Bladder cancer
Bladder cancer (BC) is the most common malignancy of
the urinary tract. In 2018, there were ~549,000 new cases of BC and
200,000 deaths related to BC globally (1). BC is the eighth most common cancer
among men in the US (2) and is
reported to affect men more frequently than women, with a ratio of
3.2:0.9 (1,2). In addition, the incidence of BC
increases with age (3). According
to the European Association of Urology, BC can be classified into
two divergent phenotypes: Non-muscle-invasive BC (NMIBC) and
muscle-invasive BC (MIBC). Furthermore, BC can be categorized into
the following subtypes: Urothelial carcinoma, squamous epithelial
carcinoma and adenocarcinoma (1–3).
Urothelial carcinomas account for an overwhelming 90% of worldwide
BC cases (4). Risk factors for BC
include occupational factors, age, sex, race, socioeconomic status,
personal health, diet and infection by pathogens (5–7). It
is well-established that the progression of a normal cell to a
cancer cell is a multistep process involving the accumulation of
genetic alterations, referred to as carcinogenesis. NMIBC generally
involves the mutation of fibroblast growth factor receptor 3
(FGFR3), giving rise to low-grade cancer that frequently
recurs but seldom becomes invasive or progresses. By contrast, MIBC
and carcinoma in situ exhibit deletions or mutations of
TP53, RB transcriptional corepressor 1 (RB1), erb-b2
receptor tyrosine kinase 2 or PTEN, leading to high-grade
and metastatic cancer (8). The
emergence of high-throughput transcriptome sequencing techniques
has assisted in the identification of versatile BC biomarkers,
including long non-coding RNAs (lncRNAs), the aberrant expression
of which can contribute to tumorigenesis in bladder tissues
(9). lncRNA abhydrolase-domain
containing 11 antisense RNA 1 (ABHD11-AS1) and lncRNA
hypoxia-inducible factor 1α antisense RNA 2 (HIF1A-AS2) have been
reported to be upregulated in BC tissues and cells, and their
expression levels in tissues have been shown to be positively
associated with advanced pathological grade and TNM classification
(10,11).
Recently reported molecular subtype
classification of BC
Classification systems for cancer are mainly based
on pathological parameters, such as stage and grade. Such
classification provides predictive prognostic information; however,
for BC, recurrence and progression vary widely from patient to
patient, greatly affecting monitoring and treatment (12). The development of advanced
techniques, such as sequencing and mass spectrometry (MS), and
their implementation in omics, has provided better diagnostic and
therapeutic information for the treatment of BC (13–16).
Molecular subtyping, which is based on genetic characteristics, has
made particularly notable progress in BC and is of increasing
interest (17). The current
molecular subtypes of BC share various characteristics, such as
molecular features. However, these classifications can also vary,
with two to seven distinct subtypes (18–20).
As a result of this diversity, molecular classifications are not
feasible for use in the clinical setting. This also highlights the
need for a consensus on a single set of molecular subtypes that is
applicable to clinical use. Nevertheless, the understanding of the
biology of BC has been substantially improved by key achievements
in molecular classification; for example, associations have been
identified between molecular subtypes and urothelial
differentiation, and similarities have been noted between BC
subtypes and other types of cancer.
The first study into BC subtyping using molecular
signatures was conducted at the University of Lund. This previous
study examined the transcriptomes from 308 BC samples, identifying
five different subtypes: Urobasal A, genomically unstable, urobasal
B, squamous cell carcinoma-like and infiltrated (2). In addition, two subtypes of
high-grade MIBC were identified by examining published data from
262 BC samples (18). The
expression levels of keratin and CD44 were analyzed, which
were found to be related to differentiation of the urinary
epithelium. Owing to similarities with expression profiles in
breast cancer, these two molecular subtypes were named luminal-like
and basal-like. Case studies of the luminal-like subtype revealed
that it had much higher disease-specific and overall survival rates
compared with the basal-like subtype, and the following
transcription factors were enriched: FGFR3 and tubercular
sclerosis 1. Specific changes were also noted in various pathways,
including deletion mutations in the RB1 pathway and amplification
of cyclin D1, E2F transcription factor 3 and cyclin E1 (18). In 2014, the MD Anderson Cancer
Center analyzed mRNA expression patterns in 73 MIBC samples using
molecular signatures identified from breast cancer studies. This
led to the identification of three BC subtypes: Luminal, p53-like
and basal (19). In addition to
the aforementioned studies, The Cancer Genome Atlas (TCGA) has
greatly improved biological databases and led to the updating of
subtypes. Through genetic analysis of 129 patients with MIBC, TCGA
identified four molecular BC subtypes: Clusters I, II, III and IV
(20). This classification system
was updated to include luminal, immune undifferentiated, luminal
immune and basal subtypes based on genetic signatures, such as
uroplakins and immune infiltration. Upon analysis of an additional
412 MIBC cases, TCGA classification of BC was further updated and
consolidated into five subtypes: Luminal, luminal-infiltrated,
basal-squamous, neural and luminal-papillary (21).
Interstitial cystitis
Interstitial cystitis (IC) is a chronic condition of
unknown etiology with long-term notable pelvic/suprapubic pain and
urinary storage symptoms, such as urgency, nocturia and frequency
(22). The advent of cystoscopy
led to major findings in IC, including bladder glomerulations
during hydrodistention and Hunner's lesions (23). Although the epidemiology of IC is
difficult to monitor due to its plethora of symptoms, recent
studies have suggested an estimated prevalence of 100–300 per
100,000 women, and the prevalence rate is ≥10–20% lower in men
(24,25).
IC is generally diagnosed through exclusion;
however, several attempts have been made to define standard
diagnostic criteria. Recent guidelines set by the European Society
for the Study of Interstitial Cystitis and the American Urological
Association are currently being used worldwide to treat IC
(26). Although treatment options
for IC are limited and include hydrodistention, several oral
pharmaceutical drugs have been approved by the US Food and Drug
Administration, including pentosan polysulfate (elmiron),
antihistamines, tricyclic antidepressants and immune modulators
(27).
Owing to its unknown etiology, and large variability
in sites of occurrence and symptom severity, IC is difficult to
subtype. However, there is still an urgent need for a
well-established and precise subtyping system. A recent study
revealed that IC with Hunner's lesions displayed completely
different histology, gene expression and prognoses compared to
other forms of IC (28). IC can
also be defined as a distinct non-inflammatory disorder
characterized by preservation of the urothelium layer and symptom
spread beyond the bladder without lesions (29).
Metabolomics and lipidomics
Metabolomics
Metabolomics is defined as the large-scale study of
small molecules and metabolites involved in the regulation of
metabolic pathways and their networks. Compared with genomics and
proteomics, metabolomics is more closely linked to phenotypes;
therefore, it can detect subtle changes in biological pathways
under different physiological conditions and abnormal pathological
processes. For the purposes of this review, we will focus on the
application of metabolomics and lipidomics to BC.
The aim of precision medicine is to create novel
approaches to prevent disease and update clinical strategies to
consider each individual's variability in terms of environment,
lifestyle, genetics and molecular phenotypes (30). Metabolomics holds much promise for
precision medicine and can be used to measure all metabolites in
biological specimens (31).
However, metabolomics presents significant analytical challenges
over genomics and proteomics; it aims to measure molecules that
range in polarity, from organic water-soluble acids to nonpolar
lipids, which have disparate physical properties (32). As a complement to other omics
techniques, metabolomics serves as a critical component of systems
biology. Moreover, the study of metabolites and molecules is
closely related to phenotypes and can improve understanding of
intracellular metabolic alterations (31). The main aim of metabolomics is to
identify altered metabolic pathways and biomarkers (33). Recent developments in metabolomics
and statistical capabilities have improved the ability to
investigate cancer metabolism and better understand cancer-related
changes in metabolism, such as the conversion of glucose into the
macromolecules needed for tumor cell proliferation and
vascularization (34–36).
Lipidomics
Lipids are essential building blocks in the body
that have several critical cellular functions and can provide
information regarding ongoing lipid metabolism. The lipidome is the
total lipid content in a cell (37). The emergence of lipidomics allows
for the complete characterization of the cellular metabolome.
Lipidomics may be the potential key to numerous metabolic diseases
and can be utilized in several research areas, as well as in the
development of diagnostic tools, drugs and therapeutic strategies
(38). Lipidomics combined with
bioinformatics can serve as a powerful tool for better
understanding the biochemical mechanisms underlying lipid-related
diseases by quantifying alterations in the levels of individual
lipids, subclasses and molecular species, and identifying changes
in pathways and networks (37).
The emergence of metabolomics and lipidomics has enabled improved
definition of differential metabolites in pathological conditions.
Over the past two decades, metabolomics and lipidomics have seen
significant advances, facilitated by rapid developments in novel
analysis strategies, approaches, instruments and techniques
(39).
Emerging technologies
Current development of
methodologies
The physicochemical properties of all metabolites
add additional complexity to metabolomics studies. To overcome
these restrictions, various methods have been applied to overcome
this complexity and challenges. MS and nuclear magnetic resonance
(NMR) are the most frequently applied analytical approaches in
metabolomics studies.
NMR spectroscopy
NMR is a nondestructive, nonbiased, easily
quantifiable, fast and reproducible spectroscopy technique based on
the principle that nuclei absorb and emit electromagnetic signals
based on changes in the external magnetic field. NMR has several
unique advantages in metabolomics (40). Metabolomics profiling by NMR is a
powerful tool that can be used to diagnose a variety of diseases.
NMR is based on the fact that nuclei, such as 1H,
13C and 31P, have nuclear spins and are able
to exist at different energy levels in a magnetic field. Thus,
these nuclei can generate valuable and identifiable information
about metabolites. 1H NMR is the most commonly used
technique in metabolomics since 1H is naturally abundant
in biological samples. 13C and 31P NMR are
used less frequently but can provide additional information on
specific metabolites (40).
MS
MS-based metabolomics offers quantitative analysis
of metabolites, ranging from measurement of a single molecule to
thousands, with high selectivity and sensitivity. The combination
of MS with separation techniques reduces the complexity of mass
spectra by separating metabolites based on time, providing isobar
separation and delivering additional information regarding
physicochemical properties. To calculate the mass-to-charge ratio
(m/z), MS acquires spectral data and relative intensity of the
measured compounds. One potential drawback of MS-based techniques
is the need for sample preparation, which can lead to potential
loss of metabolites, changes in experimental conditions,
discrimination of specific metabolite classes and other
consequences (41,42).
MS can effectively analyze small molecules separated
by techniques, such as gas chromatography (GC), liquid
chromatography (LC) and capillary electrophoresis. LC-MS and GC-MS
can provide large amounts of chemical information for metabolomics
studies. GC-MS uses the gaseous phase and achieves better
metabolite separation than LC; however, unlike LC, GC typically
requires chemical derivatization of the metabolic species prior to
analysis. GC-MS is widely used in metabolomics studies as it can
detect a wide range of intact metabolites with no need for chemical
modification. For the separation of nonpolar to slightly polar
molecules, traditional reverse-phase chromatography is used.
Hydrophilic interaction LC is the technique of choice for
separating strongly to slightly polar metabolites (41,42).
Advantages and disadvantages
For metabolomics studies, each analytical technique
has its own advantages and limitations. No single instrument or
method can detect all metabolites accurately. Therefore, multiple
methods and instruments are recommended to detect the greatest
number of metabolites. For example, the Phenome Centre Birmingham
utilizes LC-MS and NMR spectroscopy for metabolomic profiling and
is able to detect a higher number of metabolites compared with
using a single method alone (https://www.birmingham.ac.uk/research/activity/phenome-centre/about/index.aspx).
Owing to the complexity of the metabolome, no single analytical
method can fully discern the metabolome. NMR and MS each have their
own strengths and weaknesses, which are described in previous
publications (41–43).
Applying metabolomics and lipidomics to
BC
Applying metabolomics to BC
To diagnose initial or recurrent BC, two standard
diagnostic procedures are used: Cystoscopy and urine cytology.
However, there are several limitations (43). As a result, there is an urgent need
for a noninvasive, highly-sensitive, specific and convenient method
for BC diagnosis. Urine is particularly suited for diagnostic
purposes due to its availability, easy sample collection and
storage in the malignant bladder (42,44).
Both MS and NMR are used to analyze the metabolic
profile and have become critical techniques for quantitatively and
qualitatively measuring the metabolome. Both techniques allow for
extensive and rapid analysis of small-molecule metabolites
(45). Metabolomics can be useful
in cancer research, demonstrating its potential for not only
identifying candidate biomarkers but also elucidating the
mechanisms underlying cancer pathogenesis. Metabolomics has already
been applied to several cancer types with encouraging results,
including breast, prostate, lung and liver cancer (46–50).
Sahu et al (51) identified
metabolic signatures, including those for glucose, the
tricarboxylic acid (TCA) cycle, lipids, amino acids and nucleotide
pathways, by profiling the global metabolome using GC-MS and LC-MS.
The results of this previous study revealed alterations in numerous
pathways between normal urothelium and high-grade urothelial
carcinoma at different stages. Recently, novel analytical methods
have been developed using reverse-phase-high performance LC coupled
with triple quadrupole MS for quantitatively determining and
validating previously identified BC metabolites (52,53).
Clinical validation has previously been performed using urine
samples from 40 patients with BC and matched controls, and
suggested that the recovery and precision values were within the
ranges set by FDA guidelines (52). Jin et al (53) performed LC-MS-based profiling of
metabolites and identified distinctive metabolites in 138 BC
samples and 121 controls. This previous study identified 12
putative glycolysis- and β-oxidation-related markers. Multivariate
regression analysis was then applied to confirm the association
between the metabolic profiles and survival.
Applying lipidomics to BC
Current technologies allow for lipidomic analysis of
a wide variety of biological specimens derived from animal models
and clinical samples (54). The
most appropriate analytical technique is selected based on the
characteristics of the biological sample and the chemical
properties of the targeted lipids. NMR and MS are accepted as the
most powerful tools for phospholipid structure identification
(55). Owing to structural
diversity across phospholipid classes, analytical methods for
lipidomics are continuously being improved. Notable progress has
been made in lipid research by coupling MS with chromatographic
separations. Soft ionization techniques, including matrix-assisted
laser desorption/ionization and electrospray ionization (ESI), are
good examples (56). Dill et
al (57) used desorption
ESI-imaging MS to investigate lipid species as diagnostic
biomarkers of human BC compared to adjacent normal bladder tissue
samples. The results revealed significant differences in the levels
of glycerophosphoinositols, glycerophosphoserines, and fatty acids
in tumor tissues compared with those in normal samples. Our group
previously used ultra-performance LC-MS (UPLC-MS) to identify 1,864
differentially expressed lipids in cisplatin-resistant BC cells
(58). Another of our lipidomics
studies on cisplatin resistance of BC demonstrated that acyl-CoA
synthetase short chain family member 2 inhibition perturbed lipid
metabolism, suggesting that cisplatin-resistant BC may have a
specific lipidomic profile (58).
Previously reported metabolomic biomarker candidates are summarized
in Table I.
 | Table I.List of metabolomic biomarkers in
BC. |
Table I.
List of metabolomic biomarkers in
BC.
| First author,
year | Biomarker | Method | Sample size | Sensitivity
(%) | Specificity
(%) | AUC | Notes | (Refs.) |
|---|
| Pasikanti et
al, 2013 |
2,5-furandicarboxylic acid, ribitol and
ribonic acid | GCxGC/TOFMS | 38 BC, 61
Controls | 71 | 100 | – | Decreased | (70) |
| Wittmann et
al, 2014 | Taurine | MS | 95 BC, 345
Controls | – | – | – | Increased | (68) |
| Srivastava et
al, 2010 | Taurine | NMR
spectroscopy | 33 BC, 37
Controls | – | – | – | Increased | (65) |
| Jin et al,
2014 | Glycolysis and
acylcarnitines | LC-QTOFMS | 138 BC, 121
Controls | 85-91.3 | 85-92.5 | 0.93 | Increased | (53) |
Applying metabolomics and lipidomics to
IC
Applying metabolomics to IC
Chronic bladder pain is a hallmark of IC.
Metabolomics studies can be used to analyze the characteristics of
the disease state and identify novel approaches for reducing
symptoms (41). Kind et al
(59) performed global
metabolomics profiling using various platforms, including NMR and
LC-MS. Utilizing urine from patients with IC, this previous study
profiled 490 metabolites, including histidine, erythronic acid and
tartaric acid, and identified those with the highest fold changes.
The identified metabolites were found to be associated with IC,
suggesting its possible clinical use in urinary IC diagnosis. Using
an MS-based metabolomics approach, the central clinical protocol of
the Multidisciplinary Approach to the Study of Chronic Pelvic Pain
(MAPP) Research Network, the Trans-MAPP Epidemiology and
Phenotyping discovered urinary biomarkers in female patients with
IC who underwent extensive urologic and non-urologic phenotyping
(60). Parker et al
(61) used LC-MS to identify
molecular correlates of IC from urine obtained from female
patients. This previous study identified a novel biomarker,
etiocholan-3α-ol-17-one sulfate (Etio-S), a steroid metabolite, as
being associated with a phenotypic subgroup of highly symptomatic
IC. To the best of our knowledge, there are no reports in the
literature involving the use of lipidomics to identify lipid
compounds associated with IC.
Human specimens-based metabolomics and
lipidomics biomarkers for BC
Diagnosis of BC is dependent on several variables,
including sensitivity and specificity of the methods, the
invasiveness of the procedures and cost. Currently, cystoscopy and
urine cytology are the most commonly used methods; however, both
have several critical drawbacks, the most important being their
limitations for detecting early BC (62). Overall survival in BC is highly
dependent on early detection (62). The discovery of clinically relevant
BC biomarkers will provide clinical value for prognostication,
stratification, and identification of patients at higher risk for
recurrence and progression. It is only through these outcomes that
better management and treatment of patients with BC can be achieved
(50). Zhang et al
(63) compiled the results of
previous metabolomics studies to discover BC biomarkers using
urine, blood, tissue and cell lines. However, there is still a lack
of consensus surrounding the pathophysiology of BC. Thus, there is
a great need for noninvasive markers to differentially diagnose BC
(64).
BC biomarkers
Human urine
Numerous studies have reported that biomarkers can
be identified using metabolomics, and recent studies have
identified biomarkers that are capable of detecting early BC and
predicting response to chemotherapy or relapse (53,65–70).
Some diagnostic biomarker studies have already compared the
metabolic profiles of urine samples from patients with BC and
healthy controls. Using 1H NMR, Srivastava et al
(65) revealed significant
differences in the urine concentrations of hippurate, citrate and
taurine in patients with BC compared with those in healthy
controls. Jin et al (53)
hypothesized that patients with BC could be distinguished from
healthy controls based on metabolic profiles. This previous study
revealed that the metabolic components of glycolysis and
acylcarnitines were increased in MIBC compared with those in NMIBC.
Citrate levels, a key metabolite of the TCA cycle, are altered in
BC (66). Other urinary
metabolites, including citrate, succinate and hippurate, have also
been shown to be reduced in BC compared with those in healthy
controls (67,68). Shen et al (69) also identified three upregulated and
downregulated metabolites in BC: Nicotinuric acid, trehalose and
AspAspGlyTrp were upregulated, whereas inosinic acid,
ureidosuccinic acid and GlyCysAlaLys were downregulated. Several
other studies have confirmed these results (70). The major findings from these
studies are listed in Table I.
Tissue samples
Putluri et al (71) identified LC-MS-based metabolomic
signatures using BC tissue samples, benign adjacent tissues and
healthy controls. This previous study aimed to identify potential
biomarker candidates and identify biology-related processes in BC
carcinogenesis. A total of 50 metabolites with significant
differences between BC and healthy controls were detected. Tripathi
et al (72) performed
high-resolution magic angle spinning NMR analysis of benign and BC
tissues. The results revealed that BC exhibited more metabolic
abnormalities compared with benign or healthy samples. These
results identified 22 clearly differentiated metabolites. Using the
same tissue samples, these results were cross-validated via
targeted GC-MS analysis, demonstrating the potential of these
biomarkers in clinically diagnosing BC. Yang et al (73) examined 48 BC tissue samples treated
with gemcitabine as well as adjacent normal tissues from 12 of
those patients. Based on UPLC-Q-Exactive-MS analysis, 34
significantly altered metabolites were found to be associated with
BC.
Blood serum samples
Blood serum-based studies have established ways of
distinguishing patients with BC from healthy controls. Cao et
al (74) examined the serum
profiles of patients with high- or low-grade BC; in addition,
patients with urinary calculi (hematuria) were included in the
control group. Statistical analysis revealed that the serum
profiles of patients with BC differed from those of healthy
controls and those of patients with calculi. Serum metabolic
profiles also allowed for classification of low- and high-grade BC.
The levels of isoleucine/leucine, tyrosine, phenylalanine, choline,
lactate, glycine and citrate were also shown to be significantly
lower in patients with BC compared with those in healthy controls,
whereas lipid and glucose levels were higher in patients with BC.
Notably, additional comparisons of metabolite levels between
patients with low- and high-grade BC revealed that the levels of
tyrosine, phenylalanine, lactate and glycine were comparatively
higher in low-grade patients, whereas glucose levels were lower. In
addition, Bansal et al (75) used blood serum samples from
patients with low-grade and high-grade BC and healthy controls. A
total of six metabolites, dimethylamine, malonate, lactate,
glutamine, histidine and valine, were significantly altered in BC
samples compared with those in controls. Notably, external
validation via a double-blind study consisting of 106 patients with
suspected BC confirmed the utility of these metabolites for early
diagnosis of BC.
Human specimens-based metabolomics markers
in IC
IC can present as a long continuum of mild to severe
symptoms. In recent years, novel metabolomic techniques have been
applied to gain a better understanding of disease mechanisms and
uncover novel biomarkers (76). A
previous study applied UPLC-MS-based metabolomics to examine urine
samples from 10 patients with IC and 10 healthy controls.
Phenylacetylglutamine was identified as a urinary marker of IC and
was revealed to be elevated in the urine of patients with
mild-to-moderate IC (77). In a
separate study, Parker et al (61) used LC-MS to profile the metabolomes
of urine samples from 40 patients with IC and matched controls. The
results identified six metabolites as being closely associated with
IC pathogenesis; one of which was Etio-S. Further analysis
demonstrated that elevated Etio-S was a good predictor of IC, with
sensitivity of 91.2%, specificity of 87.4%, and area under the
curve of 0.92. Longitudinal analysis of women in this cohort
demonstrated that the differences in Etio-S persisted, indicating
that these changes were long-lasting.
Taking an untargeted comprehensive metabolomic
profiling approach, Kind et al (59) performed GC-MS analysis on urine
specimens from patients with IC and healthy donors, and identified
a total of 490 differentially expressed metabolites. Furthermore,
Lamale et al (78) used
urine samples from 40 women with IC and 29 healthy controls
collected within a 24-h time frame. They discovered higher
expression of three inflammatory markers, histamine,
methylhistamine and IL-6, in patients with IC compared with those
in the controls. In our previous biomarker discovery study,
NMR-based global metabolomics analysis was applied to urine samples
obtained from female patients with IC and matched healthy controls.
The levels of tyramine and 2-oxoglutarate were significantly
elevated in the IC urine specimens (76). Furthermore, in another of our
previous studies, comprehensive solid-phase
microextraction-GC-time-of-flight-MS profiling combined with
bioinformatics analysis revealed that levels of volatile urinary
metabolites, including menthol, were significantly reduced in
patients with IC compared with those in normal controls (79). Previously reported
metabolomics-based IC biomarker candidates are presented in
Table II.
 | Table II.List of metabolomic biomarkers in
IC. |
Table II.
List of metabolomic biomarkers in
IC.
| First author,
year | Biomarker | Method | Sample size | Sensitivity
(%) | Specificity
(%) | AUC | Notes | (Refs.) |
|---|
| Parker et
al, 2016 | Etiocholan-3α-ol-1
7-one sulfate | MS | 40 IC, 40
Controls | 87.4 | 0.92 | 0.92 | Increased | (61) |
| Kind et al,
2016 | Erythronic acid,
histidine and tartaric acid | GC/MS | 42 IC, 21
Controls | – | – | 0.9 | Increased | (59) |
| Wen et al,
2015 | tyramine and
2-oxoglutarate | NMR | 43 IC, 21
Controls | – | – | – | Increased | (76) |
| Shahid et
al, 2018 | Menthol | GC-TOF-MS | 10 IC, 10
Controls | – | – | – | Decreased | (79) |
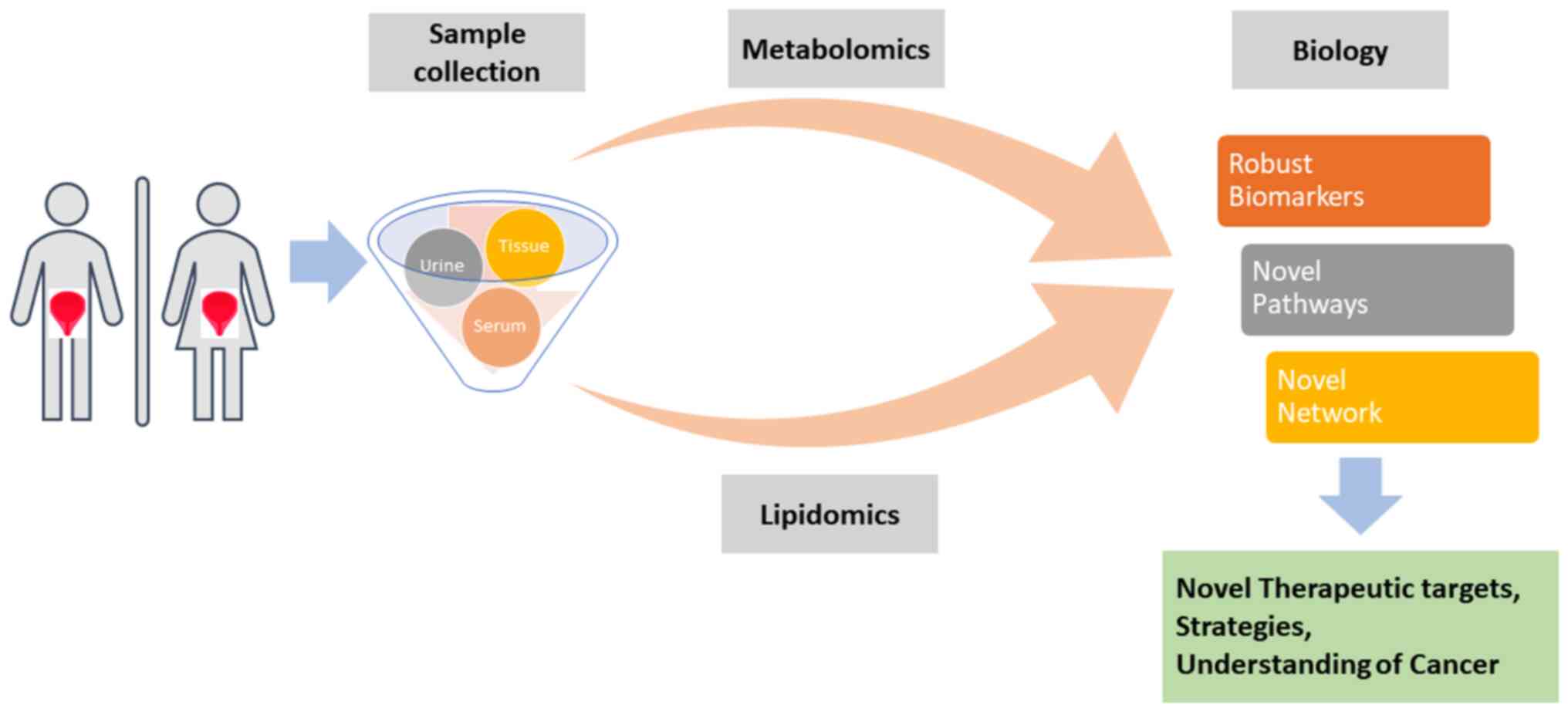
Conclusion
The present review aimed to understand the current
development of biomarkers for bladder diseases based on various
bioresources. In recent years, metabolomics and lipidomics has been
widely used to understand the clinicopathology of the bladder and
to discover the key differentially expressed metabolites or lipids
specifically associated with bladder diseases. To determine the
biological implications of metabolomic lipidomic signatures,
bioinformatics tools, such as network and pathway enrichment
analyses have been applied. The present review provided an overall
summary of the metabolomics and lipidomic-based biomarker
candidates for IC and BC (Fig.
1).
Acknowledgements
Not applicable.
Funding
The authors acknowledge support from National
Institutes of Health grants (grant nos. 1U01DK103260, 1R01DK100974,
U24 DK097154 and NIH NCATS UCLA CTSI UL1TR000124), Department of
Defense grants (grant nos. W81XWH-15-1-0415 and W81XWH-19-1-0109),
Centers for Disease Controls and Prevention (grant no.
1U01DP006079), IMAGINE NO IC Research Grant, the Steven Spielberg
Discovery Fund in Prostate Cancer Research Career Development
Award, and the U.S.-Egypt Science and Technology Joint Fund. This
research was partly supported by the Samuel Oschin Comprehensive
Cancer Institute at Cedars-Sinai Medical Center through 2019 Lucy
S. Gonda Award (to J.K.). In addition, this article is derived from
the Subject Data funded in whole or part by National Academies of
Sciences, Engineering, and Medicine (NAS) and The United States
Agency for International Development (USAID). Any opinions,
findings, conclusions, or recommendations expressed in this article
are those of the authors alone, and do not necessarily reflect the
views of USAID or NAS.
Availability of data and materials
Not applicable.
Authors' contributions
Research conception and design: JK. Data
acquisition, data analysis and interpretation: MS and AY. Drafting
of the manuscript: MS and AY. Critical revision of the manuscript:
JK. Supervision: JK. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
BC
|
bladder cancer
|
|
ESI
|
electrospray ionization
|
|
FGFR3
|
fibroblast growth factor receptor
3
|
|
GC
|
gas chromatography
|
|
IC
|
interstitial cystitis
|
|
LC
|
liquid chromatography
|
|
MAPP
|
Multidisciplinary Approach to the
Study of Chronic Pelvic Pain
|
|
MIBC
|
muscle-invasive BC
|
|
MS
|
mass spectrometry
|
|
NMIBC
|
non-muscle-invasive BC
|
|
NMR
|
nuclear magnetic resonance
|
|
TCA
|
tricarboxylic acid
|
|
TCGA
|
The Cancer Genome Atlas
|
|
UPLC
|
ultra-performance LC-MS
|
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sjodahl G, Lauss M, Lovgren K, Chebil G,
Gudjonsson S, Veerla S, Patschan O, Aine M, Fernö M, Ringnér M, et
al: A molecular taxonomy for urothelial carcinoma. Clin Cancer Res.
18:3377–3386. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
European Association of Urology (EAU), .
EAU Guidelines on Urological Infections. EAU Guidelines Office;
Arnhem: 2018, simplehttps://uroweb.org/wp-content/uploads/EAU-Guidelines-on-Urological-Infections-2018-large-text.pdf
|
|
5
|
Czerniak B, Dinney C and McConkey D:
Origins of bladder cancer. Annu Rev Pathol. 11:149–174. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gruber K: Coffee consumption and bladder
cancer are linked, analysis shows. BMJ. 350:h14772015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Markowski MC, Boorjian SA, Burton JP, Hahn
NM, Ingersoll MA, Maleki Vareki S, Pal SK and Sfanos KS: The
microbiome and genitourinary cancer: A collaborative review. Eur
Urol. 75:637–646. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Knowles MA and Hurst CD: Molecular biology
of bladder cancer: New insights into pathogenesis and clinical
diversity. Nat Rev Cancer. 15:25–41. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhang Q, Su M, Lu G and Wang J: The
complexity of bladder cancer: Long noncoding RNAs are on the stage.
Mol Cancer. 12:1012013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen M, Li J, Zhuang C and Cai Z:
Increased lncRNA ABHD11-AS1 represses the malignant phenotypes of
bladder cancer. Oncotarget. 8:28176–28186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen M, Zhuang C, Liu Y, Li J, Dai F, Xia
M, Zhan Y, Lin J, Chen Z, He A, et al: Tetracycline-inducible shRNA
targeting antisense long non-coding RNA HIF1A-AS2 represses the
malignant phenotypes of bladder cancer. Cancer Lett. 376:155–164.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lopez-Beltran A, Henriques V, Montironi R,
Cimadamore A, Raspollini MR and Cheng L: Variants and new entities
of bladder cancer. Histopathology. 74:77–96. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Boormans JL, Zwarthoff EC, Black PC,
Goebell PJ, Kamat AM, Nawroth R, Seiler R, Williams SB and
Schmitz-Dräger BJ: New horizons in bladder cancer research. Urol
Oncol. March 7–2019.(Epub ahead of print). View Article : Google Scholar
|
|
14
|
Kim S, Kim Y, Kong J, Kim E, Choi JH, Yuk
HD, Lee H, Kim HR, Lee KH, Kang M, et al: Epigenetic regulation of
mammalian Hedgehog signaling to the stroma determines the molecular
subtype of bladder cancer. Elife. 8:e430242019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen LM, Chang M, Dai Y, Chai KX, Dyrskjøt
L, Sanchez-Carbayo M, Szarvas T, Zwarthoff EC, Lokeshwar V,
Jeronimo C, et al: External validation of a multiplex urinary
protein panel for the detection of bladder cancer in a multicenter
cohort. Cancer Epidemiol Biomarkers Prev. 23:1804–1812. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Loras A, Suarez-Cabrera C, Martinez-Bisbal
MC, Quintás G, Paramio JM, Martínez-Máñez R, Gil S and Ruiz-Cerdá
JL: Integrative metabolomic and transcriptomic analysis for the
study of bladder cancer. Cancers (Basel). 11:6862019. View Article : Google Scholar
|
|
17
|
Sjodahl G, Jackson CL, Bartlett JM,
Siemens DR and Berman DM: Molecular profiling in muscle-invasive
bladder cancer: More than the sum of its parts. J Pathol.
247:563–573. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Damrauer JS, Hoadley KA, Chism DD, Fan C,
Tiganelli CJ, Wobker SE, Yeh JJ, Milowsky MI, Iyer G, Parker JS and
Kim WY: Intrinsic subtypes of high-grade bladder cancer reflect the
hallmarks of breast cancer biology. Proc Natl Acad Sci USA.
111:3110–3115. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Choi W, Porten S, Kim S, Willis D, Plimack
ER, Hoffman-Censits J, Roth B, Cheng T, Tran M, Lee IL, et al:
Identification of distinct basal and luminal subtypes of
muscle-invasive bladder cancer with different sensitivities to
frontline chemotherapy. Cancer Cell. 25:152–165. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cancer Genome Atlas Research Network, .
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Robertson AG, Kim J, Al-Ahmadie H,
Bellmunt J, Guo G, Cherniack AD, Hinoue T, Laird PW, Hoadley KA,
Akbani R, et al: Comprehensive molecular characterization of
muscle-invasive bladder cancer. Cell. 174:10332018. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Davis NF, Brady CM and Creagh T:
Interstitial cystitis/painful bladder syndrome: Epidemiology,
pathophysiology and evidence-based treatment options. Eur J Obstet
Gynecol Reprod Biol. 175:30–37. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Fall M, Baranowski AP, Elneil S, Engeler
D, Hughes J, Messelink EJ, Oberpenning F and de C Williams AC;
European Association of Urology, : EAU guidelines on chronic pelvic
pain. Eur Urol. 57:35–48. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hanno P, Lin A, Nordling J, van Ophoven A,
Ueda T and Wein A; Bladder Pain Syndrome Committee of the
International Consultation on Incontinence, : Bladder pain syndrome
committee of the international consultation on incontinence.
Neurourol Urodyn. 29:191–198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Clemens JQ, Mullins C, Ackerman AL,
Bavendam T, van Bokhoven A, Ellingson BM, Harte SE, Kutch JJ, Lai
HH, Martucci KT, et al: Urologic chronic pelvic pain syndrome:
Insights from the MAPP research network. Nat Rev Urol. 16:187–200.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
van de Merwe JP, Nordling J, Bouchelouche
P, Bouchelouche K, Cervigni M, Daha LK, Elneil S, Fall M,
Hohlbrugger G, Irwin P, et al: Diagnostic criteria, classification,
and nomenclature for painful bladder syndrome/interstitial
cystitis: An ESSIC proposal. Eur Urol. 53:60–67. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hanno PM, Burks DA, Clemens JQ, Dmochowski
RR, Erickson D, Fitzgerald MP, Forrest JB, Gordon B, Gray M, Mayer
RD, et al: AUA guideline for the diagnosis and treatment of
interstitial cystitis/bladder pain syndrome. J Urol. 185:2162–2170.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Akiyama Y, Maeda D, Katoh H, Morikawa T,
Niimi A, Nomiya A, Sato Y, Kawai T, Goto A, Fujimura T, et al:
Molecular taxonomy of interstitial cystitis/bladder pain syndrome
based on whole transcriptome profiling by next-generation RNA
sequencing of bladder mucosal biopsies. J Urol. 202:290–300. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Peters KM, Killinger KA, Mounayer MH and
Boura JA: Are ulcerative and nonulcerative interstitial
cystitis/painful bladder syndrome 2 distinct diseases? A study of
coexisting conditions. Urology. 78:301–308. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Seyhan AA and Carini C: Are innovation and
new technologies in precision medicine paving a new era in patients
centric care? J Transl Med. 17:1142019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Clish CB: Metabolomics: An emerging but
powerful tool for precision medicine. Cold Spring Harb Mol Case
Stud. 1:a0005882015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kuehnbaum NL and Britz-McKibbin P: New
advances in separation science for metabolomics: Resolving chemical
diversity in a post-genomic era. Chem Rev. 113:2437–2468. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Griffin JL and Shockcor JP: Metabolic
profiles of cancer cells. Nat Rev Cancer. 4:551–561. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Vander Heiden MG: Targeting cancer
metabolism: A therapeutic window opens. Nat Rev Drug Discov.
10:671–684. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pereira MM, Shori DK, Dormer RL and
McPherson MA: Studies on phosphorylation of calcineurin. Biochem
Soc Trans. 18:4471990. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gatenby RA and Gillies RJ: Why do cancers
have high aerobic glycolysis? Nat Rev Cancer. 4:891–899. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Han X: Lipidomics for studying metabolism.
Nat Rev Endocrinol. 12:668–679. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lydic TA and Goo YH: Lipidomics unveils
the complexity of the lipidome in metabolic diseases. Clin Transl
Med. 7:42018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Blanksby SJ and Mitchell TW: Advances in
mass spectrometry for lipidomics. Annu Rev Anal Chem (Palo Alto
Calif). 3:433–465. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Emwas AH, Roy R, McKay RT, Tenori L,
Saccenti E, Gowda GAN, Raftery D, Alahmari F, Jaremko L, Jaremko M
and Wishart DS: NMR spectroscopy for metabolomics research.
Metabolites. 9:1232019. View Article : Google Scholar
|
|
41
|
Fiehn O and Kim J: Metabolomics insights
into pathophysiological mechanisms of interstitial cystitis. Int
Neurourol J. 18:106–114. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen Z and Kim J: Urinary proteomics and
metabolomics studies to monitor bladder health and urological
diseases. BMC Urol. 16:112016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhu CZ, Ting HN, Ng KH and Ong TA: A
review on the accuracy of bladder cancer detection methods. J
Cancer. 10:4038–4044. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim J, Kim WT and Kim WJ: Advances in
urinary biomarker discovery in urological research. Investig Clin
Urol. 61 (Suppl 1):S8–S22. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Nicholson JK, Connelly J, Lindon JC and
Holmes E: Metabonomics: A platform for studying drug toxicity and
gene function. Nat Rev Drug Discov. 1:153–161. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sreekumar A, Poisson LM, Rajendiran TM,
Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al:
Metabolomic profiles delineate potential role for sarcosine in
prostate cancer progression. Nature. 457:910–914. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jobard E, Pontoizeau C, Blaise BJ,
Bachelot T, Elena-Herrmann B and Tredan O: A serum nuclear magnetic
resonance-based metabolomic signature of advanced metastatic human
breast cancer. Cancer Lett. 343:33–41. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Carrola J, Rocha CM, Barros AS, Gil AM,
Goodfellow BJ, Carreira IM, Bernardo J, Gomes A, Sousa V, Carvalho
L and Duarte IF: Metabolic signatures of lung cancer in biofluids:
NMR-based metabonomics of urine. J Proteome Res. 10:221–230. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen F, Xue J, Zhou L, Wu S and Chen Z:
Identification of serum biomarkers of hepatocarcinoma through
liquid chromatography/mass spectrometry-based metabonomic method.
Anal Bioanal Chem. 401:1899–1904. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Rodrigues D, Jeronimo C, Henrique R, Belo
L, de Lourdes Bastos M, de Pinho PG and Carvalho M: Biomarkers in
bladder cancer: A metabolomic approach using in vitro and ex vivo
model systems. Int J Cancer. 139:256–268. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sahu D, Lotan Y, Wittmann B, Neri B and
Hansel DE: Metabolomics analysis reveals distinct profiles of
nonmuscle-invasive and muscle-invasive bladder cancer. Cancer Med.
6:2106–2120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yumba Mpanga A, Siluk D, Jacyna J, Szerkus
O, Wawrzyniak R, Markuszewski M, Matuszewski M, Kaliszan R and
Markuszewski MJ: Targeted metabolomics in bladder cancer: From
analytical methods development and validation towards application
to clinical samples. Anal Chim Acta. 1037:188–199. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Jin X, Yun SJ, Jeong P, Kim IY, Kim WJ and
Park S: Diagnosis of bladder cancer and prediction of survival by
urinary metabolomics. Oncotarget. 5:1635–1645. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang K and Han X: Lipidomics: Techniques,
applications, and outcomes related to biomedical sciences. Trends
Biochem Sci. 41:954–969. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ellis DI, Dunn WB, Griffin JL, Allwood JW
and Goodacre R: Metabolic fingerprinting as a diagnostic tool.
Pharmacogenomics. 8:1243–1266. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wang C, Wang M and Han X: Applications of
mass spectrometry for cellular lipid analysis. Mol Biosyst.
11:698–713. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Dill AL, Eberlin LS, Costa AB, Zheng C,
Ifa DR, Cheng L, Masterson TA, Koch MO, Vitek O and Cooks RG:
Multivariate statistical identification of human bladder carcinomas
using ambient ionization imaging mass spectrometry. Chemistry.
17:2897–2902. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lee MY, Yeon A, Shahid M, Cho E, Sairam V,
Figlin R, Kim KH and Kim J: Reprogrammed lipid metabolism in
bladder cancer with cisplatin resistance. Oncotarget.
9:13231–13243. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Kind T, Cho E, Park TD, Deng N, Liu Z, Lee
T, Fiehn O and Kim J: Interstitial cystitis-associated urinary
metabolites identified by mass-spectrometry based metabolomics
analysis. Sci Rep. 6:392272016. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Clemens JQ, Mullins C, Kusek JW, Kirkali
Z, Mayer EA, Rodríguez LV, Klumpp DJ, Schaeffer AJ, Kreder KJ,
Buchwald D, et al: The MAPP research network: A novel study of
urologic chronic pelvic pain syndromes. BMC Urol. 14:572014.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Parker KS, Crowley JR, Stephens-Shields
AJ, van Bokhoven A, Lucia MS, Lai HH, Andriole GL, Hooton TM,
Mullins C and Henderson JP: Urinary metabolomics identifies a
molecular correlate of interstitial cystitis/bladder pain syndrome
in a multidisciplinary approach to the study of chronic pelvic pain
(MAPP) research network cohort. EBioMedicine. 7:167–174. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Mbeutcha A, Lucca I, Mathieu R, Lotan Y
and Shariat SF: Current status of urinary biomarkers for detection
and surveillance of bladder cancer. Urol Clin North Am. 43:47–62.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang WT, Zhang ZW, Guo YD, Wang LS, Mao
SY, Zhang JF, Liu MN and Yao XD: Discovering biomarkers in bladder
cancer by metabolomics. Biomark Med. 12:1347–1359. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Kuo HC: Potential urine and serum
biomarkers for patients with bladder pain syndrome/interstitial
cystitis. Int J Urol. 21 (Suppl 1):S34–S41. 2014. View Article : Google Scholar
|
|
65
|
Srivastava S, Roy R, Singh S, Kumar P,
Dalela D, Sankhwar SN, Goel A and Sonkar AA: Taurine-a possible
fingerprint biomarker in non-muscle invasive bladder cancer: A
pilot study by 1H NMR spectroscopy. Cancer Biomark. 6:11–20. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Anderson NM, Mucka P, Kern JG and Feng H:
The emerging role and targetability of the TCA cycle in cancer
metabolism. Protein Cell. 9:216–237. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Pasikanti KK, Esuvaranathan K, Ho PC,
Mahendran R, Kamaraj R, Wu QH, Chiong E and Chan EC: Noninvasive
urinary metabonomic diagnosis of human bladder cancer. J Proteome
Res. 9:2988–2995. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Wittmann BM, Stirdivant SM, Mitchell MW,
Wulff JE, McDunn JE, Li Z, Dennis-Barrie A, Neri BP, Milburn MV,
Lotan Y and Wolfert RL: Bladder cancer biomarker discovery using
global metabolomic profiling of urine. PLoS One. 9:e1158702014.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Shen C, Sun Z, Chen D, Su X, Jiang J, Li
G, Lin B and Yan J: Developing urinary metabolomic signatures as
early bladder cancer diagnostic markers. OMICS. 19:1–11. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Pasikanti KK, Esuvaranathan K, Hong Y, Ho
PC, Mahendran R, Raman Nee Mani L, Chiong E and Chan EC: Urinary
metabotyping of bladder cancer using two-dimensional gas
chromatography time-of-flight mass spectrometry. J Proteome Res.
12:3865–3873. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Putluri N, Shojaie A, Vasu VT, Vareed SK,
Nalluri S, Putluri V, Thangjam GS, Panzitt K, Tallman CT, Butler C,
et al: Metabolomic profiling reveals potential markers and
bioprocesses altered in bladder cancer progression. Cancer Res.
71:7376–7386. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Tripathi P, Somashekar BS, Ponnusamy M,
Gursky A, Dailey S, Kunju P, Lee CT, Chinnaiyan AM, Rajendiran TM
and Ramamoorthy A: HR-MAS NMR tissue metabolomic signatures
cross-validated by mass spectrometry distinguish bladder cancer
from benign disease. J Proteome Res. 12:3519–3528. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Yang C, Sun X, Wang H, Lu T, Wu K, Guan Y,
Tang J, Liang J, Sun R, Guo Z, et al: Metabolomic profiling
identifies novel biomarkers and mechanisms in human bladder cancer
treated with submucosal injection of gemcitabine. Int J Mol Med.
44:1952–1962. 2019.PubMed/NCBI
|
|
74
|
Cao M, Zhao L, Chen H, Xue W and Lin D:
NMR-based metabolomic analysis of human bladder cancer. Anal Sci.
28:451–456. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Bansal N, Gupta A, Mitash N, Shakya PS,
Mandhani A, Mahdi AA, Sankhwar SN and Mandal SK: Low- and
high-grade bladder cancer determination via human serum-based
metabolomics approach. J Proteome Res. 12:5839–5850. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Wen H, Lee T, You S, Park SH, Song H,
Eilber KS, Anger JT, Freeman MR, Park S and Kim J: Urinary
metabolite profiling combined with computational analysis predicts
interstitial cystitis-associated candidate biomarkers. J Proteome
Res. 14:541–548. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Fukui Y, Kato M, Inoue Y, Matsubara A and
Itoh K: A metabonomic approach identifies human urinary
phenylacetylglutamine as a novel marker of interstitial cystitis. J
Chromatogr B Analyt Technol Biomed Life Sci. 877:3806–3812. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Lamale LM, Lutgendorf SK, Zimmerman MB and
Kreder KJ: Interleukin-6, histamine, and methylhistamine as
diagnostic markers for interstitial cystitis. Urology. 68:702–706.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Shahid M, Lee MY, Yeon A, Cho E, Sairam V,
Valdiviez L, You S and Kim J: Menthol, a unique urinary volatile
compound, is associated with chronic inflammation in interstitial
cystitis. Sci Rep. 8:108592018. View Article : Google Scholar : PubMed/NCBI
|