyzed using the 2−ΔΔCq
method (17) to calculate the
relative expression. The PCR product was visualized by used 2%
agarose gel with ethidium bromide nucleic acid dye added ß.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR analysis. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR analysis.
| Name | Primer sequence
(5′→3′) | Product length
(bp) |
|---|
|
hsa_circ_0001380 | F:
TTCCGGCCACCCATTGATTT | 132 |
|
| R:
GGCCGTCGTCTTTTAGGAGC |
|
| GAPDH | F:
GAAGGTCGGAGTCAACGGATT | 224 |
|
| R:
CCTGGAAGATGGTGATGGGATT |
|
Function prediction of
hsa_circ_0001380
The CircBank database (http://www.circbank.cn/) was used to search circRNA
basic information, retrieve circRNA sequence, and predict circRNA
protein coding potential, circRNA conservation and circRNA
modification. The interactions between circRNA and miRNAs were
predicted using the CircBank and Circinteractome databases
(https://circinteractome.nia.nih.gov/).
Statistical analysis
Receiver operating characteristic (ROC) curve
analysis was conducted to assess the clinical diagnostic ability of
hsa_circ_0001380. Each value is represented as the mean ± SEM. Data
analysis was performed using the Student's t-test or one-way ANOVA
with Tukey's test for continuous variables and χ2
test for discontinuous variables, and P<0.05 was considered to
indicate a statistically significant difference. The statistical
analyses were performed using GraphPad Prism (version 5.0; GraphPad
Software, Inc.) and SPSS software (version 17.0; SPSS, Inc.).
Results
Clinical characteristics of all study
subjects
In the present study, prospective clinical data were
collected from 32 patients with APTB and 31 HV. The demographic and
clinical characteristics of all study subjects are displayed in
Table II. No statistically
significant age- and sex-related differences were observed between
patients with APTB and HV (P>0.05). Among patients with APTB,
Ziehl-Neelsen acid fast staining analysis of the sputum smears
revealed that 78.12% of samples were Mtb positive.
 | Table II.Clinical data for all study
subjects. |
Table II.
Clinical data for all study
subjects.
| Characteristic | APTB group
(n=32) | HV group
(n=31) |
|---|
| Age, years; mean ±
SD (range) | 36.03±13.96 | 33.26±14.56 |
|
| (17–60) | (20–65) |
| Sex, n |
|
|
|
Male | 22 | 20 |
|
Female | 10 | 11 |
| Sputum smear,
n |
|
|
|
Positive | 25 | 0 |
|
Negative | 7 | 31 |
Validation of hsa_circ_0001380
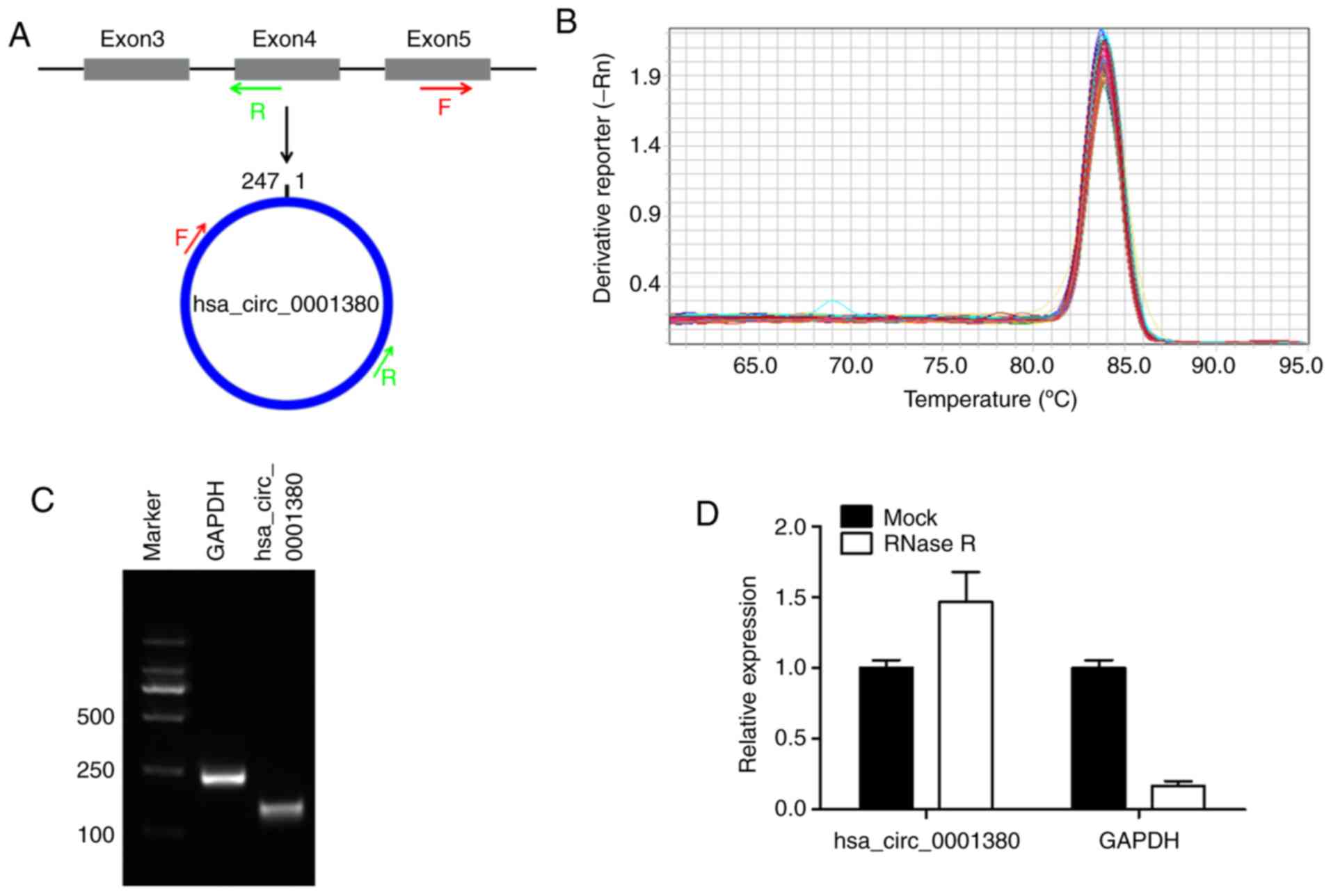
hsa_circ_0001380 was derived from exon 3–5 of the
UBXN7 gene locus by back-splicing. In order to detect its specific
expression, divergent primers crossing the back-splice junction
were designed (Fig. 1A). The
melting curve analysis of RT-qPCR revealed a single peak,
demonstrating the specificity of the amplification product
(Fig. 1B). The size of the
amplification product was evaluated by agarose gel electrophoresis
and was consistent with its full size (Fig. 1C). Subsequently, RNase R was used
to detect the stability of hsa_circ_0001380, and the results
indicated that hsa_circ_0001380 was stable in tolerance to RNase
enzyme treatment. However, the control linear mRNA GAPDH could not
tolerate RNAase treatment (Fig.
1D). These results demonstrated that hsa_circ_0001380 in human
PBMCs could be specifically amplified by RT-qPCR. Additionally,
before RT-qPCR analysis, the RNA integrity was evaluated and
visualized using 2% agarose gel added ethidium bromide dye (data
not shown).
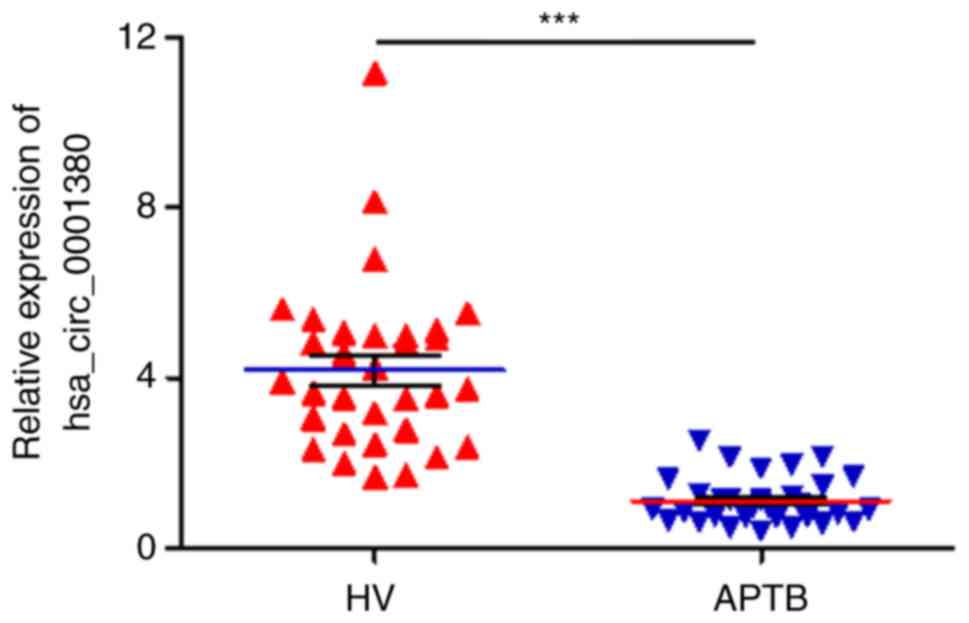
hsa_circ_0001380 was downregulated in
APTB
In our previous circRNA sequencing study, a large
number of differential circRNAs were identified in the PBMCs of
patients with APTB (14). Of note,
hsa_circ_0001380 was revealed as one of these differential
circRNAs, and the fold-change between healthy participants and APTB
patients was found to be ~1.6. In order to validate the expression
level of hsa_circ_0001380 in APTB, PBMCs from 32 patients with APTB
and 31 HV were collected and analyzed by RT-qPCR, using GAPDH as an
internal control. The results demonstrated that hsa_circ_0001380
expression was significantly downregulated in the PBMCs of patients
with APTB as compared with the HV group (P<0.001; Fig. 2). This finding suggested that
hsa_circ_0001380 may be related to tuberculosis infection.
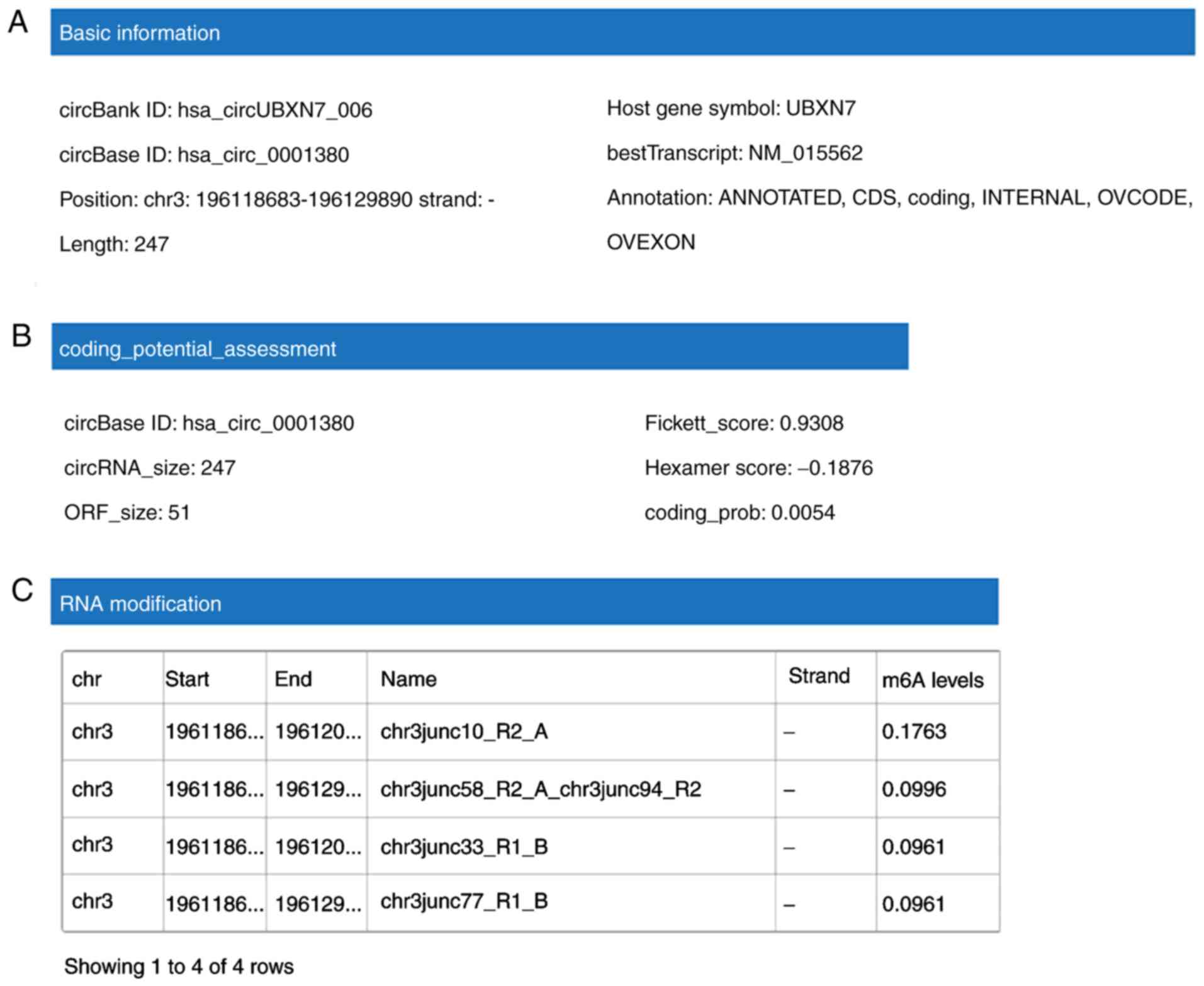
Functions of hsa_circ_0001380
predicted in silico
CircBank database analysis indicated that
hsa_circ_0001380 is located in chr3 (196,118,683-196,129,890), has
a length of 247 nt, consists of three exons (exons 3–5) from the
UBXN7 gene locus and its CircBank ID is hsa_circUBXN7_006 (Fig. 3A). Since circRNAs are able to exert
their biological functions through multiple pathways, the functions
of hsa_circ_0001380 were predicted in silico. Although
certain circRNAs are capable of translating proteins or
polypeptides, hsa_circ_0001380 has little translation potential
(Fig. 3B) according to the
prediction of coding potential assessment based on the CircBank
database. The N6-adenosine methylation (m6A) modification serves an
important role in RNA modification (18), thus m6A expression in
hsa_circ_0001380 was also predicted. The results demonstrated that
hsa_circ_0001380 has four m6A modification sites (Fig. 3C). To investigate the potential
microRNAs (miRNAs/miRs) associated with hsa_circ_0001380, the
CircBank and Circinteractome databases were used based on the
TargetScan and miRanda algorithms (Fig. 3D). The two most potentially
complementary binding miRNAs, miR-136 and miR-622, are presented in
Fig. 3E.
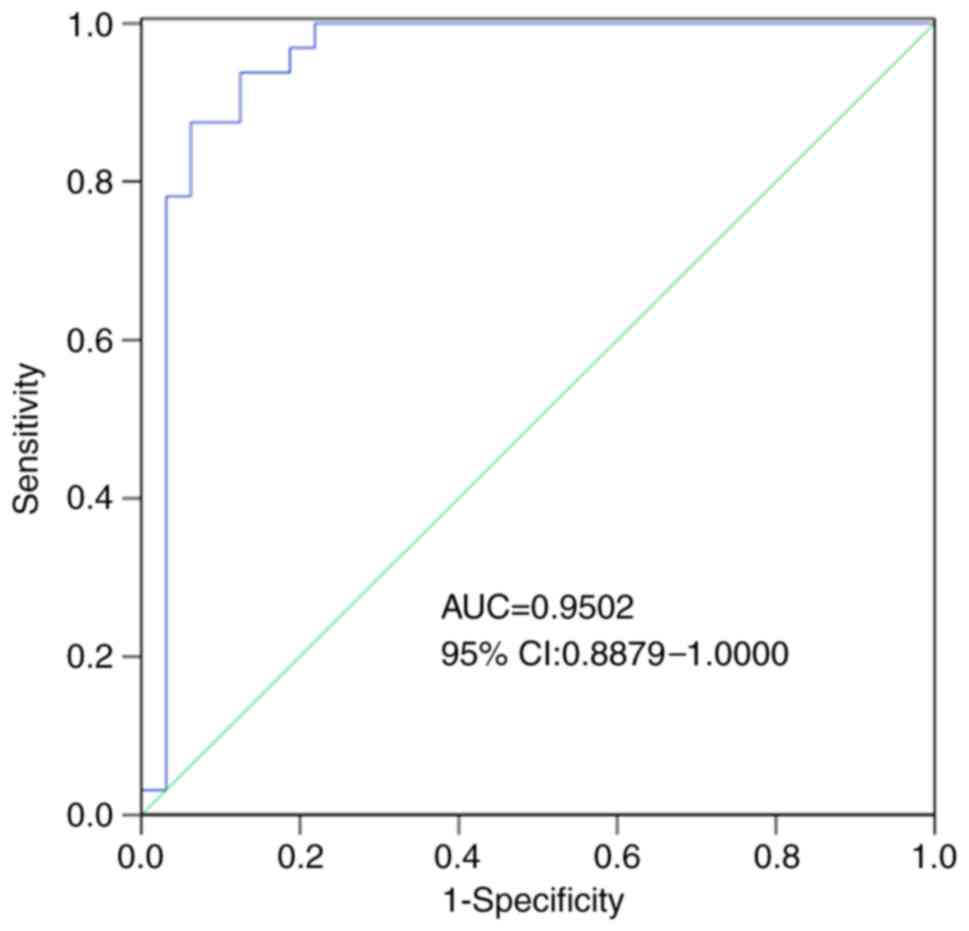
Diagnostic potential of
hsa_circ_0001380 in APTB
Numerous previous studies have reported that
circRNAs may be effective biomarkers for disease diagnosis
(11–13). The results indicated that
hsa_circ_0001380 was downregulated in APTB and was more stable than
linear RNA. Therefore, hsa_circ_0001380 may act as a promising
biomarker for APTB. To assess the diagnostic value of
hsa_circ_0001380, ROC analysis was conducted. The results revealed
that the area under the ROC curve (AUC) was 0.9502 (95% confidence
interval, 0.8879–1.0000), which indicated the high diagnostic value
of hsa_circ_0001380 in APTB. The sensitivity, specificity, total
consistent rate, and Youden's index were 93.75, 87.50, 90.62% and
0.81, respectively (Fig. 4 and
Table III).
 | Table III.Relevant indices of diagnostic
experiment. |
Table III.
Relevant indices of diagnostic
experiment.
| circRNA | AUC | Sensitivity | Specificity | Total consistent
rate | Youden index |
|---|
|
Hsa_circ_0001380 | 0.95 | 93.75% | 87.50% | 90.62% | 0.81 |
Discussion
Tuberculosis, which is caused by Mtb, remains
a major public health problem worldwide. The emergence of
drug-resistant tuberculosis, tuberculosis concomitant with HIV
infection, and the current lack of techniques for its early
diagnosis have led to the global spread of tuberculosis (1). Accurate and early diagnosis and
detection of drug-sensitive and drug-resistant tuberculosis is
essential for achieving tuberculosis control worldwide (19). In addition to traditional methods
based on Mtb detection, molecular biological techniques
based on Mtb-specific genomic DNA and immunological methods,
a number of novel techniques have also been utilized to detect
Mtb (20). For example,
electrochemical (21), optical
(22) and magnetic (23) detection techniques based on
biosensing technology have been used to detect Mtb. However,
these methods fail to meet clinical needs due to their low
sensitivity and specificity.
Previous studies have indicated that circRNAs are
abundant and stable non-coding RNAs lacking 5′ and 3′ termini that
are widely present in diverse bodily fluids, including plasma/serum
(24), blood (25) and saliva (26). Therefore, circRNAs may represent a
potential diagnostic biomarker for human diseases. Numerous studies
have indicated that circRNAs are efficient biomarkers for human
diseases, including bladder cancer (27). hepatocellular carcinoma (28). glioma (29) and systemic lupus erythematosus
(30).
In the present study, the potential diagnostic value
of hsa_circ_0001380 was explored in the PBMCs of patients with
APTB. The expression of hsa_circ_0001380 was significantly
decreased in APTB. A ROC curve was constructed to assess the
diagnostic value of hsa_circ_0001380, and the results showed that
the AUC was 0.9502 and sensitivity and specificity were 93.75 and
87.50%, respectively. These results suggested that hsa_circ_0001380
may serve as an efficient biomarker for APTB. Future studies with
larger clinical sample sizes and additional disease control groups
are required to further understand the role of
hsa_circ_0001380.
circRNAs may exert their biological functions in
various ways, affecting the occurrence and development of human
diseases. Numerous studies have proposed that circRNAs act as
competing endogenous RNA, which compete with miRNA binding to
mRNAs. For example, Cheng et al (4) reported that circRNA VMA21 prevented
intervertebral disc degeneration by targeting miR-200c and the
X-linked inhibitor of apoptosis protein. In the present study,
hsa_circ_0001380 was matched with hsa-miR-622 and hsa-miR-136-5p
based on TargetScan and miRanda algorithms. In addition, Zhou et
al (18) demonstrated that the
m6A modification is widespread in circRNAs and m6A circRNAs are
expressed in cell-type-specific patterns. The bioinformatics
results showed that hsa_circ_0001380 has four m6A modification
sites. Hence, hsa_circ_0001380 may exert its biological function
via m6A modification. Moreover, certain circRNAs have short open
reading frames and/or internal ribosome entry sites, and are able
to translate proteins or polypeptides (31–33).
However, hsa_circ_0001380 was found to have little translation
potential in silico.
The present study has several limitations. First,
the sample size was limited. Nevertheless, since the statistical
analysis results were considered statistically significant, the
sample size was not expanded. Secondly, the present study was only
a preliminary exploration of hsa_circ_0001380 as a diagnostic
marker for APTB. At present, to the best of the authors' knowledge,
there is no similar study on hsa_circ_0001380 in TB. In the future,
its specific molecular function should be further explored by
loss-of-function and gain-of-function analyses in TB.
In conclusion, the present study suggested that the
expression of hsa_circ_0001380 was significantly downregulated in
the PBMCs of patients with APTB as compared with HV. The
dysregulation of hsa_circ_0001380 expression indicated that it may
be involved in the occurrence and development of APTB. More
importantly, hsa_circ_0001380 may serve as a novel potential
diagnostic biomarker for APTB.
Acknowledgements
Not applicable.
Funding
The present study was supported by The National
Natural Science Foundation of China (grant nos. 81870016, 81570009
and 81273237), The Natural Science Foundation of Guangdong Province
(grant no. 2015A030313513) and The Science and Technology
Innovation Fund of Guangdong Medical University (grant nos.
STIF201110 and B2012078).
Availability of data and materials
The datasets used and/or analyzed in the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
JX, HLL, YP, JAZ, BZ and JH conceived and designed
the study. HLL, YP and HL performed the experiments. HLL, YP, GL,
HX, GH, YS, JXZ and JX performed data collection and
interpretation. HLL, YP, JXZ and BZ prepared the figures and
drafted the manuscript. JX, JXZ and BZ revised the manuscript. HLL,
JX and JH reviewed and edited the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethical
Committee of Dongguan Sixth Hospital, Guangdong Medical University.
Experimental procedures were implemented in accordance with the
guidelines and regulations of Guangdong Medical University and
written informed consent was obtained from all patients and healthy
blood donors.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
WHO, . Global tuberculosis report 2019.
Geneva: World Health Organization; 2017
|
|
2
|
Floyd K, Glaziou P, Zumla A and Raviglione
M: The global tuberculosis epidemic and progress in care,
prevention, and research: An overview in year 3 of the End TB era.
Lancet Respir Med. 6:299–314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang L, Han B, Zhang Y, Bai Y, Chao J, Hu
G and Yao H: Engagement of circular RNA HECW2 in the nonautophagic
role of ATG5 implicated in the endothelial-mesenchymal transition.
Autophagy. 14:404–418. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cheng X, Zhang L, Zhang K, Zhang G, Hu Y,
Sun X, Zhao C, Li H, Li YM and Zhao J: Circular RNA VMA21 protects
against intervertebral disc degeneration through targeting miR-200c
and X linked inhibitor-of-apoptosis protein. Ann Rheum Dis.
77:770–779. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z,
Xu W, Zhang E, Wang J, Fang T, et al: The circular RNA circPRKCI
promotes tumor growth in lung adenocarcinoma. Cancer Res.
78:2839–2851. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen L, Huang C, Wang X and Shan G:
Circular RNAs in eukaryotic cells. Curr Genomics. 16:312–318. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Suzuki H and Tsukahara T: A view of
pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci.
15:9331–9342. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Salzman J, Chen RE, Olsen MN, Wang PL and
Brown PO: Cell-type specific features of circular RNA expression.
PLoS Genet. 9:e10037772013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bao X, Zheng S, Mao S, Gu T, Liu S, Sun J
and Zhang L: A potential risk factor of essential hypertension in
case-control study: Circular RNA hsa_circ_0037911. Biochem Biophys
Res Commun. 498:789–794. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin
G, Hu Z, Dai J and Shen H: A novel plasma circular RNA circFARSA is
a potential biomarker for non-small cell lung cancer. Cancer Med.
7:2783–2791. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao K, Zhao Q, Guo Z, Chen Z, Hu Y, Su J,
Chen L, He Z, Cai X, Chen M, et al: Hsa_Circ_0001275: A potential
novel diagnostic biomarker for postmenopausal osteoporosis. Cell
Physiol Biochem. 46:2508–2516. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhuang ZG, Zhang JA, Luo HL, Liu GB, Lu
YB, Ge NH, Zheng BY, Li RX, Chen C, Wang X, et al: The circular RNA
of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new
diagnostic biomarker and therapeutic target of active pulmonary
tuberculosis. Mol Immunol. 90:264–272. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
National Health Commission of the People's
Republic of China, . WS 288–2017 diagnosis for pulmonary
tuberculosis[S]. Beijing: China Standards Press; 2017
|
|
16
|
Zeng JC, Lin DZ, Yi LL, Liu GB, Zhang H,
Wang WD, Zhang JA, Wu XJ, Xiang WY, Kong B, et al: BTLA exhibits
immune memory for alphabeta T cells in patients with active
pulmonary tuberculosis. Am J Transl Res. 6:494–506. 2014.PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) methods. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou C, Molinie B, Daneshvar K, Pondick
JV, Wang J, Van Wittenberghe N, Xing Y, Giallourakis CC and Mullen
AC: Genome-Wide maps of m6A circRNAs identify widespread and
cell-type-specific methylation patterns that are distinct from
mRNAs. Cell Rep. 20:2262–2276. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Walzl G, Mcnerney R, du Plessis N, Bates
M, McHugh TD, Chegou NN and Zumla A: Tuberculosis: Advances and
challenges in development of new diagnostics and biomarkers. Lancet
Infect Dis. 18:e199–e210. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gupta S and Kakkar V: Recent technological
advancements in tuberculosis diagnostics-A review. Biosens
Bioelectron. 115:14–29. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pavlou AK, Magan N, Jones JM, Brown J,
Klatser P and Turner AP: Detection of Mycobacterium tuberculosis
(TB) in vitro and in situ using an electronic nose in combination
with a neural network system. Biosens Bioelectron. 20:538–544.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Duman M and Piskin E: Detection of
Mycobacterium tuberculosis complex and Mycobacterium gordonae on
the same portable surface plasmon resonance sensor. Biosens
Bioelectron. 26:908–912. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lee H, Yoon TJ and Weissleder R:
Ultrasensitive detection of bacteria using core-shell nanoparticles
and an NMR-filter system. Angew Chem Int Ed Engl. 48:5657–5660.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Koh W, Pan W, Gawad C, Fan HC, Kerchner
GA, Wyss-Coray T, Blumenfeld YJ, El-Sayed YY and Quake SR:
Noninvasive in vivo monitoring of tissue-specific global gene
expression in humans. Proc Natl Acad Sci USA. 111:7361–7366. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e1412142015. View Article : Google Scholar
|
|
26
|
Bahn JH, Zhang Q, Li F, Chan TM, Lin X,
Kim Y, Wong DT and Xiao X: The landscape of microRNA,
Piwi-interacting RNA, and circular RNA in human saliva. Clin Chem.
61:221–230. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yang C, Yuan W, Yang X, Li P, Wang J, Han
J, Tao J, Li P, Yang H, Lv Q and Zhang W: Circular RNA circ-ITCH
inhibits bladder cancer progression by sponging miR-17/miR-224 and
regulating p21, PTEN expression. Mol Cancer. 17:192018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen G, Shi Y, Liu M and Sun J: circHIPK3
regulates cell proliferation and migration by sponging miR-124 and
regulating AQP3 expression in hepatocellular carcinoma. Cell Death
Dis. 9:1752018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li G, Yang H, Han K, Zhu D, Lun P and Zhao
Y: A novel circular RNA, hsa_circ_0046701, promotes carcinogenesis
by increasing the expression of miR-142-3p target ITGB8 in glioma.
Biochem Biophys Res Commun. 498:254–261. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang X, Zhang C, Wu Z, Chen Y and Shi W:
CircIBTK inhibits DNA demethylation and activation of AKT signaling
pathway via miR-29b in peripheral blood mononuclear cells in
systemic lupus erythematosus. Arthritis Res Ther. 20:1182018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang M, Huang N, Yang X, Luo J, Yan S,
Xiao F, Chen W, Gao X, Zhao K, Zhou H, et al: A novel protein
encoded by the circular form of the SHPRH gene suppresses glioma
tumorigenesis. Oncogene. 37:1805–1814. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of circRNAs. Mol
Cell. 66:9–21 e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37. 2017. View Article : Google Scholar : PubMed/NCBI
|