Introduction
Accounting for 6.6% (307,471 cases) of all cancer
mortality in men, prostate cancer (PCa) is the most common solid
malignancy in men worldwide and the second highest cause of
cancer-associated mortality in western countries (1,2). The
early diagnosis of PCa may improve the overall survival and
progression-free survival. Currently, a serum prostate-specific
antigen (PSA) test and digital rectal examination are the main
screening strategies for PCa in the clinic (3). However, the PSA level may be
influenced by confounding factors, such as urinary tract infection,
inflammation or transurethral operation (4). Androgen deprivation therapy is the
first-line therapeutic method for reducing circulating androgen
levels and tumor growth; however, usually after 2–3 years, the
patients develop hormone-refractory PCa and progresses to
castration-resistant PCa (CRPC) (1,5).
Therefore, determining the potential mechanisms of PCa progression
and identifying novel biomarkers is important for the diagnosis and
treatment of PCa.
Telomeric repeat binding factor 1 (TERF1) is an
important telomeric binding protein and is vital for the protection
and maintenance of telomere DNA in mammalian cells (6). Several studies have reported the
downregulation of TERF1 in numerous types of cancer; therefore, it
may be a potential marker for cancer diagnosis (7,8).
Nevertheless, the mechanism of TERF1 in PCa remains unclear. Thus,
investigations into the function of TERF1 may provide novel
insights into the molecular mechanisms of PCa. MicroRNAs
(miRNAs/miRs) are a set of small non-coding RNA molecules of 18–25
nucleotides in length that negatively mediate gene expression
through binding to the 3′-untranslated region (3′-UTR) of target
mRNAs (9,10). Multiple studies have demonstrated
that miRNAs are involved in the regulation of various biological
processes such as cancer invasion or migration by targeting the
majority of protein-coding genes (11,12).
Accumulating data has suggested that the abnormal expression of
miRNAs was implicated in the progression of PCa through numerous
signaling pathways, including Notch and HIF-1α pathways (13–16).
Hence, it is necessary to determine the functions of miRNAs in PCa,
which may reveal a novel mechanism of PCa progression.
Bioinformatics analysis in the present study using
online tool miRWalk and data from the The Cancer Genome Atlas
database indicated that TERF1 may serve as a tumor suppressor in
PCa. Moreover, miR-155 was discovered to directly bind to the
3′-UTR of TERF1 (17). The present
study demonstrated an essential role of the miR-155/TERF1 axis in
the progression of PCa.
Materials and methods
Cell culture and transfection
The human prostate cancer cell line, PC3, was
purchased from the American Type Culture Collection (ATCC). PC3
cells were cultured in RPMI-1640 medium (Thermo Fisher Scientific,
Inc.) supplemented with 10% FBS, (Gibco; Thermo Fisher Scientific,
Inc), and maintained at 37°C in a humidified atmosphere of 5%
CO2.
PC3 cells (2×105 cells/well) were
cultured until 60% confluence and transiently transfected with 50
nmol/l agomiR-155 (Guangzhou RiboBio Co., Ltd.) or agomiR-NC
(Guangzhou RiboBio Co., Ltd.) to overexpress miR-155, antagomiR-155
(Guangzhou RiboBio Co., Ltd.) or antagomiR-NC (Guangzhou RiboBio
Co., Ltd.) to downregulate miR-155 expression, TERF1-siRNA
(5′-GGAAGUUACUUAAGAUAAUCU-3′; Guangzhou RiboBio Co., Ltd.) or siRNA
NC (5′-AATTCTCCGAACGTGTCACGT-3′; Guangzhou RiboBio Co., Ltd.) to
downregulate TERF1 expression levels, or pcDNA3.1-TERF1
(Invitrogen; Thermo Fisher Scientific, Inc) overexpression plasmid
to overexpress TERF1 using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). Empty vector was used
as the negative control. Transfected cells were cultured at 37°C
for 6 h prior to the replacement of complete medium. Following
24–72 h, the transfected cells were harvested for in vitro
experiments depending on the different assay. The transfection
efficiencies were analyzed using reverse transcription-quantitative
PCR (RT-qPCR).
The cells were divided into the following groups: i)
Mock control group (without transfection); ii) TERF1-small
interfering RNA (siRNA) negative control (NC) group (transfected
with NC siRNA); iii) TERF1-siRNA group (transfected with TERF1
siRNA); iv) agomiR-155 NC group (transfected with agomiR-155 NC);
v) agomiR-155 group (transfected with agomiR-155); vi)
antagomiR-155 NC group (transfected with antagomiR-155 NC); vii)
antagomiR-155 group (transfected with antagomiR-155); viii)
agomiR-155 + TERF1 group (transfected with agomiR-155 and
pcDNA3.1-TERF1 plasmid); and ix) antagomiR-155 + TERF1-siRNA group
(transfected with antagomiR-155 and TERF1-siRNA).
Flow cytometric analysis of apoptosis. To determine
the levels of apoptosis, PC3 cells (after transfection for 48 h)
were washed with PBS and centrifuged at 300 × g at 4°C for 4 min 3
days post-transfection. The cells were resuspending in 100 µl
binding buffer and incubated with 5 µl Annexin V-FITC and 10 µl
propidium iodide (PI) in the dark at room temperature for 30 min.
Following the incubation, 400 µl binding buffer was added and the
apoptotic cells were analyzed by flow cytometry (FACSCanto II; BD
Biosciences) and software FlowJo (version 7.6.3; FlowJo LLC). The
percentage of early apoptotic cells were calculated to evaluate the
difference between groups.
Wound healing assay
PC3 cells (after transfection for 24 h) were
trypsinized at 37°C until the cell layer was suspended and then
centrifuged at 300 × g at room temperature for 5 min. The cells
were resuspended in RPMI-1640 medium and seeded in a 6-well cell
culture plate at a density of 4×105 cells/well. Upon the
cells reaching 90–100% confluence, vertical linear scratches were
made in the cell monolayer using a sterile 200-µl pipette tip. The
suspended cells were removed by washing with PBS and the adhered
cells were subsequently incubated with serum-free RPMI-1640 medium.
Images of the scratches were photographed under an inverted light
microscope with 10× magnification at 0 and 48 h. The percentage of
the healed wound area was measured by ImageJ (version 5.0; Bio-Rad
Laboratories, Inc.).
Matrigel invasion assay
To analyze cell invasion, Transwell chambers were
precoated with 100 µl Matrigel diluted to 50 µg/ml with DMEM at
37°C for 2 h. PC3 cells (after transfection for 24 h) were
harvested by centrifuging at 300 × g at room temperature for 5 min
and 2×104 cells were seeded into the upper chamber in a
serum-free RPMI-1640 medium. RPMI-1640 medium supplemented with 10%
FBS was added to the lower chambers. Following incubation for 48 h
at 37°C, non-invasive cells on the upper surface of the filter were
removed with a cotton swab. The remaining invasive cells were fixed
by submerging in 10% formalin for 10 min at room temparature, then
washed with PBS once. Fixed cells were stained with 0.5%
hematoxylin for 30 min at room temperature and counted under a
light microscope (magnification, ×200) in three randomly selected
fields of view.
MTT assay
Cells in the exponential phase were harvested by
centrifuging at 300 × g at room temperature for 5 min and
resuspended in RPMI-1640 medium. The cells were plated into a
96-well cell culture plate at a density of 1×103
cells/well and cultured at 37°C and 5% CO2 for 1–7 days.
The cells were then incubated for 4 h with 5% MTT solution (20
µl/well) in the dark at 37°C. Plates were then treated with 100 µl
DMSO/well in the dark to dissolve the purple formazan crystals.
Subsequently, a microplate reader (SAF-680T; Jiangsu Baju
Pharmaceutical Co. Ltd.) was used to measure the optical density
(OD) at a wavelength of 490 nm. Growth curves of the cells were
plotted with the OD value on the y-axis and time on the x-axis. The
assays were conducted three times independently.
Dual luciferase reporter assay
The putative wild-type (WT) miR-155 complementary
binding site in the 3′-UTR of TERF1 was amplified by PCR. A mutant
(Mut) construct in the miR-155 binding site of the TERF1 3′-UTR
region was also generated using a Quick-Change Site-Directed
Mutagenesis kit (Agilent Technologies, Inc.) and named Mut-TERF1
3′-UTR. The 3′-UTR of TERF1 or its Mut sequence were cloned into
the pmiRRB-REPORT vector (Guangzhou RiboBio Co., Ltd.). Then,
WT-TERF1 3′-UTR or Mut-TERF1 3′-UTR were co-transfected using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) into 293T cells (ATCC; 1×105
cells/well) with agomiR-155 or antagomiR-155 in 48-well plates. The
relative luciferase activity was measured using a Dual-Luciferase
Reporter assay system (Promega Corporation) following transfection
for 48 h at 37°C following 48 h of transfection. All data in dual
luciferase reporter assay were normalized to Renilla luciferase
activity. Each assay was independently repeated three times.
RT-qPCR
Total RNA was extracted from PC3 cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA (2 µg) was reverse transcribed into cDNA using
First Strand cDNA Synthesis kit (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C for 15 min, and cDNA was incubated at
85°C for 5 sec to inactivate the reverse transcriptase. qPCR was
subsequently performed using a SYBR Premix Ex Taq II kit (Takara
Bio, Inc.). The following thermocycling conditions were used for
the qPCR: Initial denaturation at 94°C for 5 min; followed by 40
cycles at 94°C for 45 sec, 55°C for 30 sec and 72°C for 45 sec. The
primer sequences used for the qPCR are listed in Table I. Expression levels were quantified
using the 2−ΔΔCq method (18). mRNA expression levels were
normalized to the internal loading control, GAPDH, while miRNA
expression levels were normalized to the internal loading control,
U6. All samples were run in triplicate.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Primer sequence
(5′→3′) |
|---|
| GAPDH | F:
ATGGCCTTCCGTGTTCCTAC |
|
| R:
CTTTACAAAGTTGTCGTTGA |
| Telomeric repeat
binding factor 1 | F:
CACCTCCTAACACAGGCTGG |
|
| R:
TTGCCGCTGCCTTCATTAGA |
| E-cadherin | F:
TGGCTTCCCTCTTTCATC |
|
| R:
GTTCCGCTCTGTCTTTGG |
| Vimentin | F:
TGATTAAGACGGTTGAAACTAG |
|
| R:
AGAAAGGCACTTGAAAGCT |
| N-cadherin | F:
TTTGATGGAGGTCTCCTAACACC |
|
| R:
ACGTTTAACACGTTGGAAATGTG |
| U6 | F:
CTCGCTTCGGCAGCAC |
|
| R:
AACGCTTCACGAATTTGCGT |
| MicroRNA-155 | F:
ACACTCCAGCTGGGTTAATGCTAATCGTGA |
|
| R:
TGGTGTCGTGGAGTCG |
Western blotting
Cells were cultured in complete RPMI-1640 medium at
37°C for 3 days and total protein was extracted using 120 µl
ice-cold RIPA buffer (Sangon Biotech Co., Ltd., Shanghai, China).
Protein concentration was quantified using a Bradford protein assay
kit (Beyotime Institute of Biotechnology) and 50 µg protein/lane
was separated via 10% SDS-PAGE. The separated proteins were
subsequently transferred onto PVDF membranes (Invitrogen, Thermo
Fisher Scientific, Inc.) for 2 h and then blocked with 5% BSA
(Gibco, Thermo Fisher Scientific, Inc.)at room temperature for 1 h.
The membranes were then incubated with the following primary
antibodies at 4°C overnight: Anti-TERF1 (ab10579; Abcam; 1:1,000),
anti-E-cadherin (cat. no. ab40772, Abcam; 1:500), anti-N-cadherin
(cat. no. ab98952, Abcam; 1:500), anti-Vimentin (cat. no. ab217673,
Abcam; 1:1,000) and anti-GAPDH (cat. no. ab181603, Abcam; 1:1,000).
Following the primary antibody incubation, the membranes were
washed three times with 0.1% PBS-Tween-20 (PBST) and then incubated
with HRP-conjugated goat anti-rabbit or -mouse IgG secondary
antibody for 1 h at room temperature. Protein bands were visualized
using an ECL reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
and a Fusion FX5 image analysis system (Vilber Lourmat) after the
final wash with PBST three times was completed. The densitometric
analysis of protein was measured using ImageJ (version 5.0; Bio-Rad
Laboratories, Inc.).
Bioinformatics prediction
The online bioinformatics analysis website miRWalk
(mirwalk.umm.uni-heidelberg.de)
(19) was used to predict the
complementary binding sites for miR-155 in the 3′-UTR of target
genes. The UALCAN database (http://ualcan.path.uab.edu/index.html) (20) was used to determine the expression
levels of TERF1 between normal prostate tissue and primary PCa
tissue. The assigned Gleason score in the UALCAN database was
referenced from The 2014 International Society of Urological
Pathology Consensus Conference on Gleason Grading of Prostatic
Carcinoma (21).
Statistical analysis
All data from at ≥3 independent experiments are
presented as the mean ± SEM. A two-tailed unpaired Student's t-test
was used to compare the statistical differences between two groups,
while a one-way ANOVA and a Bonferroni's post hoc test were used to
compare the statistical differences among multiple groups. Analysis
was performed using SPSS (version 20.0; IBM Corp.). P<0.05 was
considered to indicate a statistically significant difference.
Results
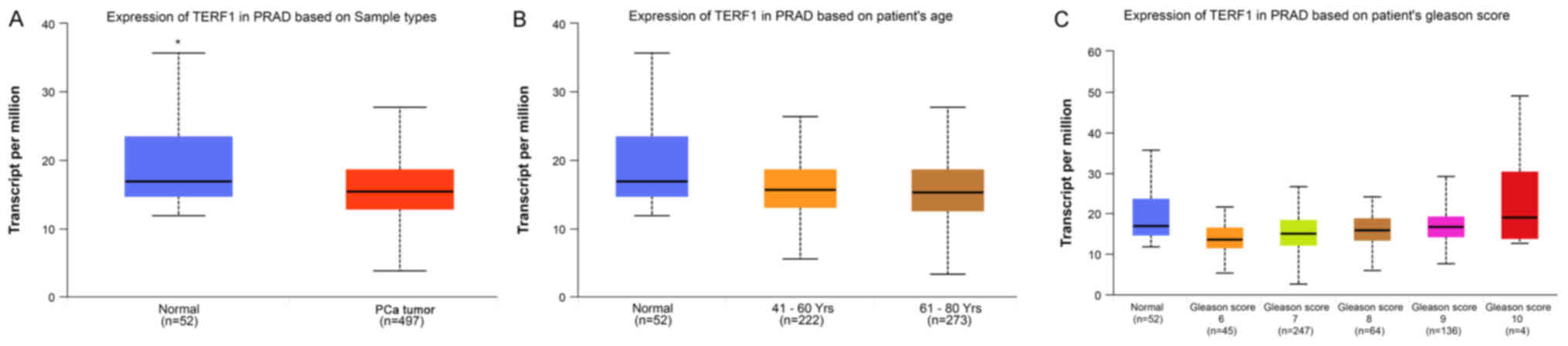
TERF1 expression levels are
downregulated in PCa
To investigate the role of TERF1 in human PCa, the
UALCAN database was used to analyze the expression levels of
TERF1in 52 normal prostate tissue samples and 497 primary PCa
tissue samples. The results revealed that the expression levels of
TERF1 were significantly downregulated in primary PCa compared with
normal prostate tissue samples (Fig.
1A). Nevertheless, both the patient's age and Gleason score
exhibited no significant association with TERF1 (Fig. 1B and C).
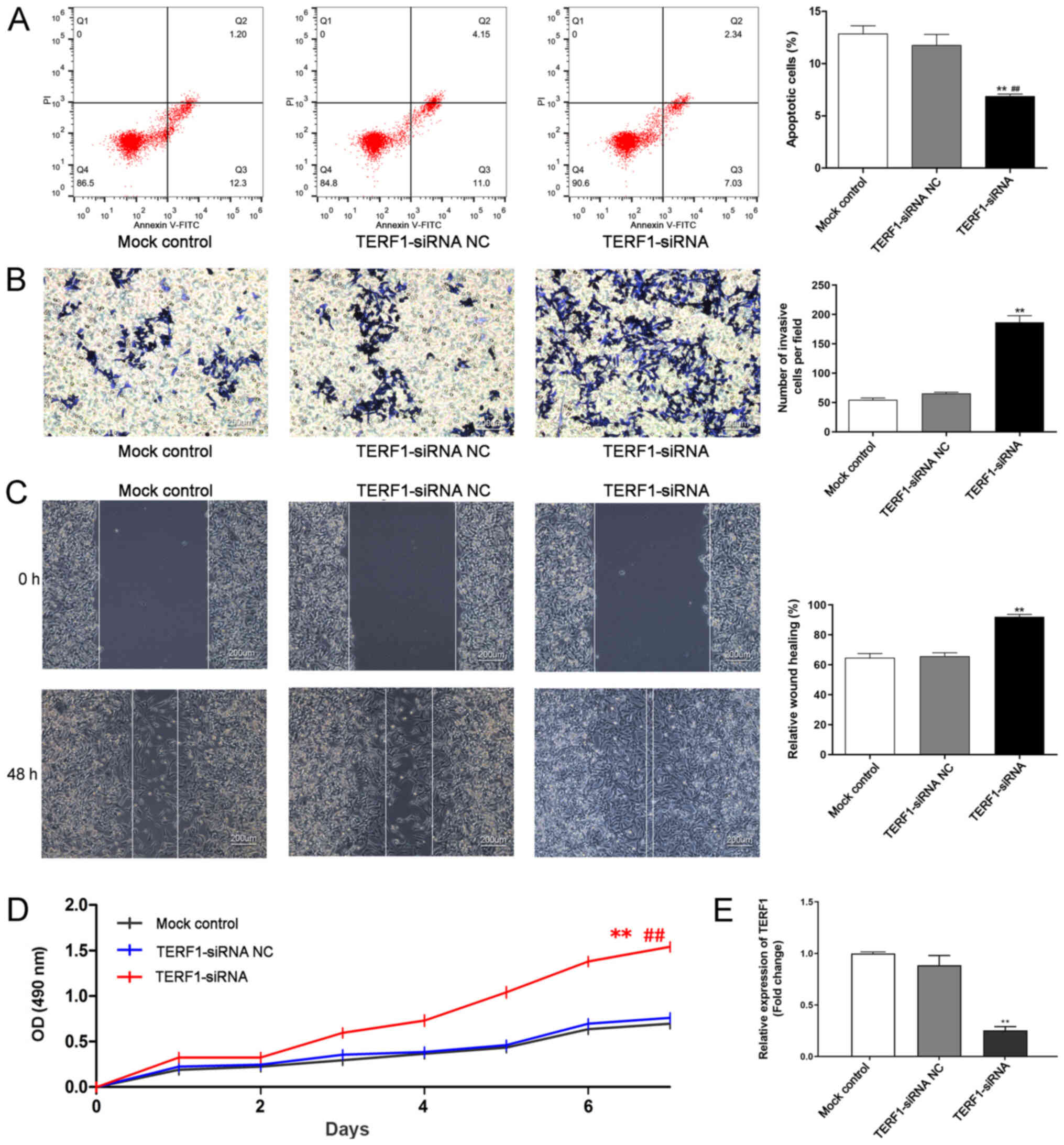
Downregulation of TERF1 expression
levels promotes the progression of PCa in vitro
To investigate the functions of TERF1 in PCa in
vitro, TERF1-siRNA was transfected into PC3 cells. The
transfection efficiency of the knockdown of TERF1 using siRNA in
PC3 cells was initially confirmed by RT-qPCR (Fig. 2E). Flow cytometric analysis of
apoptosis revealed that the knockdown of TERF1 significantly
inhibited the apoptosis of PC3 cells compared with the mock control
and TERF1-siRNA NC groups (Fig.
2A). The effect of TERF1 knockdown on cell invasion and
migration was analyzed using Transwell and wound healing assays,
respectively; the results indicated that the number of invasive and
migratory cells were significantly increased in the TERF1-siRNA
group compared with the mock control and TERF1-siRNA NC groups
(Fig. 2B and C). Furthermore, an
MTT assay demonstrated that the knockdown of TERF1 expression
significantly increased the cell viability of PC3 cells compared
with the TERF1-siRNA NC and mock control groups at days 1–7
(Fig. 2D).
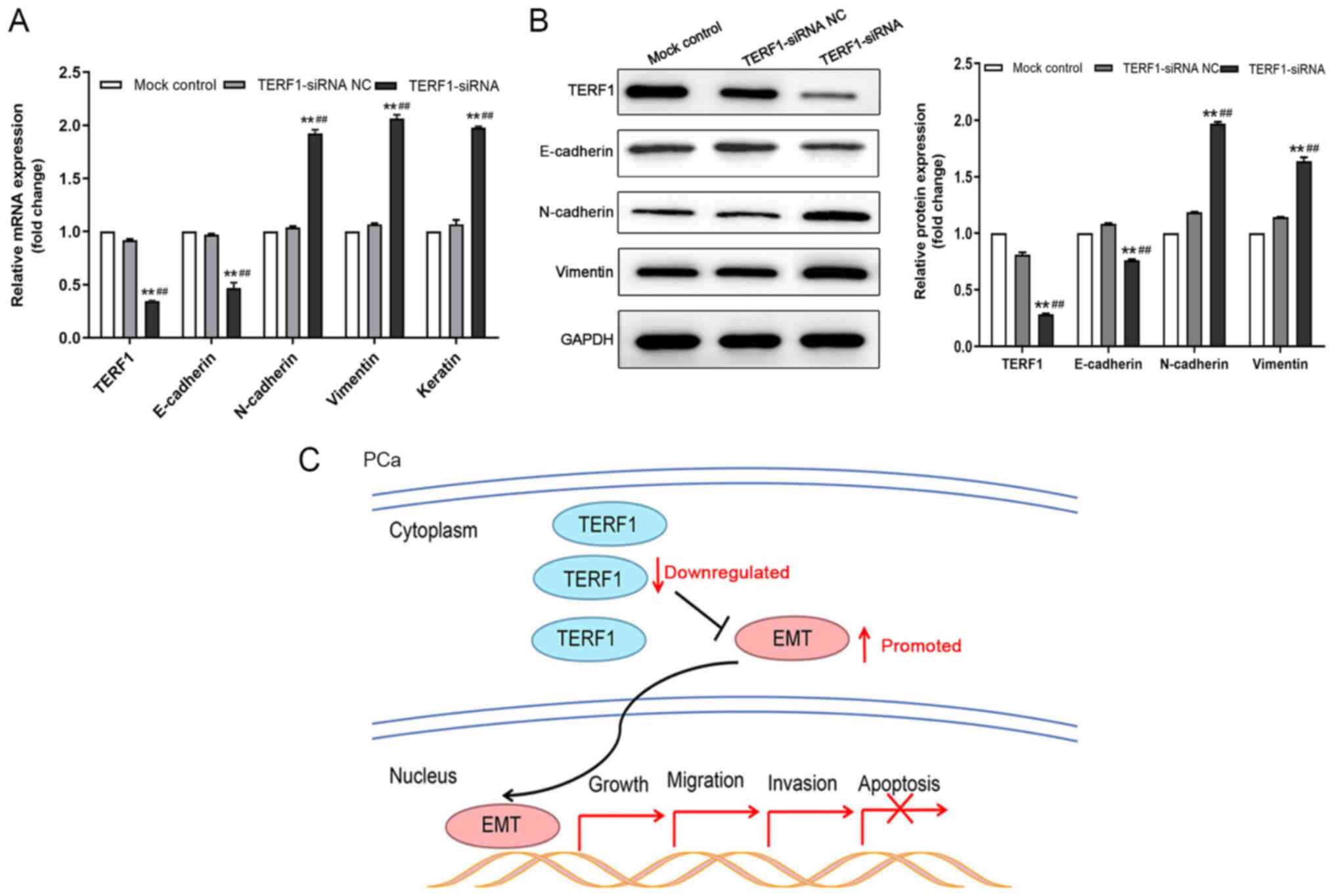
Downregulation of TERF1 expression
levels promotes epithelial-mesenchymal transition (EMT) in PCa
cells
Due to the suppressive role of TERF1 on the
progression of cultured PC3 cells, the present study subsequently
investigated whether the EMT pathway participated in this process.
RT-qPCR analysis revealed that the transfection of PC3 cells with
TERF1-siRNA resulted in significantly upregulated expression levels
of N-cadherin and vimentin compared with the TERF1-siRNA NC and
mock control groups (Fig. 3A).
However, the expression levels of E-cadherin were significantly
downregulated following the transfection with TERF1-siRNA compared
with the TERF1-siRNA NC and mock control groups (Fig. 3A). Western blotting analysis
revealed an identical trend at the protein level (Fig. 3B). In summary, these findings
suggested that the knockdown of TERF1 expression levels may promote
EMT in PCa, and therefore may serve a role in PCa metastasis
(Fig. 3C).
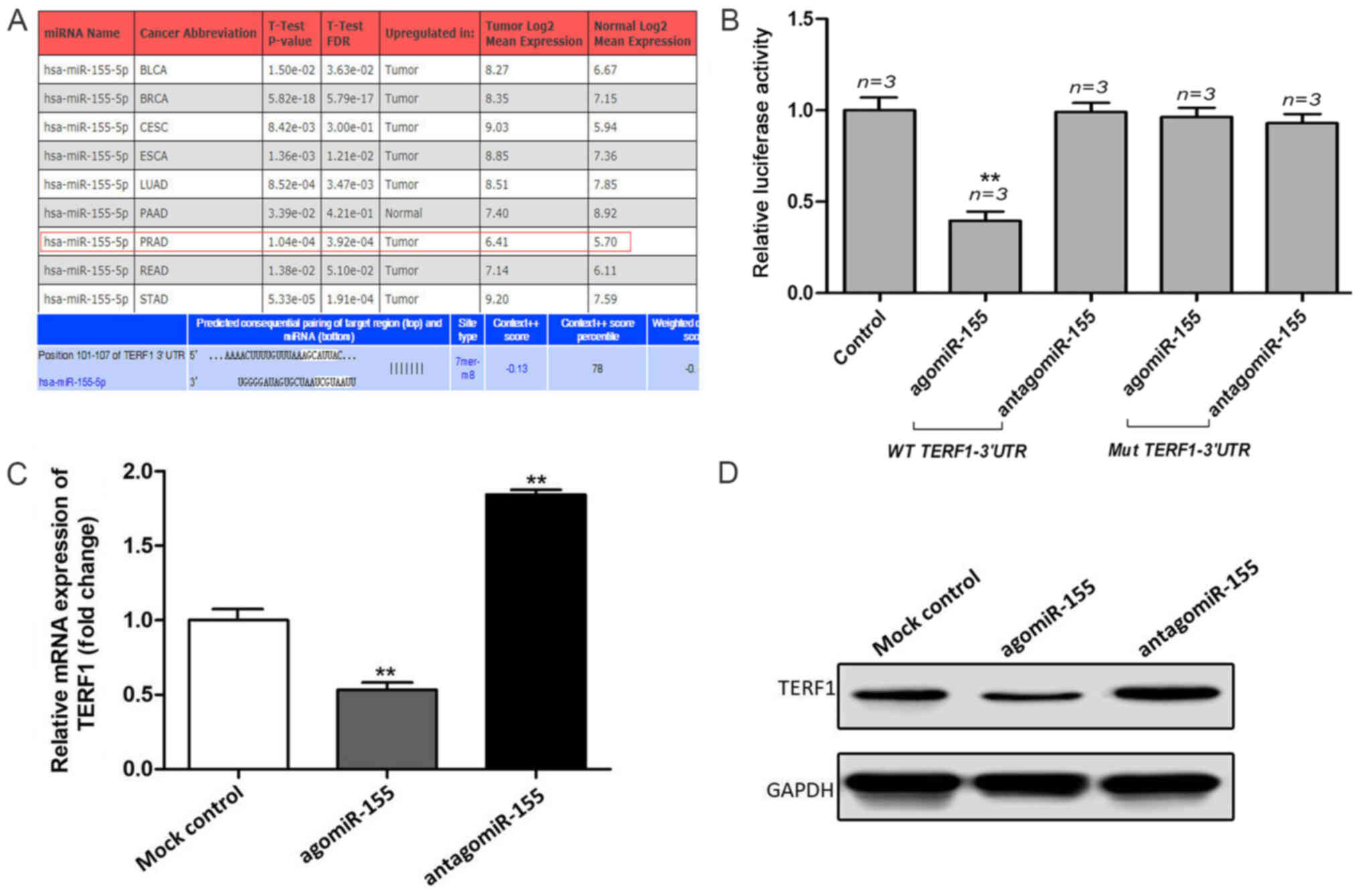
TERF1 is a direct target of
miR-155
To further investigate the mechanism of TERF1 in
promoting PCa progression, bioinformatics analysis using miRWalk
was used to predict the potential miRNAs that regulate TERF1
(Fig. 4A). miR-155 was selected as
a candidate, which has been reported as an oncogenic-associated
molecule in other types of cancer (17). Subsequently, a dual luciferase
reporter assay was used to verify whether miR-155 could directly
bind to the TERF1 3′-UTR. Compared with mock control group, the
results revealed that the relative luciferase activity of the
WT-TERF1 3′-UTR in PC3 cells was significantly repressed following
the co-transfection with the agomiR-155 (Fig. 4B). However, there was no
statistical difference observed in the relative luciferase activity
when PC3 cells were co-transfected with the WT-TERF1 3′-UTR and
antagomiR-155. When compared with mock control group, the dual
luciferase reporter assay also indicated that the upregulation of
miR-155 did not affect the relative luciferase activity of the
Mut-TERF1 3′-UTR in PC3 cells (Fig.
4B). In addition, a negative association was identified between
the expression levels of miR-155 and TERF1. The expression levels
of TERF1 were significantly upregulated following the knockdown of
miR-155 compared with the mock control group, while the expression
levels of TERF1 were significantly downregulated following the
overexpression of miR-155 compared with the mock control group
(Fig. 4C and D). These results
indicated that miR-155 may directly target the TERF1 3′-UTR to
participate in the progression of PCa.
miR-155 promotes the migration and
invasion of PCa by targeting TERF1
To confirm the association between miR-155 and TERF1
and their role in PCa metastasis, the expression levels of miR-155
in PC3 cells were overexpressed and knocked down by the
transfection with an agomiR-155 or antagomiR-155, respectively.
Furthermore, pcDNA3.1-TERF1 plasmids and TERF1-siRNA
oligonucleotides were used to regulate the expression levels of
TERF1 in rescue experiments. Flow cytometric analysis of apoptosis
revealed that the overexpression of miR-155 significantly inhibited
the apoptosis of PC3 cells compared with the mock control and
agomiR-155 NC groups (Fig. 5A). By
contrast, the cells transfected with antagomiR-155 had
significantly increased levels of cell apoptosis compared with the
mock control and anatgomiR-155 NC groups (Fig. 6A). The transfection efficiencies
were confirmed by RT-qPCR (Figs.
5E and 6E).
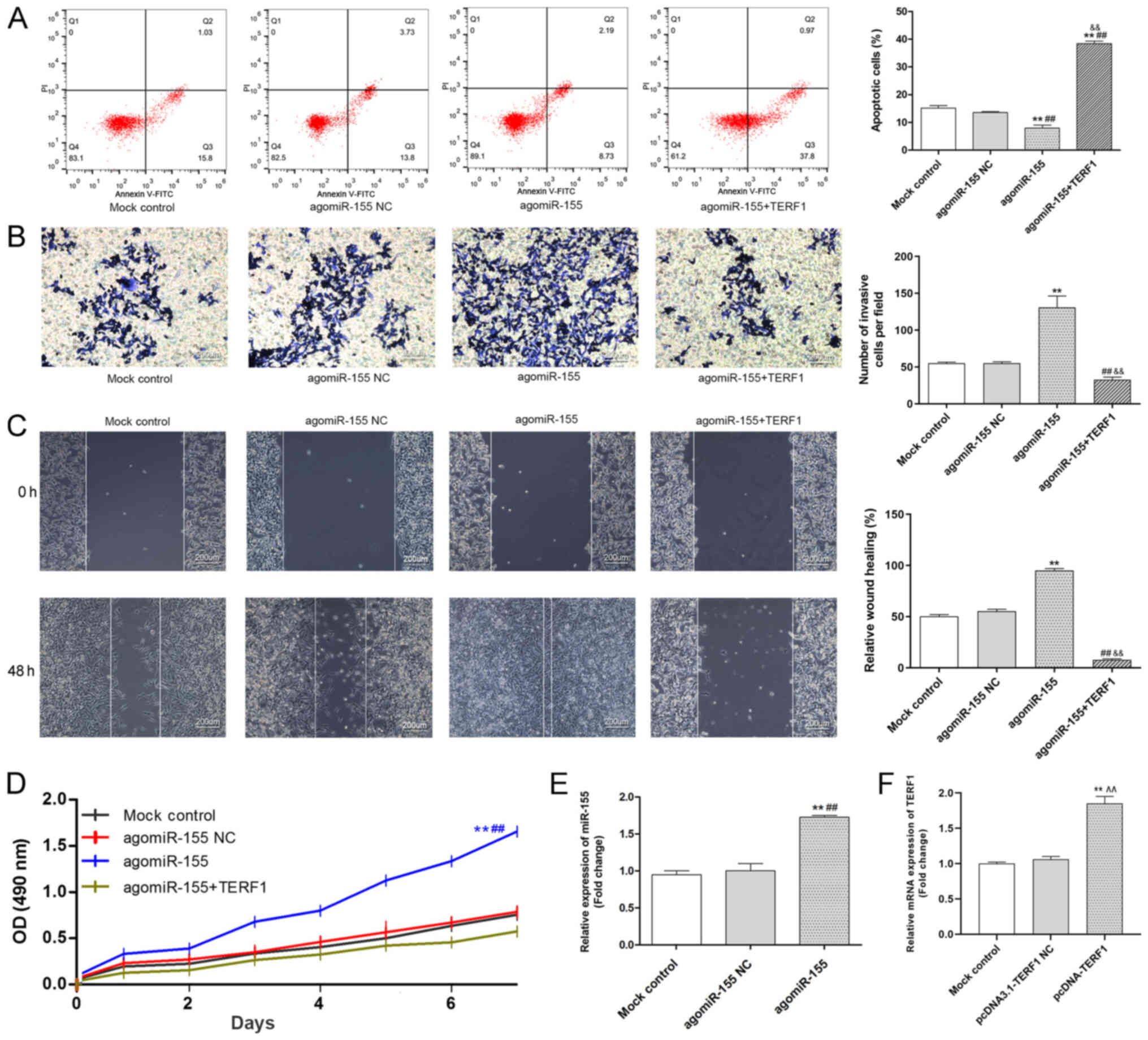 | Figure 5.Overexpression of miR-155 promotes
the viability, migration and invasion of the PC3 cell line, while
inhibiting apoptosis, by targeting TERF1. (A) Levels of apoptosis
in PC3 cells in each group were analyzed by flow cytometry. (B)
Invasive ability of PC3 cells in each group was analyzed using a
Transwell assay. Scale bar, 200-µm. (C) Migratory ability of PC3
cells in each group was analyzed using a wound healing assay. Scale
bar, 200-µm. (D) Viability of PC3 cells in each group was analyzed
using an MTT assay. (E) Transfection efficiency of the agomiR-155
in PC3 cells was analyzed using RT-qPCR. (F) Transfection
efficiency of pcDNA3.1-TERF1 in PC3 cells was analyzed using
RT-qPCR. **P<0.01 vs. mock control; ##P<0.01 vs.
agomiR-155 NC; ^^P<0.01 vs. pcDNA3.1-TERF1 NC.
&&P<0.01 agomiR-155 vs. agomiR-155+TERF1.
TERF1, telomeric repeat binding factor 1; miR, microRNA; RT-qPCR,
reverse transcription-quantitative PCR; PI, propidium iodide; NC,
negative control; OD, optical density. |
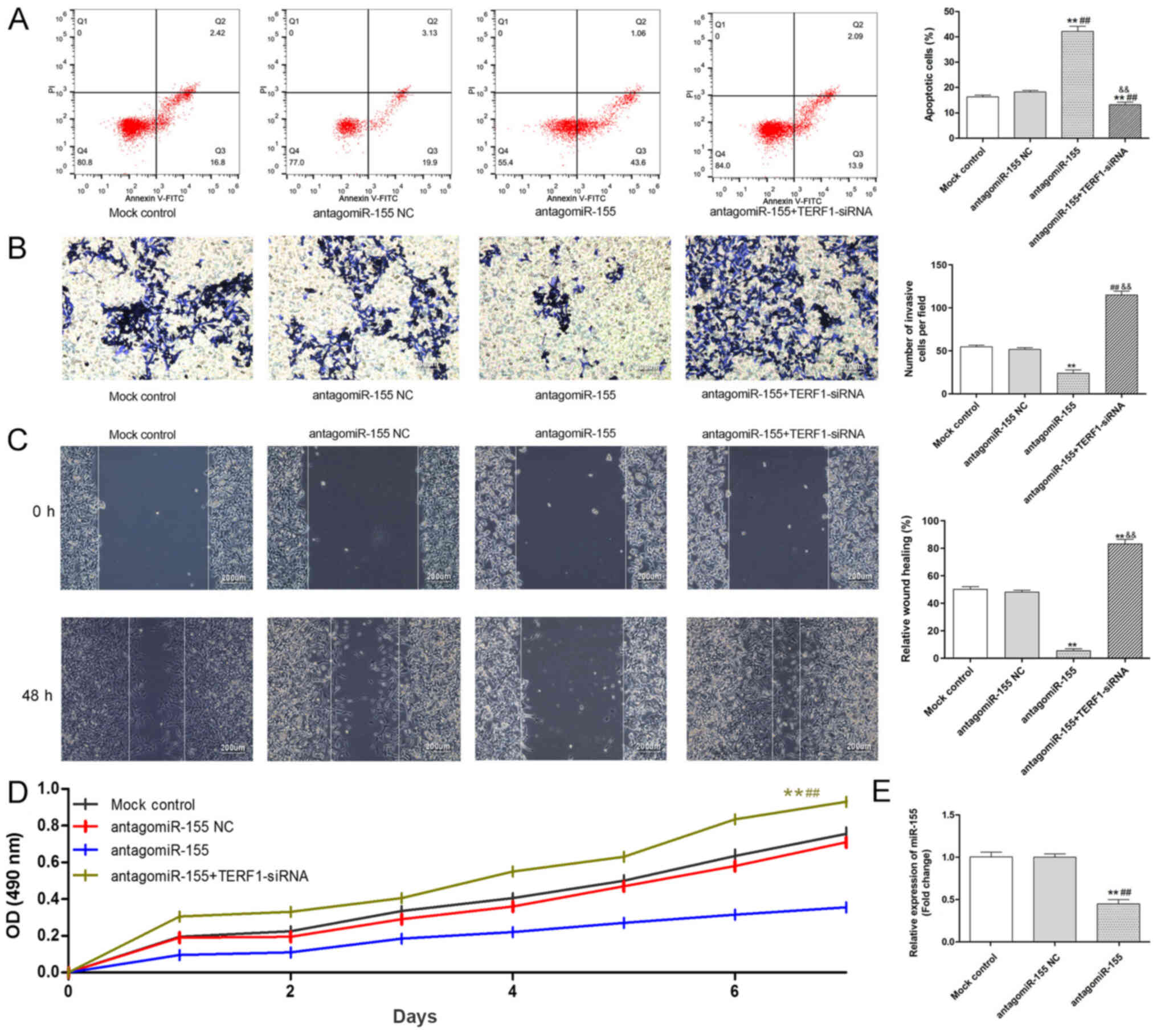 | Figure 6.Knockdown of miR-155 inhibits the
viability, migration and invasion of the PC3 cell line, while
promoting apoptosis, by targeting TERF1. (A) Levels of apoptosis in
PC3 cells in each group were analyzed by flow cytometry. (B)
Invasive ability of PC3 cells in each group was analyzed using a
Transwell assay. Scale bar, 200-µm. (C) Migratory ability of PC3
cells in each group was analyzed using a wound healing assay. Scale
bar, 200-µm. (D) Viability of PC3 cells in each group was analyzed
using an MTT assay. (E) Transfection efficiency of antagomiR-155 in
PC3 cells was analyzed using RT-qPCR. **P<0.01 vs. mock control;
##P<0.01 vs. antagomiR-155 NC;
&&P<0.01 antagomiR-155 vs.
antagomiR-155+TERF1-siRNA.TERF1, telomeric repeat binding factor 1;
miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR; PI,
propidium iodide; NC, negative control; siRNA, small interfering
RNA; OD, optical density. |
To investigate the effects of miR-155 on PCa
invasion and migration, Transwell and wound healing assays,
respectively, were performed; the results revealed that the
upregulation of miR-155 significantly increased the number of
invasive and migratory cells compared with the mock control and
agomiR-155 NC groups (Fig. 5B and
C). The data from the MTT assay illustrated that the
overexpression of miR-155 could significantly promote the viability
of PC3 cells compared with the mock control group at days 3–7
(Fig. 5D). Notably, TERF1
overexpression could reverse the promotive effects of upregulated
miR-155 expression on the invasion, migration and viability of PC3
cells (Fig. 5B-D). In addition,
the overexpression of TERF1 could reverse the inhibitory effects of
the agomiR-155 on apoptosis (Fig.
5A). On the contrary, the knockdown of TERF1 expression
reversed the inhibitory effects of the antagomiR-155 on the
cellular behaviors of PC3 cells (invasion, migration and
viability), while reversing the promoting effects of the
antagomiR-155 on apoptosis (Fig.
6A-D). Taken together, the present data indicated that miR-155
may promote the progression of PCa by directly binding to
TERF1.
Discussion
The main causes of PCa-associated mortality and poor
prognosis in patients are castration-resistant and metastatic PCa
(1,3). The median survival time is no more
than 2 years for patients with CRPC (22). Therefore, it is necessary to
investigate the potential mechanisms of PCa progression and to
determine novel biomarkers to specifically identify cases of
aggressive PCa.
TERF1 encodes a ubiquitously expressed protein of
439 amino acids that is primarily located at chromosome ends, where
it contributes to the protection and maintenance of telomeric DNA
(23,24). Several previous studies have
reported that the expression levels of TERF1 were often
downregulated during the progression of glioblastoma and seminoma
(25–27). These results indicated that TERF1
disruption in cancer may be a general phenomenon. Nevertheless, the
molecular mechanism remains unclear. Hanahan and Weinberg (8) revealed that the maintenance of
telomeres above a minimum length is critical to maintaining the
unlimited proliferative potential of cancer cells, and telomeres
are therefore considered as potential anticancer targets. In total,
>90% of human cancers have been discovered to abnormally express
telomerase (28), while tumors
that do not express telomerase are based on recombination between
telomere sequences and activating another extension (29). The results of the present study
suggested that TERF1 may serve as a potential tumor suppressor gene
in the development of PCa, as significantly downregulated
expression levels of TERF1 were identified in PCa tissue compared
with benign prostate tissue. The downregulation of TERF1 using
siRNA in the current study was further revealed to promote the
viability, invasion and migration, while inhibiting the apoptosis,
of PCa cells. Furthermore, TERF1 knockdown downregulated the
expression levels of E-cadherin and upregulated the expression
levels of N-cadherin and vimentin, which suggested that the
downregulation of TERF1 may promote the progression of PCa
predominantly through the EMT pathway. To further analyze the
molecular mechanism of PCa progression, the interaction between
TERF1 and miRNAs was investigated.
miRNAs have been demonstrated to serve an important
role in PCa progression. For example, miR-338-3p downregulation
promoted the proliferation and invasion of PCa cells (30); and the overexpression of miR-34a
inhibited the proliferation, migration and invasion of PCa cells
(31,32). miR-765 was reported to be an
important mediator for inhibiting growth, migration and invasion in
PCa (33). Dinami et al
(17) revealed that miR-155
upregulation antagonized telomere integrity by targeting TERF1 in
breast cancer. In addition, other previous studies have indicated
that miR-155 may serve an oncogenic role in several types of
hematological malignancy and solid tumor. The overexpression of
miR-155 was previously demonstrated to be associated with cell
proliferation, cell invasion, cell death and cell survival
(34–36). Furthermore, miR-155 directly
targeted and inhibited a series of genes (e.g. FOXO3, SMARCA4 or
Ubiquilin-1) to participate in the biological process of tumor
development (37–40). To the best of our knowledge, the
present study was the first to indicate that TERF1 may be a direct
target of miR-155, as confirmed by a dual luciferase reporter
assay. Furthermore, a negative association was observed between the
expression levels of miR-155 and TERF1. The overexpression of
miR-155 was discovered to promote PC3 cell viability, invasion and
migration, while inhibiting apoptosis. Conversely, the knockdown of
miR-155 yielded the opposite results. Therefore, miR-155 was
hypothesized to be an oncogene in PCa. The results also revealed
that TERF1 could reverse the regulatory effect of miR-155 on the
apoptosis, viability, migration and invasion of PC3 cells.
Collectively, the findings suggested that miR-155 may exert a
carcinogenic role in PCa by targeting and downregulating TERF1.
The local hypoxic environment is important during
the progression of PCa. A number of signaling pathways participated
in the progression of prostate cancer mediated by miR-155. Hypoxia
inducible factor 1α (HIF-1α) induces the expression of VEGF, which
has been reported to promote neovascularization in PCa (41). This pathological process has been
suggested to promote the proliferation and metastasis of PCa
targeted by miRNAs (42). Previous
studies have illustrated that the overexpression of HIF-1α
increased the risk of CRPC and the metastasis of PCa. In addition,
it was also suggested that HIF-1α may be a potential molecular
target of PCa (43). The Notch
signaling pathway was discovered to serve an important role in the
PCa cell proliferation and apoptosis (44). Nevertheless, there are few studies
focusing on the role of the Notch signaling pathway on the invasion
and metastasis of PCa. Previous findings have highlighted the
involvement of Notch signaling in prostate development and in the
maintenance of prostate homeostasis (45). Guo et al (46) reported contradictory roles of Notch
signaling, where it served as a tumor-promoting role in human acute
lymphoblastic leukaemia or tumor-suppressive role in basal cell
carcinoma. Furthermore, studies have reported synergistic positive
results in PCa cells when Notch signaling inhibitors were combined
with androgen deprivation therapy (47,48).
However, the present study only used one cell line
to investigate the potential mechanisms of TERF1 in PCa, which is a
limitation of the study. In order to validate the results, both
androgen-sensitive and androgen-resistant PCa cell lines should be
used in further investigations.
In conclusion, the results of the present study
suggested that TERF1 may function as a tumor suppressor in PCa,
which suppresses the migration and invasion of PC3 cells through
the EMT pathway. Furthermore, miR-155 was discovered to serve an
important role in the progression of PCa via negatively regulating
TERF1. Therefore, TERF1 and miR-155 may serve as potential
diagnostic biomarkers and prognostic markers in PCa.
Acknowledgements
The authors would like to thank Professor Wanlong
Tan and Professor Fei Li (Nanfang Hospital, Southern Medical
University) for their help in guiding experiments and quality
control strategies.
Funding
The present study was supported by the Sichuan
Science and Technology Program (grant no. 2018JY0670), the Key
Project of Zigong Science and Technology (grant nos. 2016SF07 and
2018SHFZ06), the Scientific Research Subject of Sichuan Medical
Association (grant no. S16087), the Science Foundation of Health
Commission of Sichuan Province (grant no. 18PJ459) and the Science
Foundation of Health Commission of Zigong (grant nos. 2018WJWZD02
and 2018WJWZC08).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
WC and LNH conceptualized the study, performed the
experiments and wrote the manuscript. YL and XZ contributed
significantly to data analysis and manuscript preparation. CPW, MQS
and JHL analyzed the data. YC analyzed data, and prepared the
figures and tables.. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Subudhi SK: New approaches to
immunotherapy for metastatic castration-resistant prostate cancer.
Clin Adv Hematol Oncol. 17:283–286. 2019.PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
He BM, Chen R, Sun TQ, Yang Y, Zhang CL,
Ren SC, Gao X and Sun YH: Prostate cancer risk prediction models in
Eastern Asian populations: Current status, racial difference, and
future directions. Asian J Androl. 22:158–161. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koo K and Hyams ES: Assessment of men's
risk thresholds to proceed with prostate biopsy for the early
detection of prostate cancer. Int Urol Nephrol. 51:1297–1302. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dalla Via J, Daly RM, Owen PJ, Mundell NL,
Rantalainen T and Fraser SF: Bone mineral density, structure,
distribution and strength in men with prostate cancer treated with
androgen deprivation therapy. Bone. 127:367–375. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Grun LK, Teixeira NDR Jr, Mengden LV, de
Bastiani MA, Parisi MM, Bortolin R, Lavandoski P, Pierdoná V, Alves
LB, Moreira JCF, et al: TRF1 as a major contributor for telomeres'
shortening in the context of obesity. Free Radic Biol Med.
129:286–295. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hu J, Sun L, Wang LM and Jiang SJ:
Analysis of the nuclear localization signal of TRF1 in non-small
cell lung cancer. Biol Res. 42:217–222. 2009.PubMed/NCBI
|
|
8
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Takahashi RU, Prieto-Vila M, Kohama I and
Ochiya T: Development of miRNA-based therapeutic approaches for
cancer patients. Cancer Sci. 110:1140–1147. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu G and Li B: Role of miRNA in
transformation from normal tissue to colorectal adenoma and cancer.
J Cancer Res Ther. 15:278–285. 2019.PubMed/NCBI
|
|
11
|
Vos PD, Leedman PJ, Filipovska A and
Rackham O: Modulation of miRNA function by natural and synthetic
RNA-binding proteins in cancer. Cell Mol Life Sci. 76:3745–3752.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kogure A, Kosaka N and Ochiya T:
Cross-talk between cancer cells and their neighbors via miRNA in
extracellular vesicles: An emerging player in cancer metastasis. J
Biomed Sci. 26:72019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang M, Jiang S, Zhou L, Yu F, Ding H, Li
P, Zhou M and Wang K: Potential Mechanisms of Action of Curcumin
for Cancer Prevention: Focus on Cellular Signaling Pathways and
miRNAs. Int J Biol Sci. 15:1200–1214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu Z, Wang Z, Li F, Yang J and Tang L: miR
138 modulates prostate cancer cell invasion and migration via Wnt/β
catenin pathway. Mol Med Rep. 17:3140–3145. 2018.PubMed/NCBI
|
|
15
|
Li L, Tang P, Li S, Qin X, Yang H, Wu C
and Liu Y: Notch signaling pathway networks in cancer metastasis: A
new target for cancer therapy. Med Oncol. 34:1802017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen CH, Li SX, Xiang LX, Mu HQ, Wang SB
and Yu KY: HIF-1α induces immune escape of prostate cancer by
regulating NCR1/NKp46 signaling through miR-224. Biochem Biophys
Res Commun. 503:228–234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dinami R, Ercolani C, Petti E, Piazza S,
Ciani Y, Sestito R, Sacconi A, Biagioni F, le Sage C, Agami R, et
al: miR-155 drives telomere fragility in human breast cancer by
targeting TRF1. Cancer Res. 74:4145–4156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dweep H, Sticht C, Pandey P and Gretz N:
miRWalk - database: Prediction of possible miRNA binding sites by
‘walking’ the genes of three genomes. J Biomed Inform. 44:839–847.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A Portal for Facilitating Tumor Subgroup Gene
Expression and Survival Analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Epstein JI, Egevad L, Amin MB, Delahunt B,
Srigley JR and Humphrey PA; Grading Committee, : The 2014
International Society of Urological Pathology (ISUP) Consensus
Conference on Gleason Grading of Prostatic Carcinoma: Definition of
Grading Patterns and Proposal for a New Grading System. Am J Surg
Pathol. 40:244–252. 2016.PubMed/NCBI
|
|
22
|
Blanker MH and Bangma CH: Presence of
prostate cancer, but absence of active treatment. Ned Tijdschr
Geneeskd. 163:D36982019.(In Dutch). PubMed/NCBI
|
|
23
|
Wang L, Tu Z, Liu C, Liu H, Kaldis P, Chen
Z and Li W: Dual roles of TRF1 in tethering telomeres to the
nuclear envelope and protecting them from fusion during meiosis.
Cell Death Differ. 25:1174–1188. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lee YW, Arora R, Wischnewski H and Azzalin
CM: TRF1 participates in chromosome end protection by averting
TRF2-dependent telomeric R loops. Nat Struct Mol Biol. 25:147–153.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bejarano L, Schuhmacher AJ, Méndez M,
Megías D, Blanco-Aparicio C, Martínez S, Pastor J, Squatrito M and
Blasco MA: Inhibition of TRF1 Telomere Protein Impairs Tumor
Initiation and Progression in Glioblastoma Mouse Models and
Patient-Derived Xenografts. Cancer Cell. 32:590–607.e4. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu Q, Wang G, Lyu Y, Bai M, Jiapaer Z,
Jia W, Han T, Weng R, Yang Y, Yu Y, et al: The miR-590/Acvr2a/Terf1
Axis Regulates Telomere Elongation and Pluripotency of Mouse iPSCs.
Stem Cell Reports. 11:88–101. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chan FL, Vinod B, Novy K, Schittenhelm RB,
Huang C, Udugama M, Nunez-Iglesias J, Lin JI, Hii L, Chan J, et al:
Aurora Kinase B, a novel regulator of TERF1 binding and telomeric
integrity. Nucleic Acids Res. 45:12340–12353. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kim NW, Piatyszek MA, Prowse KR, Harley
CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL and Shay
JW: Specific association of human telomerase activity with immortal
cells and cancer. Science. 266:2011–2015. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bryan TM, Englezou A, Dalla-Pozza L,
Dunham MA and Reddel RR: Evidence for an alternative mechanism for
maintaining telomere length in human tumors and tumor-derived cell
lines. Nat Med. 3:1271–1274. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cai C, Zhi Y, Wang K, Zhang P, Ji Z, Xie C
and Sun F: CircHIPK3 overexpression accelerates the proliferation
and invasion of prostate cancer cells through regulating
miRNA-338-3p. OncoTargets Ther. 12:3363–3372. 2019. View Article : Google Scholar
|
|
31
|
Zhu M, Zheng Z, Huang J, Ma X, Huang C, Wu
R, Li X, Liang Z, Deng F, Wu J, et al: Modulation of miR-34a in
curcumin-induced antiproliferation of prostate cancer cells. J Cell
Biochem. 120:15616–15624. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Li N, Zhang LY, Qiao YH and Song RJ: Long
noncoding RNA LINC00662 functions as miRNA sponge to promote the
prostate cancer tumorigenesis through targeting miR-34a. Eur Rev
Med Pharmacol Sci. 23:3688–3698. 2019.PubMed/NCBI
|
|
33
|
Leung YK, Chan QK, Ng CF, Ma FM, Tse HM,
To KF, Maranchie J, Ho SM and Lau KM: Correction: Hsa-miRNA-765 as
a Key Mediator for Inhibiting Growth, Migration and Invasion in
Fulvestrant-Treated Prostate Cancer. PLoS One. 14:e02141842019.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Witten LW, Cheng CJ and Slack FJ: miR-155
drives oncogenesis by promoting and cooperating with mutations in
the c-Kit oncogene. Oncogene. 38:2151–2161. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Witten L and Slack FJ: miR-155 as a novel
clinical target for hematological malignancies. Carcinogenesis.
41:2–7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jurkovicova D, Magyerkova M, Kulcsar L,
Krivjanska M, Krivjansky V, Gibadulinova A, Oveckova I and Chovanec
M: miR-155 as a diagnostic and prognostic marker in hematological
and solid malignancies. Neoplasma. 61:241–251. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhang Y, Zhao H and Zhang L:
Identification of the tumor suppressive function of circular RNA
FOXO3 in non small cell lung cancer through sponging miR-155. Mol
Med Rep. 17:7692–7700. 2018.PubMed/NCBI
|
|
38
|
Yang YP, Nguyen PNN, Ma HI, Ho WJ, Chen
YW, Chien Y, Yarmishyn AA, Huang PI, Lo WL, Wang CY, et al: Tumor
Mesenchymal Stromal Cells Regulate Cell Migration of Atypical
Teratoid Rhabdoid Tumor through Exosome-Mediated miR155/SMARCA4
Pathway. Cancers (Basel). 11:E7202019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yadav S, Singh N, Shah PP, Rowbotham DA,
Malik D, Srivastav A, Shankar J, Lam WL, Lockwood WW and Beverly
LJ: MIR155 Regulation of Ubiquilin1 and Ubiquilin2: Implications in
Cellular Protection and Tumorigenesis. Neoplasia. 19:321–332. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang Y, Yan L, Zhang L, Xu H, Chen T, Li
Y, Wang H, Chen S, Wang W, Chen C, et al: NT21MP negatively
regulates paclitaxel-resistant cells by targeting miR 155 3p and
miR 155-5p via the CXCR4 pathway in breast cancer. Int J Oncol.
53:1043–1054. 2018.PubMed/NCBI
|
|
41
|
Deep G, Kumar R, Nambiar DK, Jain AK,
Ramteke AM, Serkova NJ, Agarwal C and Agarwal R: Silibinin inhibits
hypoxia-induced HIF-1α-mediated signaling, angiogenesis and
lipogenesis in prostate cancer cells: In vitro evidence and in vivo
functional imaging and metabolomics. Mol Carcinog. 56:833–848.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hasan D, Gamen E, Abu Tarboush N, Ismail
Y, Pak O and Azab B: PKM2 and HIF-1α regulation in prostate cancer
cell lines. PLoS One. 13:e02037452018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chen B, Zhang M, Xing D and Feng Y:
Atorvastatin enhances radiosensitivity in hypoxia-induced prostate
cancer cells related with HIF-1α inhibition. Biosci Rep.
37:BSR201703402017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mohamed AA, Tan SH, Xavier CP, Katta S,
Huang W, Ravindranath L, Jamal M, Li H, Srivastava M, Srivatsan ES,
et al: Synergistic activity with NOTCH inhibition and androgen
ablation in ERG-positive prostate cancer cells. Mol Cancer Res.
15:1308–1317. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Deng G, Ma L, Meng Q, Ju X, Jiang K, Jiang
P and Yu Z: Notch signaling in the prostate: Critical roles during
development and in the hallmarks of prostate cancer biology. J
Cancer Res Clin Oncol. 142:531–547. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Guo H, Lu Y, Wang J, Liu X, Keller ET, Liu
Q, Zhou Q and Zhang J: Targeting the Notch signaling pathway in
cancer therapeutics. Thorac Cancer. 5:473–486. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kita Y, Goto T, Akamatsu S, Yamasaki T,
Inoue T, Ogawa O and Kobayashi T: Castration-resistant prostate
cancer refractory to second-generation androgen receptor
axis-targeted agents: Opportunities and challenges. Cancers
(Basel). 10:3452018. View Article : Google Scholar
|
|
48
|
Domingo-Domenech J, Vidal SJ,
Rodriguez-Bravo V, Castillo-Martin M, Quinn SA, Rodriguez-Barrueco
R, Bonal DM, Charytonowicz E, Gladoun N, de la Iglesia-Vicente J,
et al: Suppression of acquired docetaxel resistance in prostate
cancer through depletion of notch- and hedgehog-dependent
tumor-initiating cells. Cancer Cell. 22:373–388. 2012. View Article : Google Scholar : PubMed/NCBI
|